Unraveling the Complexity of Chikungunya Virus Infection Immunological and Genetic Insights in Acute and Chronic Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Ethics Statement
2.2. Study Design and Sample Collection
2.3. RNA Extraction and mRNA Library Preparation
2.4. Quality Control and Transcriptome Reconstruction
2.5. Differential Expression Analysis
2.6. Gene Coexpression and Functional Enrichment Pathway Analysis
3. Results
3.1. Social Vulnerability Marks Chikungunya Infection in Chronic Patients
3.2. A Complex Symptomatological Scenario
3.3. Clinical Data Suggests Possible Neuropathic Disorder in CHIKV Chronic
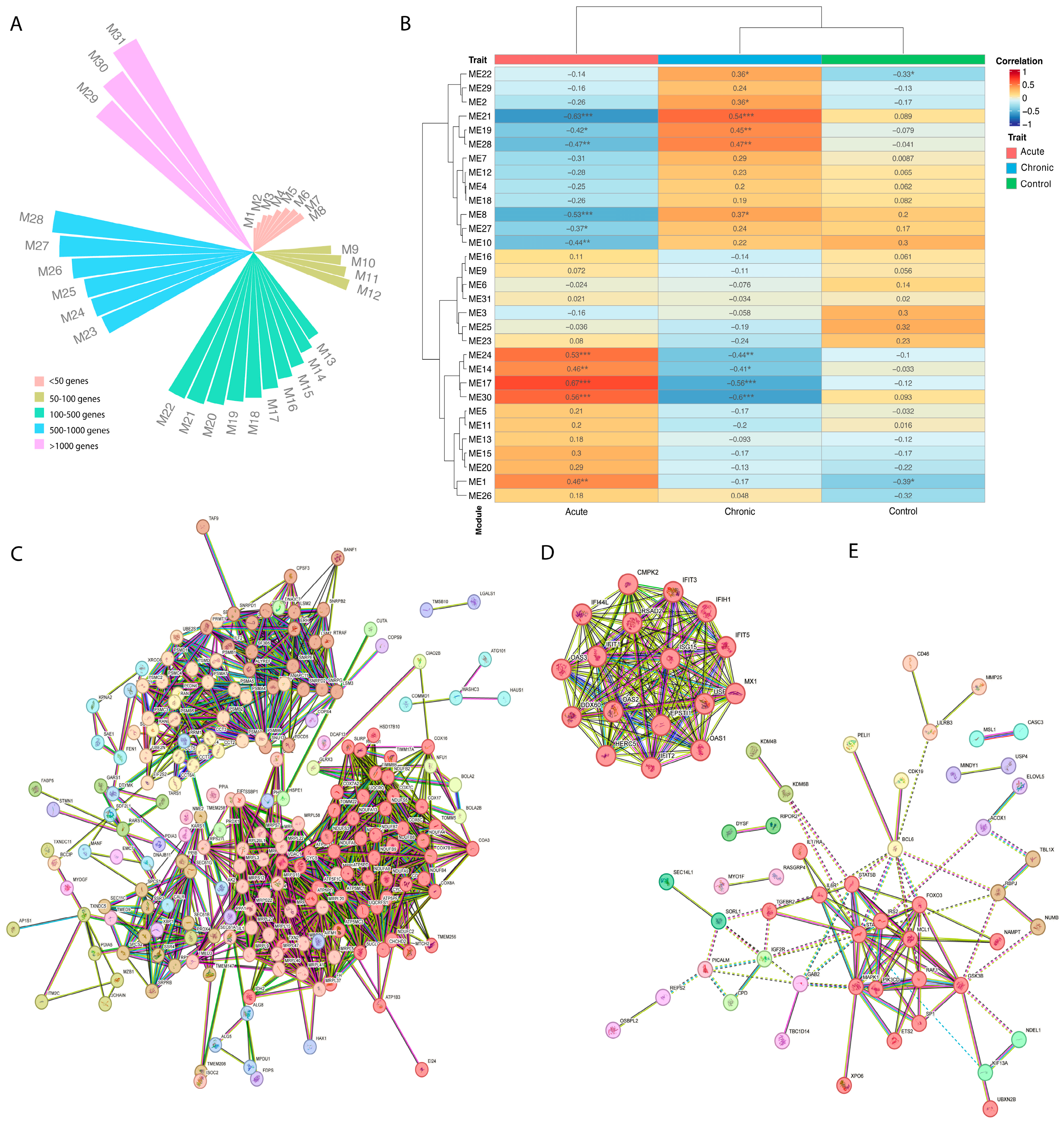
3.4. Expression Profile and Coexpression Analysis Identified Six Different Gene Sets
3.5. CHIKV Acute Individuals Appear to Show Increase in Mitochondrial Respiratory Chain Genes, Besides Antiviral Expression Profile
3.6. CHIKV Chronic Individuals Seem to Show Differentiation in Histones Methylation and Th-17 Regulation Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weaver, S.C. Host Range, Amplification and Arboviral Disease Emergence. In Infectious Diseases from Nature: Mechanisms of Viral Emergence and Persistence; Peters, C.J., Calisher, C.H., Eds.; Springer-Verlag: Vienna, Austria, 2005; pp. 33–44. [Google Scholar] [CrossRef]
- Schwartz, O.; Albert, M.L. Biology and Pathogenesis of Chikungunya Virus. Nat. Rev. Microbiol. 2010, 8, 491–500. [Google Scholar] [CrossRef]
- PAHO, P.A.H. ORGANIZATION. Cases of Chikungunya Virus Disease. Available online: https://www3.paho.org/data/index.php/en/mnu-topics/chikv-en/550-chikv-weekly-en.html (accessed on 5 July 2022).
