Complete Genome Assembly of Amycolatopsis bartoniae DSM 45807T Allows the Characterization of a Novel Glycopeptide Biosynthetic Gene Cluster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequencing and Assembly of A. bartoniae DSM 45807 Genome
2.2. In Silico Analysis of Nucleic Acid and Amino Acid Sequences from A. bartoniae DSM 45807 GPA BGC
2.3. Media and Conditions for A. bartoniae DSM 45807 Cultivation
2.4. Agar Plug and Kirby-Bauer Disc Diffusion Assays
2.5. Scanning Electron Microscopy
3. Results
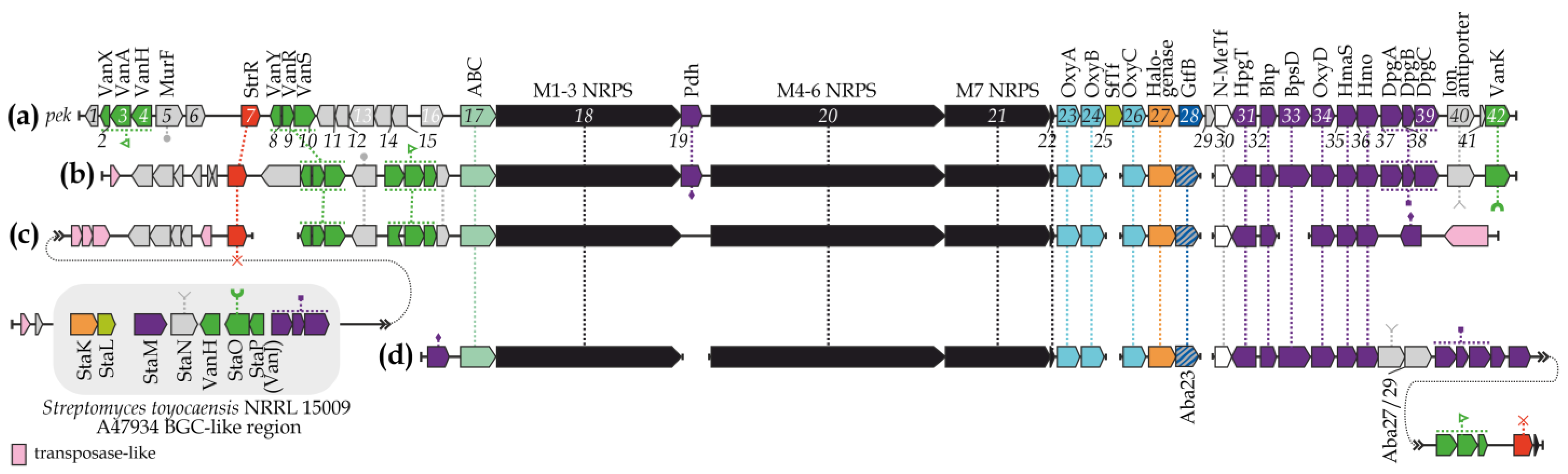
3.1. Characterization of the GPA BGC from A. bartoniae DSM 45807 and Biosynthetic Pathway Prediction
3.2. Novel aba-Encoded Glycosyltransferase Leads to a Set of Unusual GPA BGCs
3.3. aba-Encoded GPA Biosynthetic Pathway Is Inactive Under a Broad Range of Laboratory Conditions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Müller, A.; Klöckner, A.; Schneider, T. Targeting a cell wall biosynthesis hot spot. Nat. Prod. Rep. 2017, 34, 909–932. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Boddy, C.N.C.; Bräse, S.; Winssinger, N. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew. Chem. Int. Ed. 1999, 38, 2096–2152. [Google Scholar] [CrossRef]
- Stegmann, E.; Frasch, H.J.; Wohlleben, W. Glycopeptide biosynthesis in the context of basic cellular functions. Curr. Opin. Microbiol. 2010, 13, 595–602. [Google Scholar] [CrossRef]
- Chiu, H.T.; Hubbard, B.K.; Shah, A.N.; Eide, J.; Fredenburg, R.A.; Walsh, C.T.; Khosla, C. Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc. Natl. Acad. Sci. USA 2001, 98, 8548–8553. [Google Scholar] [CrossRef]
- Shawky, R.M.; Puk, O.; Wietzorrek, A.; Pelzer, S.; Takano, E.; Wohlleben, W.; Stegmann, E. The border sequence of the balhimycin biosynthesis gene cluster from Amycolatopsis balhimycina contains bbr, encoding a StrR-like pathway-specific regulator. J. Mol. Microbiol. Biotechnol. 2007, 13, 76–88. [Google Scholar] [CrossRef]
- Li, T.L.; Huang, F.; Haydock, S.F.; Mironenko, T.; Leadlay, P.F.; Spencer, J.B. Biosynthetic gene cluster of the glycopeptide antibiotic teicoplanin: Characterization of two glycosyltransferases and the key acyltransferase. Chem. Biol. 2004, 11, 107–119. [Google Scholar] [CrossRef]
- Williams, D.H. The glycopeptide story—How to kill the deadly “Superbugs”. Nat. Prod. Rep. 1996, 13, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Binda, E.; Berini, F.; Marinelli, F. Old and new glycopeptide antibiotics: Arom product to gene and back in the post-genomic era. Biotechnol. Adv. 2018, 36, 534–554. [Google Scholar] [CrossRef]
- Jeong, H.; Sim, Y.M.; Kim, H.J.; Lee, D.W.; Lim, S.K.; Lee, S.J. Genome sequence of the vancomycin-producing Amycolatopsis orientalis subsp. orientalis strain KCTC 9412T. Genome Announc. 2013, 1, e00408-13. [Google Scholar] [CrossRef] [PubMed]
- van Groesen, E.; Innocenti, P.; Martin, N.I. Recent advances in the development of semisynthetic glycopeptide antibiotics: 2014-2022. ACS Infect. Dis. 2022, 8, 1381–1407. [Google Scholar] [CrossRef] [PubMed]
