Genomic Variants and Worldwide Epidemiology of Breast Cancer: A Genome-Wide Association Studies Correlation Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. SNP Determination
2.2. Epidemiological and Genetic Data
2.3. Statistical Analysis
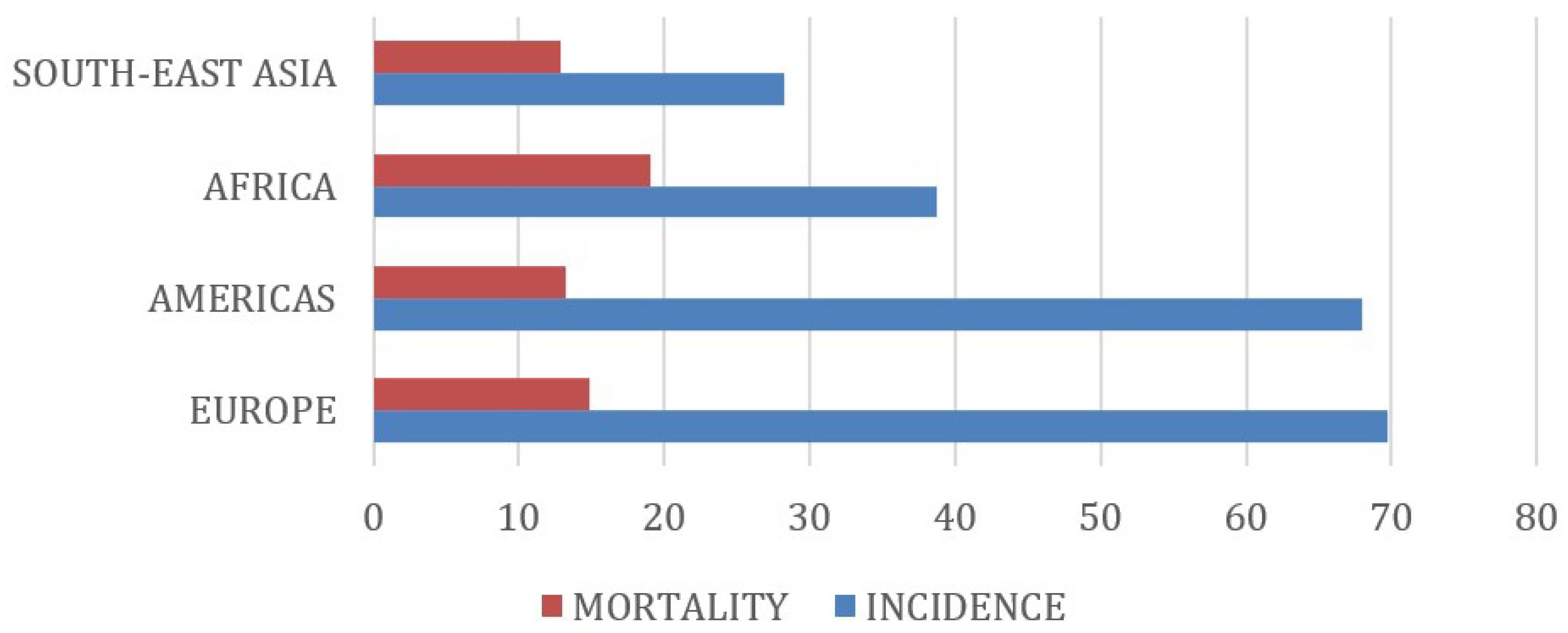
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Cancer of the Breast (Female)—Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 11 November 2023).
- Wilkinson, L.; Gathani, T. Understanding Breast Cancer as a Global Health Concern. Br. J. Radiol. 2022, 95, 20211033. [Google Scholar] [CrossRef] [PubMed]
- Zavala, V.A.; Serrano-Gomez, S.J.; Dutil, J.; Fejerman, L. Genetic Epidemiology of Breast Cancer in Latin America. Genes 2019, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. Available online: https://pubmed.ncbi.nlm.nih.gov/33471991/ (accessed on 11 November 2023).
- Use of Single-Nucleotide Polymorphisms and Mammographic Density Plus Classic Risk Factors for Breast Cancer Risk Prediction. Available online: https://pubmed.ncbi.nlm.nih.gov/29346471/ (accessed on 11 November 2023).
- Liang, B.; Ding, H.; Huang, L.; Luo, H.; Zhu, X. GWAS in cancer: Progress and challenges. Mol. Genet. Genom. 2020, 295, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Relationships between SNPs and Prognosis of Breast Cancer and Pathogenic Mechanism—He—2019—Molecular Genetics & Genomic Medicine—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/10.1002/mgg3.871 (accessed on 11 November 2023).
- World Health Organization (WHO). Global Cancer Observatory. Available online: http://gco.iarc.fr/ (accessed on 11 November 2023).