- Pialoux, G.; Gaüzère, B.-A.; Jauréguiberry, S.; Strobel, M. Chikungunya, an Epidemic Arbovirosis. Lancet Infect. Dis. 2007, 7, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Cunha, M.S.; Costa, P.A.G.; Correa, I.A.; de Souza, M.R.M.; Calil, P.T.; da Silva, G.P.D.; Costa, S.M.; Fonseca, V.W.P.; da Costa, L.J. Chikungunya Virus: An Emergent Arbovirus to the South American Continent and a Continuous Threat to the World. Front. Microbiol. 2020, 11, 1297. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Lecuit, M. Chikungunya Virus and the Global Spread of a Mosquito-Borne Disease. N. Engl. J. Med. 2015, 372, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.H.; Strauss, E.G. The Alphaviruses: Gene Expression, Replication, and Evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef]
- Powers, A.M.; Brault, A.C.; Tesh, R.B.; Weaver, S.C. Re-Emergence of Chikungunya and O’nyong-Nyong Viruses: Evidence for Distinct Geographical Lineages and Distant Evolutionary Relationships. J. Gen. Virol. 2000, 81 Pt 2, 471–479. [Google Scholar] [CrossRef]
- Powers, A.M. Genomic Evolution and Phenotypic Distinctions of Chikungunya Viruses Causing the Indian Ocean Outbreak. Exp. Biol. Med. 2011, 236, 909–914. [Google Scholar] [CrossRef]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.-C.; Lavenir, R.; Pardigon, N.; Reynes, J.-M.; Pettinelli, F.; et al. Genome Microevolution of Chikungunya Viruses Causing the Indian Ocean Outbreak. PLoS Med. 2006, 3, e263. [Google Scholar] [CrossRef]
- Bustos Carrillo, F.; Collado, D.; Sanchez, N.; Ojeda, S.; Lopez Mercado, B.; Burger-Calderon, R.; Gresh, L.; Gordon, A.; Balmaseda, A.; Kuan, G.; et al. Epidemiological Evidence for Lineage-Specific Differences in the Risk of Inapparent Chikungunya Virus Infection. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Mascarenhas, M.; Garasia, S.; Berthiaume, P.; Corrin, T.; Greig, J.; Ng, V.; Young, I.; Waddell, L. A Scoping Review of Published Literature on Chikungunya Virus. PLoS ONE 2018, 13, e0207554. [Google Scholar] [CrossRef]
- Pan American Health Organization. PLISA Health Information Platform for the Americas—Pan American Health Organization: Cases of Chikungunya Virus Disease; Pan American Health Organization: Washington, DC, USA, 2021. [Google Scholar]
- Nunes, M.R.T.; Faria, N.R.; de Vasconcelos, J.M.; Golding, N.; Kraemer, M.U.G.; de Oliveira, L.F.; da Silva Azevedo, R.o.D.; da Silva, D.E.A.; da Silva, E.V.P.; da Silva, S.P.; et al. Emergence and Potential for Spread of Chikungunya Virus in Brazil. BMC Med. 2015, 13, 102. [Google Scholar] [CrossRef]
- de Souza, T.M.A.; Ribeiro, E.D.; Corrêa, V.C.E.; Damasco, P.V.; Santos, C.C.; de Bruycker-Nogueira, F.; Chouin-Carneiro, T.; Faria, N.R.; Nunes, P.C.G.; Heringer, M.; et al. Following in the Footsteps of the Chikungunya Virus in Brazil: The First Autochthonous Cases in Amapá in 2014 and Its Emergence in Rio de Janeiro during 2016. Viruses 2018, 10, 623. [Google Scholar] [CrossRef] [PubMed]
- Brito Ferreira, M.L.; de Fatima Pessoa Militão de Albuquerque, M.; de Brito, C.A.A.; de Oliveira França, R.F.; Porto Moreira, Á.J.; de Morais Machado, M.Í.; da Paz Melo, R.; Medialdea-Carrera, R.; Dornelas Mesquita, S.; Lopes Santos, M.; et al. Neurological Disease in Adults with Zika and Chikungunya Virus Infection in Northeast Brazil: A Prospective Observational Study. Lancet Neurol. 2020, 19, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Xavier, J.; Alcantara, L.C.J.; Fonseca, V.; Lima, M.; Castro, E.; Fritsch, H.; Oliveira, C.; Guimarães, N.; Adelino, T.; Evaristo, M.; et al. Increased Interregional Virus Exchange and Nucleotide Diversity Outline the Expansion of Chikungunya Virus in Brazil. Nat. Commun. 2023, 14, 4413. [Google Scholar] [CrossRef] [PubMed]
- Souza, T.M.A.; Azeredo, E.L.; Badolato-Corrêa, J.; Damasco, P.V.; Santos, C.; Petitinga-Paiva, F.; Nunes, P.C.G.; Barbosa, L.S.; Cipitelli, M.C.; Chouin-Carneiro, T.; et al. First Report of the East-Central South African Genotype of Chikungunya Virus in Rio de Janeiro, Brazil. PLoS Curr. Influenza 2017, 9. [Google Scholar] [CrossRef]
- Xavier, J.; Fonseca, V.; Bezerra, J.F.; do Monte Alves, M.; Mares-Guia, M.A.; Claro, I.M.; de Jesus, R.; Adelino, T.; Araújo, E.; Cavalcante, K.R.L.J.; et al. Chikungunya Virus ECSA Lineage Reintroduction in the Northeasternmost Region of Brazil. Int. J. Infect. Dis. 2021, 105, 120–123. [Google Scholar] [CrossRef]