- van Wageningen, A.M.A.; Kirkpatrick, P.N.; Williams, D.H.; Harris, B.R.; Kershaw, J.K.; Lennard, N.J.; Jones, M.; Jones, S.J.M.; Solenberg, P.J. Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 1998, 5, 155–162. [Google Scholar] [CrossRef]
- Sosio, M.; Stinchi, S.; Beltrametti, F.; Lazzarini, A.; Donadio, S. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 2003, 10, 541–549. [Google Scholar] [CrossRef]
- Parenti, F.; Cavalleri, B. Proposal to name the vancomycin-ristocetin like glycopeptides as dalbaheptides. J. Antibiot. 1989, 42, 1882–1883. [Google Scholar] [CrossRef]
- Culp, E.J.; Waglechner, N.; Wang, W.; Fiebig-Comyn, A.A.; Hsu, Y.P.; Koteva, K.; Sychantha, D.; Coombes, B.K.; Van Nieuwenhze, M.S.; Brun, Y.V.; et al. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 2020, 578, 582–587. [Google Scholar] [CrossRef]
- Gavriilidou, A.; Adamek, M.; Rodler, J.-P.; Kubach, N.; Kramer, S.; Huson, D.H.; Cryle, M.J.; Stegmann, E.; Ziemert, N. Phylogenetic distance and structural diversity directing a reclassification of glycopeptide antibiotics. bioRxiv 2023. [CrossRef]
- Al Rubaye, M.; Janice, J.; Bjørnholt, J.V.; Löhr, I.H.; Sundsfjord, A.; Hegstad, K. The first vanE-type vancomycin resistant Enterococcus faecalis isolates in Norway—Phenotypic and molecular characteristics. J. Glob. Antimicrob. Resist. 2024, 36, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Wardal, E.; Żabicka, D.; Hryniewicz, W.; Sadowy, E. VanA-Enterococcus faecalis in Poland: Hospital population clonal structure and vanA mobilome. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 1245–1261. [Google Scholar] [CrossRef]
- Hotz, J.F.; Staudacher, M.; Schefberger, K.; Spettel, K.; Schmid, K.; Kriz, R.; Schneider, L.; Hagemann, J.B.; Cyran, N.; Schmidt, K.; et al. Unraveling novel mutation patterns and morphological variations in two dalbavancin-resistant MRSA strains in Austria using whole genome sequencing and transmission electron microscopy. BMC Infect. Dis. 2024, 24, 899. [Google Scholar] [CrossRef]
- Waglechner, N.; McArthur, A.G.; Wright, G.D. Phylogenetic reconciliation reveals the natural history of glycopeptide antibiotic biosynthesis and resistance. Nat. Microbiol. 2019, 4, 1862–1871. [Google Scholar] [CrossRef]
- Xu, M.; Wang, W.; Waglechner, N.; Culp, E.J.; Guitor, A.K.; Wright, G.D. Phylogeny-informed synthetic biology reveals unprecedented structural novelty in Type V glycopeptide antibiotics. ACS Cent. Sci. 2022, 8, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.H.; Adamek, M.; Iftime, D.; Petras, D.; Schuseil, F.; Grond, S.; Stegmann, E.; Cryle, M.J.; Ziemert, N. Resurrecting ancestral antibiotics: Unveiling the origins of modern lipid II targeting glycopeptides. Nat. Commun. 2023, 14, 7842. [Google Scholar] [CrossRef] [PubMed]
- Andreo-Vidal, A.; Binda, E.; Fedorenko, V.; Marinelli, F.; Yushchuk, O. Genomic insights into the distribution and phylogeny of glycopeptide resistance determinants within the Actinobacteria phylum. Antibiotics 2021, 10, 1533. [Google Scholar] [CrossRef]