- Hashemi, M.; Aftabi, S.; Moazeni-Roodi, A.; Sarani, H.; Wiechec, E.; Ghavami, S. Association of CASP8 Polymorphisms and Cancer Susceptibility: A Meta-Analysis. Eur. J. Pharmacol. 2020, 881, 173201. [Google Scholar] [CrossRef] [PubMed]
- Disulfidptosis-Associated lncRNAs Predict Breast Cancer Subtypes|Scientific Reports. Available online: https://www.nature.com/articles/s41598-023-43414-1 (accessed on 11 November 2023).
- KRAS/NRAS Mutations Associated with Distant Metastasis and BRAF/PIK3CA Mutations Associated with Poor Tumor Differentiation in Colorectal Cancer. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10503567/#cit0013 (accessed on 11 November 2023).
- Nisar, T.; Arshad, K.; Abbas, Z.; Khan, M.A.; Safdar, S.; Shaikh, R.S.; Saeed, A. Prevalence of GCKR Rs1260326 Variant in Subjects with Obesity Associated NAFLD and T2DM: A Case-Control Study in South Punjab, Pakistan. J. Obes. 2023, 2023, 6661858. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, A.; Paliwal, V.; Jain, S.; Paliwal, S.; Sharma, S. Current Insight on the Role of Glucokinase and Glucokinase Regulatory Protein in Diabetes. Mini. Rev. Med. Chem. 2023; online ahead of print. [Google Scholar] [CrossRef]
- Wang, X.; Ye, X.; Chen, Y.; Lin, J. Mechanism of M2 Type Macrophage-Derived Extracellular Vesicles Regulating PD-L1 Expression via the MISP/IQGAP1 Axis in Hepatocellular Carcinoma Immunotherapy Resistance. Int. Immunopharmacol. 2023, 124, 110848. [Google Scholar] [CrossRef] [PubMed]
- ZNF577 Methylation Levels in Leukocytes from Women with Breast Cancer Is Modulated by Adiposity, Menopausal State, and the Mediterranean Diet. Available online: https://pubmed.ncbi.nlm.nih.gov/32390948/ (accessed on 11 November 2023).
- Mechanisms of Stretch-Mediated Skin Expansion at Single-Cell Resolution. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7116042/ (accessed on 11 November 2023).
- Reed, F.; Larsuel, S.T.; Mayday, M.Y.; Scanlon, V.; Krause, D.S. MRTFA: A Critical Protein in Normal and Malignant Hematopoiesis and Beyond. J. Biol. Chem. 2021, 296, 100543. [Google Scholar] [CrossRef] [PubMed]
- Actin-Microtubule Cytoskeletal Interplay Mediated by MRTF-A/SRF Signaling Promotes Dilated Cardiomyopathy Caused by LMNA Mutations|Nature Communications. Available online: https://www.nature.com/articles/s41467-022-35639-x (accessed on 11 November 2023).
- VEGF-A Stimulates STAT3 Activity via Nitrosylation of Myocardin to Regulate the Expression of Vascular Smooth Muscle Cell Differentiation Markers|Scientific Reports. Available online: https://www.nature.com/articles/s41598-017-02907-6 (accessed on 11 November 2023).
- Sousa, J.F.; Torrieri, R.; Silva, R.R.; Pereira, C.G.; Valente, V.; Torrieri, E.; Peronni, K.C.; Martins, W.; Muto, N.; Francisco, G.; et al. Novel Primate-Specific Genes, RMEL 1, 2 and 3, with Highly Restricted Expression in Melanoma, Assessed by New Data Mining Tool. PLoS ONE 2010, 5, e13510. [Google Scholar] [CrossRef] [PubMed]
- Goedert, L.; Pereira, C.G.; Roszik, J.; Plaça, J.R.; Cardoso, C.; Chen, G.; Deng, W.; Yennu-Nanda, V.G.; Silva, W.A.; Davies, M.A.; et al. RMEL3, a Novel BRAFV600E-Associated Long Noncoding RNA, Is Required for MAPK and PI3K Signaling in Melanoma. Oncotarget 2016, 7, 36711–36718. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.; Serafim, R.B.; Kawakami, A.; Gonçalves Pereira, C.; Roszik, J.; Valente, V.; Vazquez, V.L.; Fisher, D.E.; Espreafico, E.M. The lncRNA RMEL3 Protects Immortalized Cells from Serum Withdrawal-Induced Growth Arrest and Promotes Melanoma Cell Proliferation and Tumor Growth. Pigment. Cell Melanoma Res. 2019, 32, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ma, X.; Sun, J. Update on the Relationship between the SLC4A7 Variant Rs4973768 and Breast Cancer Risk: A Systematic Review and Meta-Analysis. J. Int. Med. Res. 2023, 51, 3000605231166517. [Google Scholar] [CrossRef] [PubMed]
- Newly Discovered Breast Cancer Susceptibility Loci on 3p24 and 17q23.2. Available online: https://pubmed.ncbi.nlm.nih.gov/19330027/ (accessed on 11 November 2023).