- Périssé, A.R.S.; Souza-Santos, R.; Duarte, R.; Santos, F.; de Andrade, C.R.; Rodrigues, N.C.P.; de Andrade Schramm, J.M.; da Silva, E.D.; da Silva Viana Jacobson, L.; Lemos, M.C.F.; et al. Zika, Dengue and Chikungunya Population Prevalence in Rio de Janeiro City, Brazil, and the Importance of Seroprevalence Studies to Estimate the Real Number of Infected Individuals. PLoS ONE 2020, 15, e0243239. [Google Scholar] [CrossRef]
- Monteiro, J.D.; Valverde, J.G.; Morais, I.C.; de Medeiros Souza, C.R.; Fagundes Neto, J.C.; de Melo, M.F.; Nascimento, Y.M.; Alves, B.E.B.; de Medeiros, L.G.; Pereira, H.W.B.; et al. Epidemiologic and Clinical Investigations during a Chikungunya Outbreak in Rio Grande Do Norte State, Brazil. PLoS ONE 2020, 15, e0241799. [Google Scholar] [CrossRef]
- Fritsch, H.; Giovanetti, M.; Xavier, J.; Adelino, T.E.R.; Fonseca, V.; de Jesus, J.G.; de Jesus, R.; Freitas, C.; Peterka, C.R.L.; Campelo de Albuquerque, C.F.; et al. Retrospective Genomic Surveillance of Chikungunya Transmission in Minas Gerais State, Southeast Brazil. Microbiol. Spectr. 2022, 10, e0128522. [Google Scholar] [CrossRef]
- de Souza, W.M.; de Lima, S.T.S.; Simões Mello, L.M.; Candido, D.S.; Buss, L.; Whittaker, C.; Claro, I.M.; Chandradeva, N.; Granja, F.; de Jesus, R.; et al. Spatiotemporal Dynamics and Recurrence of Chikungunya Virus in Brazil: An Epidemiological Study. Lancet Microbe 2023, 4, e319–e329. [Google Scholar] [CrossRef]
- Jaffar-Bandjee, M.C.; Ramful, D.; Gauzere, B.A.; Hoarau, J.J.; Krejbich-Trotot, P.; Robin, S.; Ribera, A.; Selambarom, J.; Gasque, P. Emergence and Clinical Insights into the Pathology of Chikungunya Virus Infection. Expert Rev. Anti Infect. Ther. 2010, 8, 987–996. [Google Scholar] [CrossRef]
- Bastos, M.L.A.; de Abreu, F.S.; da Silva Junior, G.B. Inability to Work Due to Chikungunya Virus Infection: Impact on Public Service during the First Epidemic in the State of Ceará, Northeastern Brazil. Braz. J. Infect. Dis. 2018, 22, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Hasan, M.M.; Islam, M.S.; Islam, S.; Mozaffor, M.; Khan, M.A.S.; Ahmed, N.; Akhtar, W.; Chowdhury, S.; Arafat, S.M.Y.; et al. Chikungunya Outbreak (2017) in Bangladesh: Clinical Profile, Economic Impact and Quality of Life during the Acute Phase of the Disease. PLoS Negl. Trop. Dis. 2018, 12, e0006561. [Google Scholar] [CrossRef] [PubMed]
- Padmakumar, B.; Jayan, J.B.; Menon, R.M.; Krishnankutty, B.; Payippallil, R.; Nisha, R.S. Comparative Evaluation of Four Therapeutic Regimes in Chikungunya Arthritis: A Prospective Randomized Parallel-Group Study. Indian J. Rheumatol. 2009, 4, 94–101. [Google Scholar] [CrossRef]
- Guaraldo, L.; Wakimoto, M.D.; Ferreira, H.; Bressan, C.; Calvet, G.A.; Pinheiro, G.C.; Siqueira, A.M.; Brasil, P. Treatment of Chikungunya Musculoskeletal Disorders: A Systematic Review. Expert Rev. Anti Infect. Ther. 2018, 16, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Suhrbier, A.; Jaffar-Bandjee, M.-C.; Gasque, P. Arthritogenic Alphaviruses—An Overview. Nat. Rev. Rheumatol. 2012, 8, 420–429. [Google Scholar] [CrossRef]
- Teng, T.-S.; Foo, S.-S.; Simamarta, D.; Lum, F.-M.; Teo, T.-H.; Lulla, A.; Yeo, N.K.W.; Koh, E.G.L.; Chow, A.; Leo, Y.-S.; et al. Viperin Restricts Chikungunya Virus Replication and Pathology. J. Clin. Investig. 2012, 122, 4447–4460. [Google Scholar] [CrossRef]
- Schilte, C.; Buckwalter, M.R.; Laird, M.E.; Diamond, M.S.; Schwartz, O.; Albert, M.L. Cutting Edge: Independent Roles for IRF-3 and IRF-7 in Hematopoietic and Nonhematopoietic Cells during Host Response to Chikungunya Infection. J. Immunol. 2012, 188, 2967–2971. [Google Scholar] [CrossRef]
- Joubert, P.-E.; Werneke, S.; de la Calle, C.; Guivel-Benhassine, F.; Giodini, A.; Peduto, L.; Levine, B.; Schwartz, O.; Lenschow, D.; Albert, M.L. Chikungunya Virus–Induced Autophagy Delays Caspase-Dependent Cell Death. J. Cell Biol. 2012, 197, i5. [Google Scholar] [CrossRef]
- Schilte, C.; Staikowsky, F.; Couderc, T.; Madec, Y.; Carpentier, F.; Kassab, S.; Albert, M.L.; Lecuit, M.; Michault, A. Chikungunya Virus-Associated Long-Term Arthralgia: A 36-Month Prospective Longitudinal Study. PLoS Negl. Trop. Dis. 2013, 7, e2137. [Google Scholar] [CrossRef]
- Venugopalan, A.; Ghorpade, R.P.; Chopra, A. Cytokines in Acute Chikungunya. PLoS ONE 2014, 9, e111305. [Google Scholar] [CrossRef] [PubMed]