- Truman, A.W.; Kwun, M.J.; Cheng, J.; Yang, S.H.; Suh, J.W.; Hong, H.J. Antibiotic resistance mechanisms inform discovery: Identification and characterization of a novel Amycolatopsis strain producing ristocetin. Antimicrob. Agents Chemother. 2014, 58, 5687–5695. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wang, W.; Waglechner, N.; Culp, E.J.; Guitor, A.K.; Wright, G.D. GPAHex—A synthetic biology platform for Type IV–V glycopeptide antibiotic production and discovery. Nat. Commun. 2020, 11, 5232. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; O’Neill, K.R.; Haft, D.H.; Dicuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.; Buttner, M.; Chater, K. Practical Streptomyces Genetics, 2nd ed.; John Innes Foundation: Norwich, UK, 2000; ISBN 0-7084-0623-8. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; ISBN 0879695773. [Google Scholar]
- Yushchuk, O.; Ostash, I.; Mösker, E.; Vlasiuk, I.; Deneka, M.; Rückert, C.; Busche, T.; Fedorenko, V.; Kalinowski, J.; Süssmuth, R.D.; et al. Eliciting the silent lucensomycin biosynthetic pathway in Streptomyces cyanogenus S136 via manipulation of the global regulatory gene adpA. Sci. Rep. 2021, 11, 3507. [Google Scholar] [CrossRef]
- Mascher, T.; Zimmer, S.L.; Smith, T.A.; Helmann, J.D. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 2004, 48, 2888–2896. [Google Scholar] [CrossRef] [PubMed]
- Mascher, T.; Margulis, N.G.; Wang, T.; Ye, R.W.; Helmann, J.D. Cell wall stress responses in Bacillus subtilis: The regulatory network of the bacitracin stimulon. Mol. Microbiol. 2003, 50, 1591–1604. [Google Scholar] [CrossRef] [PubMed]
- Banik, J.J.; Brady, S.F. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Natl. Acad. Sci. USA 2008, 105, 17273–17277. [Google Scholar] [CrossRef]
- Yushchuk, O.; Ostash, B. Glycopeptide antibiotics: Genetics, chemistry, and new screening approaches. Nat. Prod. Actinomycetes Divers. Ecol. Drug Discov. 2022, 411–444. [Google Scholar] [CrossRef]
- Spohn, M.; Kirchner, N.; Kulik, A.; Jochim, A.; Wolf, F.; Muenzer, P.; Borst, O.; Gross, H.; Wohlleben, W.; Stegmann, E. Overproduction of ristomycin a by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17. Antimicrob. Agents Chemother. 2014, 58, 6185–6196. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Schneider, T.R.; Shieldrick, G.M. Crystal structure of vancomycin. Structure 1996, 4, 1509–1515. [Google Scholar] [CrossRef] [PubMed]
- Thykaer, J.; Nielsen, J.; Wohlleben, W.; Weber, T.; Gutknecht, M.; Lantz, A.E.; Stegmann, E. Increased glycopeptide production after overexpression of shikimate pathway genes being part of the balhimycin biosynthetic gene cluster. Metab. Eng. 2010, 12, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tseng, C.C.; Hubbard, B.K.; Walsh, C.T. Glycopeptide antibiotic biosynthesis: Enzymatic assembly of the dedicated amino acid monomer (S)-3,5-dihydroxyphenylglycine. Proc. Natl. Acad. Sci. USA 2001, 98, 14901–14906. [Google Scholar] [CrossRef]