- Kelvin, A.A.; Banner, D.; Silvi, G.; Moro, M.L.; Spataro, N.; Gaibani, P.; Cavrini, F.; Pierro, A.; Rossini, G.; Cameron, M.J.; et al. Inflammatory Cytokine Expression Is Associated with Chikungunya Virus Resolution and Symptom Severity. PLoS Negl. Trop. Dis. 2011, 5, e1279. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, J.-J.; Jaffar Bandjee, M.-C.; Krejbich Trotot, P.; Das, T.; Li-Pat-Yuen, G.; Dassa, B.; Denizot, M.; Guichard, E.; Ribera, A.; Henni, T.; et al. Persistent Chronic Inflammation and Infection by Chikungunya Arthritogenic Alphavirus in Spite of a Robust Host Immune Response. J. Immunol. 2010, 184, 5914–5927. [Google Scholar] [CrossRef]
- Wauquier, N.; Becquart, P.; Nkoghe, D.; Padilla, C.; Ndjoyi-Mbiguino, A.; Leroy, E.M. The Acute Phase of Chikungunya Virus Infection in Humans Is Associated with Strong Innate Immunity and T CD8 Cell Activation. J. Infect. Dis. 2011, 204, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Chow, A.; Her, Z.; Ong, E.K.S.; Chen, J.; Dimatatac, F.; Kwek, D.J.C.; Barkham, T.; Yang, H.; Rénia, L.; Leo, Y.-S.; et al. Persistent Arthralgia Induced by Chikungunya Virus Infection Is Associated with Interleukin-6 and Granulocyte Macrophage Colony-Stimulating Factor. J. Infect. Dis. 2011, 203, 149–157. [Google Scholar] [CrossRef]
- Reddy, V.; Mani, R.S.; Desai, A.; Ravi, V. Correlation of Plasma Viral Loads and Presence of Chikungunya IgM Antibodies with Cytokine/Chemokine Levels during Acute Chikungunya Virus Infection. J. Med. Virol. 2014, 86, 1393–1401. [Google Scholar] [CrossRef]
- Ng, L.F.P.; Chow, A.; Sun, Y.-J.; Kwek, D.J.C.; Lim, P.-L.; Dimatatac, F.; Ng, L.-C.; Ooi, E.-E.; Choo, K.-H.; Her, Z.; et al. IL-1beta, IL-6, and RANTES as Biomarkers of Chikungunya Severity. PLoS ONE 2009, 4, e4261. [Google Scholar] [CrossRef]
- Kittleson, M.M.; Hare, J.M. Molecular Signature Analysis: Using the Myocardial Transcriptome as a Biomarker in Cardiovascular Disease. Trends Cardiovasc. Med. 2005, 15, 130–138. [Google Scholar] [CrossRef]
- Mohr, S.; Liew, C.-C. The Peripheral-Blood Transcriptome: New Insights into Disease and Risk Assessment. Trends Mol. Med. 2007, 13, 422–432. [Google Scholar] [CrossRef]
- Liew, C.-C.; Dzau, V.J. Molecular Genetics and Genomics of Heart Failure. Nat. Rev. Genet. 2004, 5, 811–825. [Google Scholar] [CrossRef]
- Liew, C.-C.; Ma, J.; Tang, H.-C.; Zheng, R.; Dempsey, A.A. The Peripheral Blood Transcriptome Dynamically Reflects System Wide Biology: A Potential Diagnostic Tool. J. Lab. Clin. Med. 2006, 147, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. G:Profiler—A Web-Based Toolset for Functional Profiling of Gene Lists from Large-Scale Experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. G:Profiler-Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Van Dongen, S. Graph Clustering via a Discrete Uncoupling Process. SIAM. J. Matrix Anal. Appl. 2008, 30, 121–141. [Google Scholar] [CrossRef]
- Sfakianos, A.P.; Raven, R.M.; Willis, A.E. The Pleiotropic Roles of EIF5A in Cellular Life and Its Therapeutic Potential in Cancer. Biochem. Soc. Trans. 2022, 50, 1885–1895. [Google Scholar] [CrossRef]
- Wei, J.; Wu, Y.; Sun, Y.; Chen, D. Galectin-1 Regulates RNA Expression and Alternative Splicing of Angiogenic Genes in HUVECs. Front. Biosci. (Landmark Ed) 2023, 28, 74. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.K.; Yang, R.-Y.; Liu, F.-T. Galectins in Apoptosis. Meth. Enzymol. 2006, 417, 256–273. [Google Scholar] [CrossRef]
- Koul, A.; Deo, S.; Booy, E.P.; Orriss, G.L.; Genung, M.; McKenna, S.A. Impact of Double-Stranded RNA Characteristics on the Activation of Human 2′-5′-Oligoadenylate Synthetase 2 (OAS2). Biochem. Cell Biol. 2020, 98, 70–82. [Google Scholar] [CrossRef]
- Hornung, V.; Hartmann, R.; Ablasser, A.; Hopfner, K.-P. OAS Proteins and CGAS: Unifying Concepts in Sensing and Responding to Cytosolic Nucleic Acids. Nat. Rev. Immunol. 2014, 14, 521–528. [Google Scholar] [CrossRef]
- Huffman, J.E.; Butler-Laporte, G.; Khan, A.; Pairo-Castineira, E.; Drivas, T.G.; Peloso, G.M.; Nakanishi, T.; COVID-19 Host Genetics Initiative; Ganna, A.; Verma, A.; et al. Multi-Ancestry Fine Mapping Implicates OAS1 Splicing in Risk of Severe COVID-19. Nat. Genet. 2022, 54, 125–127. [Google Scholar] [CrossRef]
- Wu, Y.; Song, X.; Cui, D.; Zhang, T. IFIT3 and IFIT5 Play Potential Roles in Innate Immune Response of Porcine Pulmonary Microvascular Endothelial Cells to Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Hyde, J.L.; Diamond, M.S. Innate Immune Restriction and Antagonism of Viral RNA Lacking 2′-O Methylation. Virology 2015, 479–480, 66–74. [Google Scholar] [CrossRef]