- Pfeifer, V.; Nicholson, G.J.; Ries, J.; Recktenwald, J.; Schefer, A.B.; Shawky, R.M.; Schröder, J.; Wohlleben, W.; Pelzer, S. A polyketide synthase in glycopeptide biosynthesis. The biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J. Biol. Chem. 2001, 276, 38370–38377. [Google Scholar] [CrossRef]
- Sandercock, A.M.; Charles, E.H.; Scaife, W.; Kirkpatrick, P.N.; O’Brien, S.W.; Papageorgiou, E.A.; Spencer, J.B.; Williams, D.H. Biosynthesis of the l-p-hydroxyphenylglycine constituent of the vancomycin-group antibiotic chloroeremomycin. Chem. Commun. 2001, 7, 1252–1253. [Google Scholar] [CrossRef]
- Wu, J.; Woodard, R.W. New insights into the evolutionary links relating to the 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subfamilies. J. Biol. Chem. 2006, 281, 4042–4048. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, B.K.; Thomas, M.G.; Walsh, C.T. Biosynthesis of l-p-hydroxyphenylglycine, a non-proteinogenic amino acid constituent of peptide antibiotics. Chem. Biol. 2000, 7, 931–942. [Google Scholar] [CrossRef]
- Recktenwald, J.; Shawky, R.; Puk, O.; Pfenning, F.; Keller, U.; Wohlleben, W.; Pelzer, S. Nonribosomal biosynthesis of vancomycin-type antibiotics: A heptapeptide backbone and eight peptide synthetase modules. Microbiology 2002, 148, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Puk, O.; Bischoff, D.; Kittel, C.; Pelzer, S.; Weist, S.; Stegmann, E.; Süssmuth, R.D.; Wohlleben, W. Biosynthesis of chloro-β-hydroxytyrosine, a nonproteinogenic amino acid of the peptidic backbone of glycopeptide antibiotics. J. Bacteriol. 2004, 186, 6093–6100. [Google Scholar] [CrossRef] [PubMed]
- Mulyani, S.; Egel, E.; Kittel, C.; Turkanovic, S.; Wohlleben, W.; Süssmuth, R.D.; Van Pée, K.H. The thioesterase Bhp is involved in the formation of β-hydroxytyrosine during balhimycin biosynthesis in Amycolatopsis balhimycina. ChemBioChem 2010, 11, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, E.; Pelzer, S.; Bischoff, D.; Puk, O.; Stockert, S.; Butz, D.; Zerbe, K.; Robinson, J.; Süssmuth, R.D.; Wohlleben, W. Genetic analysis of the balhimycin (vancomycin-type) oxygenase genes. J. Biotechnol. 2006, 124, 640–653. [Google Scholar] [CrossRef]
- Hadatsch, B.; Butz, D.; Schmiederer, T.; Steudle, J.; Wohlleben, W.; Süssmuth, R.; Stegmann, E. The biosynthesis of teicoplanin-type glycopeptide antibiotics: Assignment of P450 mono-oxygenases to side chain cyclizations of glycopeptide A47934. Chem. Biol. 2007, 14, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; Wang, W.; Thaker, M.N.; Tan, S.; Wright, G.D. How to make a glycopeptide: A synthetic biology approach to expand antibiotic chemical diversity. ACS Infect. Dis. 2016, 2, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Funabashi, M.; Baba, S.; Nonaka, K.; Hosobuchi, M.; Fujita, Y.; Shibata, T.; Van Lanen, S.G. The biosynthesis of liposidomycin-like A-90289 antibiotics featuring a new type of sulfotransferase. ChemBioChem 2010, 11, 184–190. [Google Scholar] [CrossRef]
- Kaysser, L.; Siebenberg, S.; Kammerer, B.; Gust, B. Analysis of the liposidomycin gene cluster leads to the identification of new caprazamycin derivatives. ChemBioChem 2010, 11, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, K.; Petránková, B.; Petrásková, L.; Pelantová, H.; Křen, V.; Valentová, K.; Bojarová, P. New bacterial aryl sulfotransferases: Effective tools for sulfation of polyphenols. J. Agric. Food Chem. 2024, 72, 22208–22216. [Google Scholar] [CrossRef]
- Yushchuk, O.; Zhukrovska, K.; Berini, F.; Fedorenko, V.; Marinelli, F. Genetics behind the glycosylation patterns in the biosynthesis of dalbaheptides. Front. Chem. 2022, 10, 858708. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Nonaka, K.; Nie, L.; Zhang, J.; Christenson, S.D.; Bae, J.; Van Lanen, S.G.; Zazopoulos, E.; Farnet, C.M.; Yang, C.F.; et al. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem. Biol. 2005, 12, 293–302. [Google Scholar] [CrossRef]
- Yushchuk, O.; Horbal, L.; Ostash, B.; Marinelli, F.; Wohlleben, W.; Stegmann, E.; Fedorenko, V. Regulation of teicoplanin biosynthesis: Refining the roles of tei cluster-situated regulatory genes. Appl. Microbiol. Biotechnol. 2019, 103, 4089–4102. [Google Scholar] [CrossRef]
- Menges, R.; Muth, G.; Wohlleben, W.; Stegmann, E. The ABC transporter Tba of Amycolatopsis balhimycina is required for efficient export of the glycopeptide antibiotic balhimycin. Appl. Microbiol. Biotechnol. 2007, 77, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, S.; Chen, Y.; Tian, X.; Gu, Y.; Ju, J. Identification and heterologous expression of the kendomycin B biosynthetic gene cluster from Verrucosispora sp. SCSIO 07399. Mar. Drugs 2021, 19, 673. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.N.; Wang, W.; Spanogiannopoulos, P.; Waglechner, N.; King, A.M.; Medina, R.; Wright, G.D. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 2013, 31, 922–927. [Google Scholar] [CrossRef]
- Yim, G.; Wang, W.; Pawlowski, A.C.; Wright, G.D. Trichlorination of a teicoplanin-type glycopeptide antibiotic by the halogenase StaI evades resistance. Antimicrob. Agents Chemother. 2018, 62, e01540-18. [Google Scholar] [CrossRef]
- Stinchi, S.; Carrano, L.; Lazzarini, A.; Feroggio, M.; Grigoletto, A.; Sosio, M.; Donadio, S. A derivative of the glycopeptide A40926 produced by inactivation of the β-hydroxylase gene in Nonomuraea sp. ATCC39727. FEMS Microbiol. Lett. 2006, 256, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Zucchi, T.D.; Bonda, A.N.V.; Frank, S.; Kim, B.Y.; Kshetrimayum, J.D.; Goodfellow, M. Amycolatopsis bartoniae sp. nov. and Amycolatopsis bullii sp. nov., mesophilic actinomycetes isolated from arid Australian soils. Antonie Van Leeuwenhoek 2012, 102, 91–98. [Google Scholar] [CrossRef]
- Jung, H.M.; Kim, S.Y.; Prabhu, P.; Moon, H.J.; Kim, I.W.; Lee, J.K. Optimization of culture conditions and scale-up to plant scales for teicoplanin production by Actinoplanes teichomyceticus. Appl. Microbiol. Biotechnol. 2008, 80, 21–27. [Google Scholar] [CrossRef]