- Wang, Z.; Ye, J.; Dong, F.; Cao, L.; Wang, M.; Sun, G. TNS1: Emerging Insights into Its Domain Function, Biological Roles, and Tumors. Biology 2022, 11, 1571. [Google Scholar] [CrossRef]
- Hoshikawa, S.; Ogata, T.; Fujiwara, S.; Nakamura, K.; Tanaka, S. A Novel Function of RING Finger Protein 10 in Transcriptional Regulation of the Myelin-Associated Glycoprotein Gene and Myelin Formation in Schwann Cells. PLoS ONE 2008, 3, e3464. [Google Scholar] [CrossRef]
- Lavery, W.J.; Barski, A.; Wiley, S.; Schorry, E.K.; Lindsley, A.W. KMT2C/D COMPASS Complex-Associated Diseases [KCDCOM-ADs]: An Emerging Class of Congenital Regulopathies. Clin. Epigenetics 2020, 12, 10. [Google Scholar] [CrossRef]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 Lysine 4 Methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.D.; Helin, K. Role of TET Enzymes in DNA Methylation, Development, and Cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef]
- Huoman, J.; Sayyab, S.; Apostolou, E.; Karlsson, L.; Porcile, L.; Rizwan, M.; Sharma, S.; Das, J.; Rosén, A.; Lerm, M. Epigenetic Rewiring of Pathways Related to Odour Perception in Immune Cells Exposed to SARS-CoV-2 in Vivo and in Vitro. Epigenetics 2022, 17, 1875–1891. [Google Scholar] [CrossRef]
- de Aguiar, G.P.C.G.; da Silva Leite, C.M.G.; Dias, B.; Vasconcelos, S.M.M.; de Moraes, R.A.; de Moraes, M.E.A.; Vallinoto, A.C.R.; Macedo, D.S.; de Goes Cavalcanti, L.P.; Miyajima, F. Evidence for Host Epigenetic Signatures Arising from Arbovirus Infections: A Systematic Review. Front. Immunol. 2019, 10, 1207. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of Type-I- and Type-II-Interferon-Mediated Signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Y.-S. Longevity Factor FOXO3: A Key Regulator in Aging-Related Vascular Diseases. Front. Cardiovasc. Med. 2021, 8, 778674. [Google Scholar] [CrossRef]
- Widden, H.; Placzek, W.J. The Multiple Mechanisms of MCL1 in the Regulation of Cell Fate. Commun. Biol. 2021, 4, 1029. [Google Scholar] [CrossRef]
- Parola, P.; de Lamballerie, X.; Jourdan, J.; Rovery, C.; Vaillant, V.; Minodier, P.; Brouqui, P.; Flahault, A.; Raoult, D.; Charrel, R.N. Novel Chikungunya Virus Variant in Travelers Returning from Indian Ocean Islands. Emerging Infect. Dis. 2006, 12, 1493–1499. [Google Scholar] [CrossRef]
- Kam, Y.-W.; Ong, E.K.S.; Rénia, L.; Tong, J.-C.; Ng, L.F.P. Immuno-Biology of Chikungunya and Implications for Disease Intervention. Microbes Infect. 2009, 11, 1186–1196. [Google Scholar] [CrossRef]
- Rehermann, B. Hepatitis C Virus versus Innate and Adaptive Immune Responses: A Tale of Coevolution and Coexistence. J. Clin. Investig. 2009, 119, 1745–1754. [Google Scholar] [CrossRef]
- Grieder, F.B.; Nguyen, H.T. Virulent and Attenuated Mutant Venezuelan Equine Encephalitis Virus Show Marked Differences in Replication in Infection in Murine Macrophages. Microb. Pathog. 1996, 21, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.M.; Wu, L. HIV Interactions with Monocytes and Dendritic Cells: Viral Latency and Reservoirs. Retrovirology 2009, 6, 51. [Google Scholar] [CrossRef] [PubMed]
- Kou, Z.; Quinn, M.; Chen, H.; Rodrigo, W.W.S.I.; Rose, R.C.; Schlesinger, J.J.; Jin, X. Monocytes, but Not T or B Cells, Are the Principal Target Cells for Dengue Virus (DV) Infection among Human Peripheral Blood Mononuclear Cells. J. Med. Virol. 2008, 80, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Obata, H. Analgesic Mechanisms of Antidepressants for Neuropathic Pain. Int. J. Mol. Sci. 2017, 18, 2483. [Google Scholar] [CrossRef]
- Derry, S.; Bell, R.F.; Straube, S.; Wiffen, P.J.; Aldington, D.; Moore, R.A. Pregabalin for Neuropathic Pain in Adults. Cochrane Database Syst. Rev. 2019, 1, CD007076. [Google Scholar] [CrossRef]
- Foissac, M.; Javelle, E.; Ray, S.; Guérin, B.; Simon, F. Post-Chikungunya Rheumatoid Arthritis, Saint Martin. Emerging Infect. Dis. 2015, 21, 530–532. [Google Scholar] [CrossRef]
- Ribéra, A.; Degasne, I.; Jaffar Bandjee, M.C.; Gasque, P. Chronic Rheumatic Manifestations Following Chikungunya Virus Infection: Clinical Description and Therapeutic Considerations. Med. Trop. 2012, 72, 83–85. [Google Scholar]
- Bouquillard, E.; Combe, B. A Report of 21 Cases of Rheumatoid Arthritis Following Chikungunya Fever. A Mean Follow-up of Two Years. Jt. Bone Spine 2009, 76, 654–657. [Google Scholar] [CrossRef]