- Taurino, C.; Frattini, L.; Marcone, G.L.; Gastaldo, L.; Marinelli, F. Actinoplanes teichomyceticus ATCC 31121 as a cell factory for producing teicoplanin. Microb. Cell Fact. 2011, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Yushchuk, O.; Andreo-Vidal, A.; Marcone, G.L.; Bibb, M.; Marinelli, F.; Binda, E. New molecular tools for regulation and improvement of A40926 glycopeptide antibiotic production in Nonomuraea gerenzanensis ATCC 39727. Front. Microbiol. 2020, 11, 8. [Google Scholar] [CrossRef]
- Yushchuk, O.; Vior, N.M.; Andreo-Vidal, A.; Berini, F.; Rückert, C.; Busche, T.; Binda, E.; Kalinowski, J.; Truman, A.W.; Marinelli, F. Genomic-led discovery of a novel glycopeptide antibiotic by Nonomuraea coxensis DSM 45129. ACS Chem. Biol. 2021, 16, 915–928. [Google Scholar] [CrossRef]
- Thaker, M.N.; Wright, G.D. Opportunities for synthetic biology in antibiotics: Expanding glycopeptide chemical diversity. ACS Synth. Biol. 2015, 4, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Pootoolal, J.; Thomas, M.G.; Marshall, C.G.; Neu, J.M.; Hubbard, B.K.; Walsh, C.T.; Wright, G.D. Assembling the glycopeptide antibiotic scaffold: The biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. USA 2002, 99, 8962–8967. [Google Scholar] [CrossRef] [PubMed]
- McCormick, J.R.; Flärdh, K. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 2012, 36, 206–231. [Google Scholar] [CrossRef]
- Ostash, B.; Yushchuk, O.; Tistechok, S.; Mutenko, H.; Horbal, L.; Muryn, A.; Dacyuk, Y.; Kalinowski, J.; Luzhetskyy, A.; Fedorenko, V. The adpA-like regulatory gene from Actinoplanes teichomyceticus: In silico analysis and heterologous expression. World J. Microbiol. Biotechnol. 2015, 31, 1297–1301. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Binda, E.; Carrano, L.; Bibb, M.; Marinelli, F. Relationship between glycopeptide production and resistance in the actinomycete Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 2014, 58, 5191–5201. [Google Scholar] [CrossRef]
- Koshla, O.; Yushchuk, O.; Ostash, I.; Dacyuk, Y.; Myronovskyi, M.; Jäger, G.; Süssmuth, R.D.; Luzhetskyy, A.; Byström, A.; Kirsebom, L.A.; et al. Gene miaA for post-transcriptional modification of tRNAXXA is important for morphological and metabolic differentiation in Streptomyces. Mol. Microbiol. 2019, 112, 249–265. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]

| aba | Locus Tag: | Product: | Function: |
|---|---|---|---|
| aba1 | AMYBAR_003213 | StrR-like transcriptional regulator | Function unknown |
| aba2 | AMYBAR_003214 | M15 family metallopeptidase (VanY) | Cleaves d-Ala-d-Ala dipeptides, involved in GPA resistance |
| aba3 | AMYBAR_003215 | KefB membrane component of Kef-type K+ transport system | Putatively involved in inorganic ion transport |
| aba4 | AMYBAR_003216 | StrR-like transcriptional regulator | Putatively involved in the positive regulation of GPA biosynthesis |
| aba5 | AMYBAR_003217 | Chorismate mutase type I | Converts chorismate to prephenate, involved in amino acid supply for GPA biosynthesis |
| aba6 | AMYBAR_003218 | Prephenate dehydrogenase (PDH) | Converts prephenate to 4-hydroxyphenylpyruvate, involved in amino acid supply for GPA biosynthesis |