- Mathew, A.J.; Goyal, V.; George, E.; Thekkemuriyil, D.V.; Jayakumar, B.; Chopra, A.; Trivandrum COPCORD Study Group. Rheumatic-Musculoskeletal Pain and Disorders in a Naïve Group of Individuals 15 Months Following a Chikungunya Viral Epidemic in South India: A Population Based Observational Study. Int. J. Clin. Pract. 2011, 65, 1306–1312. [Google Scholar] [CrossRef]
- Chopra, A.; Anuradha, V.; Ghorpade, R.; Saluja, M. Acute Chikungunya and Persistent Musculoskeletal Pain Following the 2006 Indian Epidemic: A 2-Year Prospective Rural Community Study. Epidemiol. Infect. 2012, 140, 842–850. [Google Scholar] [CrossRef]
- Marimoutou, C.; Vivier, E.; Oliver, M.; Boutin, J.-P.; Simon, F. Morbidity and Impaired Quality of Life 30 Months after Chikungunya Infection: Comparative Cohort of Infected and Uninfected French Military Policemen in Reunion Island. Medicine 2012, 91, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Saag, K.G.; Teng, G.G.; Patkar, N.M.; Anuntiyo, J.; Finney, C.; Curtis, J.R.; Paulus, H.E.; Mudano, A.; Pisu, M.; Elkins-Melton, M.; et al. American College of Rheumatology. American College of Rheumatology 2008 Recommendations for the Use of Nonbiologic and Biologic Disease-Modifying Antirheumatic Drugs in Rheumatoid Arthritis. Arthritis Rheum. 2008, 59, 762–784. [Google Scholar] [CrossRef] [PubMed]
- Babu, N.; Mahilkar, S.; Jayaram, A.; Ibemgbo, S.A.; Mathur, G.; Shetty, U.; Sudandiradas, R.; Kumar, P.S.; Singh, S.; Pani, S.S.; et al. Cytokine Profile, Neutralisation Potential and Viral Replication Dynamics in Sera of Chikungunya Patients in India: A Cross-Sectional Study. Lancet Reg. Health Southeast Asia 2023, 19, 100269. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. The Cell Biology of Chikungunya Virus Infection. Cell. Microbiol. 2012, 14, 1354–1363. [Google Scholar] [CrossRef]
- Fox, J.M.; Diamond, M.S. Immune-Mediated Protection and Pathogenesis of Chikungunya Virus. J. Immunol. 2016, 197, 4210–4218. [Google Scholar] [CrossRef]
- Zaid, A.; Gérardin, P.; Taylor, A.; Mostafavi, H.; Malvy, D.; Mahalingam, S. Chikungunya Arthritis: Implications of Acute and Chronic Inflammation Mechanisms on Disease Management. Arthritis Rheumatol. 2018, 70, 484–495. [Google Scholar] [CrossRef]
- Ninla-Aesong, P.; Mitarnun, W.; Noipha, K. Proinflammatory Cytokines and Chemokines as Biomarkers of Persistent Arthralgia and Severe Disease After Chikungunya Virus Infection: A 5-Year Follow-Up Study in Southern Thailand. Viral Immunol. 2019, 32, 442–452. [Google Scholar] [CrossRef]
- Ferreira, A.S.; Baldoni, N.R.; Cardoso, C.S.; Oliveira, C.D.L. Biomarkers of Severity and Chronification in Chikungunya Fever: A Systematic Review and Meta-Analysis. Rev. Inst. Med. Trop. Sao Paulo 2021, 63, e16. [Google Scholar] [CrossRef]
- Soares-Schanoski, A.; Baptista Cruz, N.; de Castro-Jorge, L.A.; de Carvalho, R.V.H.; Santos, C.A.D.; da Rós, N.; Oliveira, Ú.; Costa, D.D.; Santos, C.L.S.D.; Cunha, M.D.P.; et al. Systems Analysis of Subjects Acutely Infected with the Chikungunya Virus. PLoS Pathog. 2019, 15, e1007880. [Google Scholar] [CrossRef]
- Jaffar-Bandjee, M.C.; Das, T.; Hoarau, J.J.; Krejbich Trotot, P.; Denizot, M.; Ribera, A.; Roques, P.; Gasque, P. Chikungunya Virus Takes Centre Stage in Virally Induced Arthritis: Possible Cellular and Molecular Mechanisms to Pathogenesis. Microbes Infect. 2009, 11, 1206–1218. [Google Scholar] [CrossRef]
- Sourisseau, M.; Schilte, C.; Casartelli, N.; Trouillet, C.; Guivel-Benhassine, F.; Rudnicka, D.; Sol-Foulon, N.; Le Roux, K.; Prevost, M.-C.; Fsihi, H.; et al. Characterization of Reemerging Chikungunya Virus. PLoS Pathog. 2007, 3, e89. [Google Scholar] [CrossRef] [PubMed]
- Wikan, N.; Sakoonwatanyoo, P.; Ubol, S.; Yoksan, S.; Smith, D.R. Chikungunya Virus Infection of Cell Lines: Analysis of the East, Central and South African Lineage. PLoS ONE 2012, 7, e31102. [Google Scholar] [CrossRef] [PubMed]
- Krejbich-Trotot, P.; Denizot, M.; Hoarau, J.-J.; Jaffar-Bandjee, M.-C.; Das, T.; Gasque, P. Chikungunya Virus Mobilizes the Apoptotic Machinery to Invade Host Cell Defenses. FASEB J. 2011, 25, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Her, Z.; Malleret, B.; Chan, M.; Ong, E.K.S.; Wong, S.-C.; Kwek, D.J.C.; Tolou, H.; Lin, R.T.P.; Tambyah, P.A.; Rénia, L.; et al. Active Infection of Human Blood Monocytes by Chikungunya Virus Triggers an Innate Immune Response. J. Immunol. 2010, 184, 5903–5913. [Google Scholar] [CrossRef]
- de Lima Cavalcanti, T.Y.V.; Pereira, M.R.; de Paula, S.O.; Franca, R.F.d.O. A Review on Chikungunya Virus Epidemiology, Pathogenesis and Current Vaccine Development. Viruses 2022, 14, 969. [Google Scholar] [CrossRef]