| aba7 | AMYBAR_003219 | MdlB(MsbA)-like ABC-transporter | Putatively involved in GPA export |
| abaA | AMYBAR_003220 | NRPS | GPA biosynthesis NRPS, modules 1–2 |
| abaB | AMYBAR_003221 | NRPS | GPA biosynthesis NRPS, module 3 |
| abaC | AMYBAR_003222 | NRPS | GPA biosynthesis NRPS, modules 4–5–6 |
| abaD | AMYBAR_003223 | NRPS | GPA biosynthesis NRPS, module 7 |
| aba8 | AMYBAR_003224 | MbtH-like protein | GPA biosynthesis NRPS chaperone |
| aba9 | AMYBAR_003225 | Cytochrome P450 | GPA cross-linking monooxygenase (OxyA), involved in the formation of D-O-E link |
| aba10 | AMYBAR_003226 | Cytochrome P450 | GPA cross-linking monooxygenase (OxyE), involved in the formation of F-O-G link |
| aba11 | AMYBAR_003227 | Cytochrome P450 | GPA cross-linking monooxygenase (OxyB), involved in the formation of C-O-D link |
| aba12 | AMYBAR_003228 | Sulfotransferase_1 superfamily domain protein | Putatively involved in GPA sulfation |
| aba13 | AMYBAR_003229 | Sulfotransferase_1 superfamily domain protein | Putatively involved in GPA sulfation |
| aba14 | AMYBAR_003230 | Sulfotransferase_1 superfamily domain protein | Putatively involved in GPA sulfation |
| aba15 | AMYBAR_003231 | Cytochrome P450 | GPA cross-linking monooxygenase (OxyC), involved in the formation of A-B link |
| aba16 | AMYBAR_003232 | Halogenase | Involved in GPA halogenation |
| aba17 | AMYBAR_003233 | GT2-family glycosyltransferase | Function unknown |
| aba18 | AMYBAR_003234 | GT39-family glycosyltransferase | Function unknown |
| aba19 | AMYBAR_003235 | Arylsulfotransferase (ASST) | Function unknown |
| aba20 | AMYBAR_003236 | Methyltransferase | Involved in C-methylation of the aglycone AA3 |
| aba21 | AMYBAR_003237 | Hypothetical protein | Function unknown |
| aba22 | AMYBAR_003238 | Dyp-type peroxidase family protein | Function unknown |
| aba23 | AMYBAR_003239 | GT1-family glycosyltransferase | Function unknown |
| aba24 | AMYBAR_003240 | GT39-family glycosyltransferase | Attaches d-mannose residue to GPA aglycone AA7 |
| aba25 | AMYBAR_003241 | 4-hydroxyphenylglycine transaminase (HpgT) | Converts 4-hydroxybenzoylformate to 4-hydroxyphenylglycine or 3,5-dihydroxyphenylglyoxylate to 3,5-dihydroxyphenylglycine, involved in the biosynthesis of Hpg and Dpg |
| aba26 | AMYBAR_003242 | Hydroxymandelate synthase (HmaS) | Converts 4-hydroxyphenylpyruvate to 4-hydroxymandelate, involved in Hpg biosynthesis |
| aba27 | AMYBAR_003243 | Pyrroloquinoline quinone-dependent glucose dehydrogenase | Function unknown |
| aba28 | AMYBAR_003244 | KefB membrane component of Kef-type K+ transport system | Putatively involved in inorganic ion transport |
| aba29 | AMYBAR_003245 | Pyrroloquinoline quinone-dependent glucose dehydrogenase | Function unknown |
| aba30 | AMYBAR_003246 | Type III polyketide synthase (DpgA) | Converts malonyl-CoA units into 3,5-dihydroxyphenylacetate, involved in Dpg biosynthesis |