- Shrinet, J.; Shastri, J.S.; Gaind, R.; Bhavesh, N.S.; Sunil, S. Serum Metabolomics Analysis of Patients with Chikungunya and Dengue Mono/Co-Infections Reveals Distinct Metabolite Signatures in the Three Disease Conditions. Sci. Rep. 2016, 6, 36833. [Google Scholar] [CrossRef]
- Chirathaworn, C.; Chansaenroj, J.; Poovorawan, Y. Cytokines and Chemokines in Chikungunya Virus Infection: Protection or Induction of Pathology. Pathogens 2020, 9, 415. [Google Scholar] [CrossRef]
- Chirathaworn, C.; Poovorawan, Y.; Lertmaharit, S.; Wuttirattanakowit, N. Cytokine Levels in Patients with Chikungunya Virus Infection. Asian Pac. J. Trop. Med. 2013, 6, 631–634. [Google Scholar] [CrossRef]
- Srivastava, P.; Chaudhary, S.; Malhotra, S.; Varma, B.; Sunil, S. Transcriptome Analysis of Human Macrophages upon Chikungunya Virus (CHIKV) Infection Reveals Regulation of Distinct Signaling and Metabolic Pathways during the Early and Late Stages of Infection. Heliyon 2023, 9, e17158. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. MRNA Capping: Biological Functions and Applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Igarashi, M.; Kato, H. Targeting Cap1 RNA Methyltransferases as an Antiviral Strategy. Cell Chem. Biol. 2024, 31, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.; Hons, M.; Rabah, N.; Zamarreño, N.; Arranz, R.; Reguera, J. Structural Basis and Dynamics of Chikungunya Alphavirus RNA Capping by NsP1 Capping Pores. Proc. Natl. Acad. Sci. USA 2023, 120, e2213934120. [Google Scholar] [CrossRef] [PubMed]
- Ahola, T.; Kääriäinen, L. Reaction in Alphavirus MRNA Capping: Formation of a Covalent Complex of Nonstructural Protein NsP1 with 7-Methyl-GMP. Proc. Natl. Acad. Sci. USA 1995, 92, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Stollar, V. Expression of Sindbis Virus NsP1 and Methyltransferase Activity in Escherichia Coli. Virology 1991, 184, 423–427. [Google Scholar] [CrossRef]
- Li, C.; Guillén, J.; Rabah, N.; Blanjoie, A.; Debart, F.; Vasseur, J.-J.; Canard, B.; Decroly, E.; Coutard, B. MRNA Capping by Venezuelan Equine Encephalitis Virus NsP1: Functional Characterization and Implications for Antiviral Research. J. Virol. 2015, 89, 8292–8303. [Google Scholar] [CrossRef]
- Sokoloski, K.J.; Haist, K.C.; Morrison, T.E.; Mukhopadhyay, S.; Hardy, R.W. Noncapped Alphavirus Genomic RNAs and Their Role during Infection. J. Virol. 2015, 89, 6080–6092. [Google Scholar] [CrossRef]
- Karpe, Y.A.; Aher, P.P.; Lole, K.S. NTPase and 5′-RNA Triphosphatase Activities of Chikungunya Virus NsP2 Protein. PLoS ONE 2011, 6, e22336. [Google Scholar] [CrossRef]
- Amaral, J.K.; Bilsborrow, J.B.; Schoen, R.T. Chronic Chikungunya Arthritis and Rheumatoid Arthritis: What They Have in Common. Am. J. Med. 2020, 133, e91–e97. [Google Scholar] [CrossRef]
- Ozden, S.; Huerre, M.; Riviere, J.-P.; Coffey, L.L.; Afonso, P.V.; Mouly, V.; de Monredon, J.; Roger, J.-C.; El Amrani, M.; Yvin, J.-L.; et al. Human Muscle Satellite Cells as Targets of Chikungunya Virus Infection. PLoS ONE 2007, 2, e527. [Google Scholar] [CrossRef]
- Poh, C.M.; Chan, Y.-H.; Ng, L.F.P. Role of T Cells in Chikungunya Virus Infection and Utilizing Their Potential in Anti-Viral Immunity. Front. Immunol. 2020, 11, 287. [Google Scholar] [CrossRef]
- Mapalagamage, M.; Weiskopf, D.; Sette, A.; De Silva, A.D. Current Understanding of the Role of T Cells in Chikungunya, Dengue and Zika Infections. Viruses 2022, 14, 242. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.N.; Ahmed, R. High Antigen Levels Are the Cause of T Cell Exhaustion during Chronic Viral Infection. Proc. Natl. Acad. Sci. USA 2009, 106, 8623–8628. [Google Scholar] [CrossRef] [PubMed]
- Teo, T.-H.; Chan, Y.-H.; Lee, W.W.L.; Lum, F.-M.; Amrun, S.N.; Her, Z.; Rajarethinam, R.; Merits, A.; Rötzschke, O.; Rénia, L.; et al. Fingolimod Treatment Abrogates Chikungunya Virus-Induced Arthralgia. Sci. Transl. Med. 2017, 9, eaal1333. [Google Scholar] [CrossRef] [PubMed]
- Petitdemange, C.; Wauquier, N.; Vieillard, V. Control of Immunopathology during Chikungunya Virus Infection. J. Allergy Clin. Immunol. 2015, 135, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Gardner, C.L.; Burke, C.W.; Higgs, S.T.; Klimstra, W.B.; Ryman, K.D. Interferon-Alpha/Beta Deficiency Greatly Exacerbates Arthritogenic Disease in Mice Infected with Wild-Type Chikungunya Virus but Not with the Cell Culture-Adapted Live-Attenuated 181/25 Vaccine Candidate. Virology 2012, 425, 103–112. [Google Scholar] [CrossRef]