| aba31 | AMYBAR_003247 | Enoyl-CoA hydratase (DpgB) | Converts malonyl-CoA units into 3,5-dihydroxyphenylacetate, involved in Dpg biosynthesis |
| aba32 | AMYBAR_003248 | 3,5-dihydroxyphenylacetyl-CoA 1,2-dioxygenase (DpgC) | Converts 3,5-dihydroxyphenylacetate in 3,5-dihydroxyphenylglyoxylate, involved in Dpg biosynthesis |
| aba33 | AMYBAR_003249 | Enoyl-CoA hydratase (DpgD) | Converts malonyl-CoA units into 3,5-dihydroxyphenylacetate, involved in Dpg biosynthesis |
| aba34 | AMYBAR_003250 | 3-deoxy-d-arabinoheptulosonate 7-phosphate synthase (Dahp) | Converts phosphoenolpyruvate and d-erythrose-4-phosphate into 3-deoxy-d-arabinoheptulosonate 7-phosphate, involved in amino acid supply for GPA biosynthesis |
| aba35 | AMYBAR_003251 | Thioesterase | Catalyzes Bht release from NRPS, involved in Bht biosynthesis |
| aba36 | AMYBAR_003252 | Single-modular NRPS (Bht biosynthesis) | Carries tyrosine and Bht, involved in Bht biosynthesis |
| aba37 | AMYBAR_003253 | Cytochrome P450 | Converts tyrosine to Bht, involved in Bht biosynthesis |
| aba38 | AMYBAR_003254 | d-Lactate dehydrogenase (VanH) | Converts pyruvate to d-lactate, involved in GPA resistance |
| aba39 | AMYBAR_003255 | d-Ala-d-Lac ligase (VanA) | Produces d-Ala-d-Lac depsipeptide, involved in GPA resistance |
| aba40 | AMYBAR_003256 | d,d-dipeptidase (VanX) | Cleaves d-Ala-d-Ala dipeptides, involved in GPA resistance |
| aba41 | AMYBAR_003257 | Two-component regulatory system sensor histidine kinase (VanS) | Involved in GPA sensing and resistance |
| aba42 | AMYBAR_003258 | Two-component regulatory system sensor resistance response regulator (VanR) | Involved in GPA sensing and resistance |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stepanyshyn, A.; Rückert-Reed, C.; Busche, T.; Yaruta, B.; Andreo-Vidal, A.; Marinelli, F.; Kalinowski, J.; Yushchuk, O. Complete Genome Assembly of Amycolatopsis bartoniae DSM 45807T Allows the Characterization of a Novel Glycopeptide Biosynthetic Gene Cluster. Genes 2024, 15, 1651. https://doi.org/10.3390/genes15121651
Stepanyshyn A, Rückert-Reed C, Busche T, Yaruta B, Andreo-Vidal A, Marinelli F, Kalinowski J, Yushchuk O. Complete Genome Assembly of Amycolatopsis bartoniae DSM 45807T Allows the Characterization of a Novel Glycopeptide Biosynthetic Gene Cluster. Genes. 2024; 15(12):1651. https://doi.org/10.3390/genes15121651
Chicago/Turabian StyleStepanyshyn, Anastasia, Christian Rückert-Reed, Tobias Busche, Bohdan Yaruta, Andres Andreo-Vidal, Flavia Marinelli, Jörn Kalinowski, and Oleksandr Yushchuk. 2024. "Complete Genome Assembly of Amycolatopsis bartoniae DSM 45807T Allows the Characterization of a Novel Glycopeptide Biosynthetic Gene Cluster" Genes 15, no. 12: 1651. https://doi.org/10.3390/genes15121651
APA StyleStepanyshyn, A., Rückert-Reed, C., Busche, T., Yaruta, B., Andreo-Vidal, A., Marinelli, F., Kalinowski, J., & Yushchuk, O. (2024). Complete Genome Assembly of Amycolatopsis bartoniae DSM 45807T Allows the Characterization of a Novel Glycopeptide Biosynthetic Gene Cluster. Genes, 15(12), 1651. https://doi.org/10.3390/genes15121651