- Chaaitanya, I.K.; Muruganandam, N.; Sundaram, S.G.; Kawalekar, O.; Sugunan, A.P.; Manimunda, S.P.; Ghosal, S.R.; Muthumani, K.; Vijayachari, P. Role of Proinflammatory Cytokines and Chemokines in Chronic Arthropathy in CHIKV Infection. Viral Immunol. 2011, 24, 265–271. [Google Scholar] [CrossRef]
- Lohachanakul, J.; Phuklia, W.; Thannagith, M.; Thonsakulprasert, T.; Ubol, S. High Concentrations of Circulating Interleukin-6 and Monocyte Chemotactic Protein-1 with Low Concentrations of Interleukin-8 Were Associated with Severe Chikungunya Fever during the 2009-2010 Outbreak in Thailand. Microbiol. Immunol. 2012, 56, 134–138. [Google Scholar] [CrossRef]
- Maleki, F.; Ovens, K.; McQuillan, I.; Kusalik, A.J. Size Matters: How Sample Size Affects the Reproducibility and Specificity of Gene Set Analysis. Hum. Genom. 2019, 13 (Suppl. 1), 42. [Google Scholar] [CrossRef]
- Fryett, J.J.; Morris, A.P.; Cordell, H.J. Investigation of Prediction Accuracy and the Impact of Sample Size, Ancestry, and Tissue in Transcriptome-Wide Association Studies. Genet. Epidemiol. 2020, 44, 425–441. [Google Scholar] [CrossRef]
- Li, G.; Ruan, S.; Zhao, X.; Liu, Q.; Dou, Y.; Mao, F. Transcriptomic Signatures and Repurposing Drugs for COVID-19 Patients: Findings of Bioinformatics Analyses. Comput. Struct. Biotechnol. J. 2021, 19, 1–15. [Google Scholar] [CrossRef]
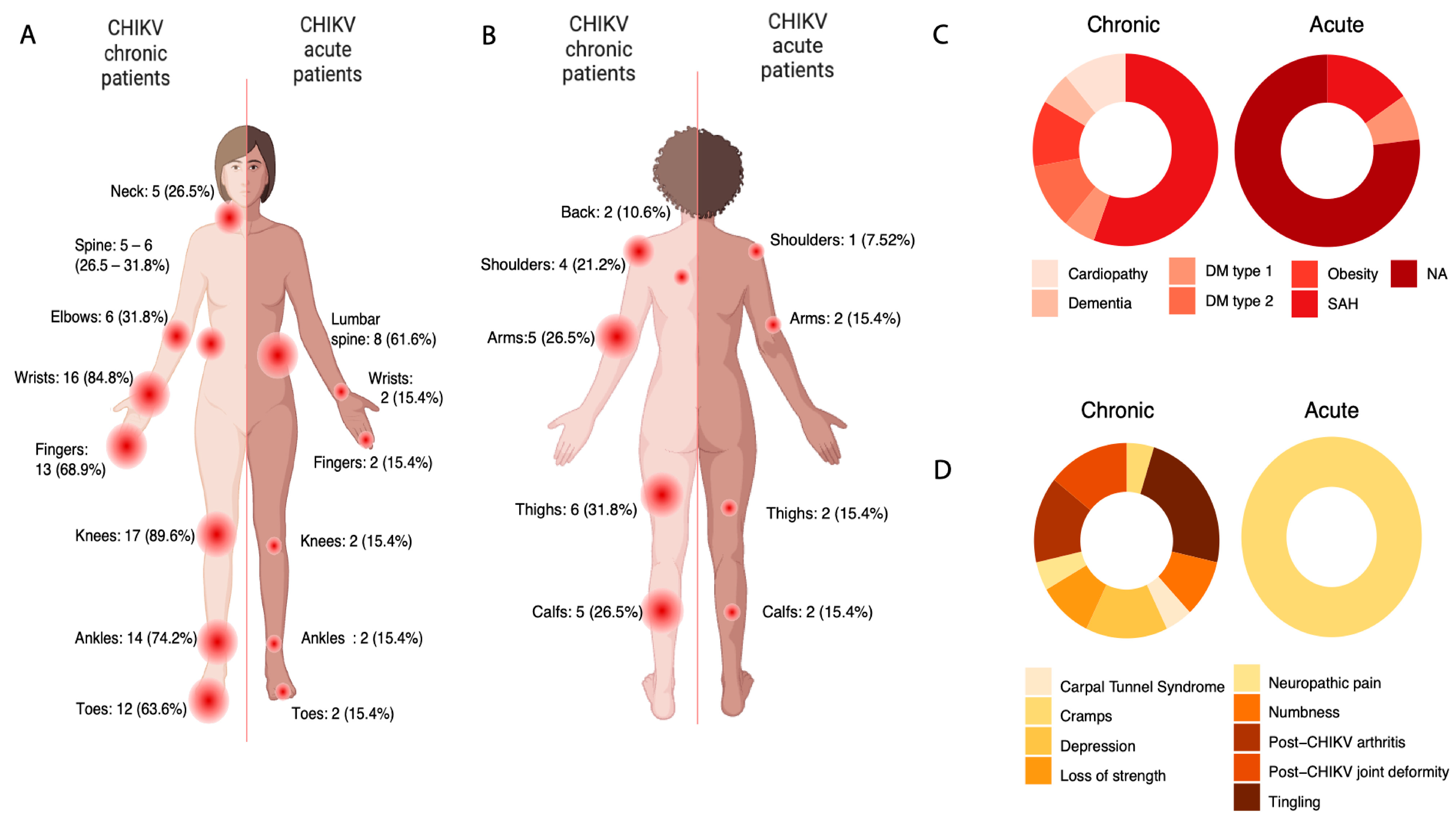
| Patients (n = 37) | |||
| Chronic (n = 19) | Acute (n = 13) | Control (n = 5) | |
| Epidemiological | |||
| Mean age (SD) | 60.4 (±13.74) | 44.0 (±20.35) | 57.8(±20.57) |
| Gender | |||
| Female | 17 (89.6%) | 7 (54%) | 1 (20%) |
| Male | 2 (10.4%) | 6 (46%) | 4 (80%) |
| NA | - | - | - |
| Onset of symptoms (SD) | - | 5 days | - |
| Schooling | |||
| NA | 1 (5.3%) | 11(84.1%) | 0 (0%) |
| Incomplete Elementary School | 2 (10.6%) | 0 (0%) | 0 (0%) |
| Complete Elementary School | 2 (10.6%) | 0 (0%) | 0 (0%) |
| Incomplete High School | 5 (26.3%) | 0 (0%) | 1 (20%) |
| Complete High School | 5 (26.3%) | 2 (15.9%) | 2 (40%) |
| Incomplete Higher Education | 2 (10.6%) | 0 (0%) | 0 (0%) |
| Complete Higher Education | 2 (10.6%) | 0 (0%) | 1 (20%) |
| Postgraduate studies | 0 (0%) | 0 (0%) | 1 (20%) |
| Household income | |||
| Less than minimum wage | 0 (0%) | 1 (7.52%) | 0 (0%) |
| From 1 to 2x minimum wages | 18 (94.8%) | 2 (15.4%) | 5 (100%) |
| From 3 to 4× minimum wages | 1 (5.3%) | 0 (0%) | 0 (0%) |
| More than 4× minimum wages | 0 (0%) | 0 (0%) | 0 (0%) |
| NA | 0 (0%) | 10 (77.08%) | 0 (0%) |
| Use of Immunosuppressive or Anti-inflammatory Medication | |||
| Yes | 9 (47.4%) | 2 (15.4%) | 0 (0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fritsch, H.; Giovanetti, M.; Clemente, L.G.; da Rocha Fernandes, G.; Fonseca, V.; de Lima, M.M.; Falcão, M.; de Jesus, N.; de Cerqueira, E.M.; Venâncio da Cunha, R.; et al. Unraveling the Complexity of Chikungunya Virus Infection Immunological and Genetic Insights in Acute and Chronic Patients. Genes 2024, 15, 1365. https://doi.org/10.3390/genes15111365
Fritsch H, Giovanetti M, Clemente LG, da Rocha Fernandes G, Fonseca V, de Lima MM, Falcão M, de Jesus N, de Cerqueira EM, Venâncio da Cunha R, et al. Unraveling the Complexity of Chikungunya Virus Infection Immunological and Genetic Insights in Acute and Chronic Patients. Genes. 2024; 15(11):1365. https://doi.org/10.3390/genes15111365
Chicago/Turabian StyleFritsch, Hegger, Marta Giovanetti, Luan Gaspar Clemente, Gabriel da Rocha Fernandes, Vagner Fonseca, Maricelia Maia de Lima, Melissa Falcão, Neuza de Jesus, Erenilde Marques de Cerqueira, Rivaldo Venâncio da Cunha, and et al. 2024. "Unraveling the Complexity of Chikungunya Virus Infection Immunological and Genetic Insights in Acute and Chronic Patients" Genes 15, no. 11: 1365. https://doi.org/10.3390/genes15111365
APA StyleFritsch, H., Giovanetti, M., Clemente, L. G., da Rocha Fernandes, G., Fonseca, V., de Lima, M. M., Falcão, M., de Jesus, N., de Cerqueira, E. M., Venâncio da Cunha, R., de Oliveira Francisco, M. V. L., de Siqueira, I. C., de Oliveira, C., Xavier, J., Ferreira, J. G. G., Queiroz, F. R., Smith, E., Tisoncik-Go, J., Van Voorhis, W. C., ... Alcantara, L. C. J. (2024). Unraveling the Complexity of Chikungunya Virus Infection Immunological and Genetic Insights in Acute and Chronic Patients. Genes, 15(11), 1365. https://doi.org/10.3390/genes15111365