- Man, Y.; Zhao, R.; Gao, X.; Liu, Y.; Zhao, S.; Lu, G.; Chan, W.-Y.; Leung, P.C.K.; Bian, Y. TOX3 Promotes Ovarian Estrogen Synthesis: An RNA-Sequencing and Network Study. Front. Endocrinol. 2020, 11, 615846. [Google Scholar] [CrossRef] [PubMed]

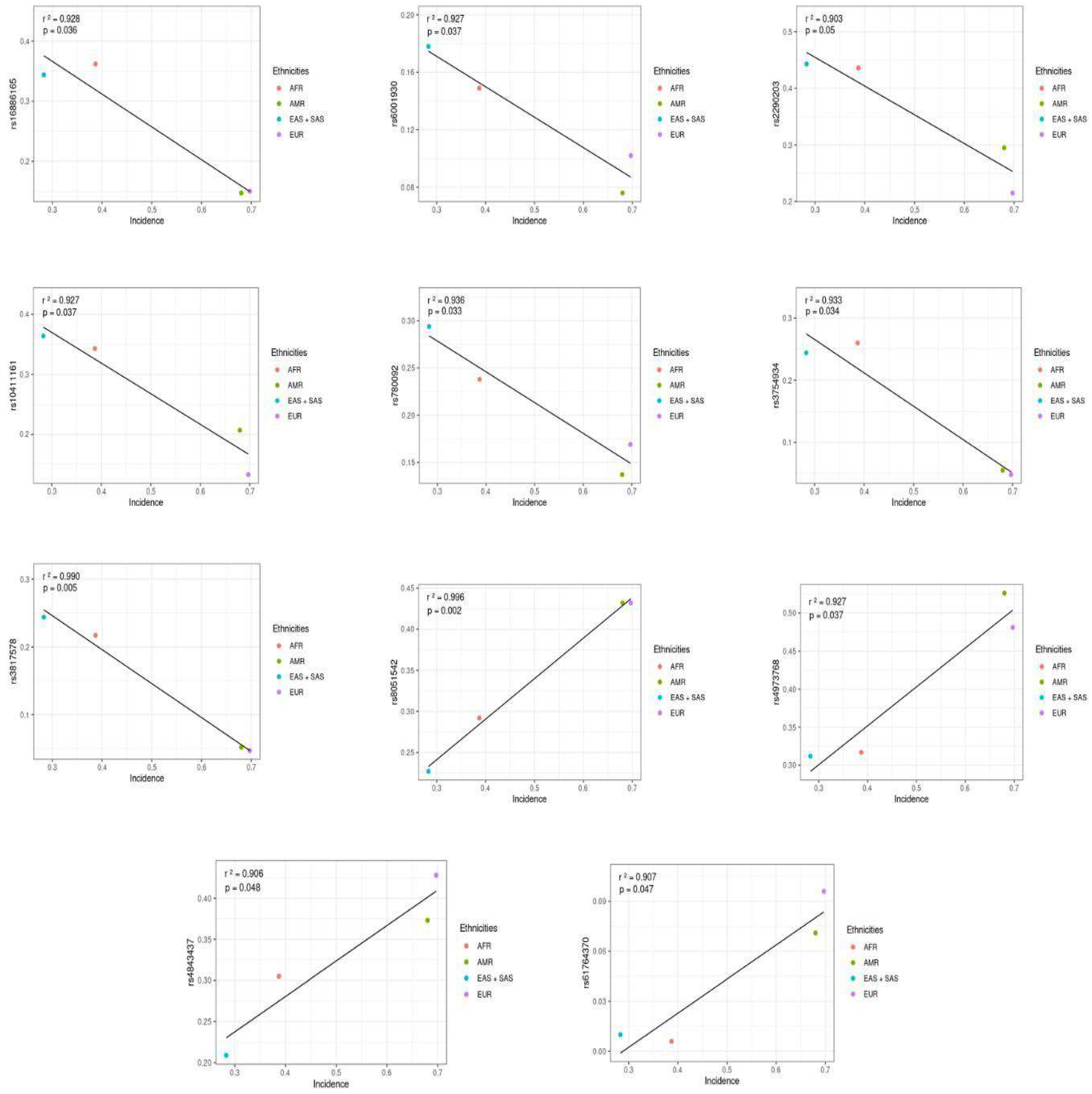
| Gene | SNP ID/Association | Most Severe Consequence | Alleles | Ancestral | p Value | Frequencies | |||
|---|---|---|---|---|---|---|---|---|---|
| AFR | AMR | EUR | SAS + EAS | ||||||
| CASP8 | rs3817578/ Incidence | Intron Variant | C/G/T | C | 0.005 | 0.217 | 0.052 | 0.047 | 0.244 |
| LINC02188 | rs4843437/ Incidence | Splice Polypyrimidine Tract Variant | T/A/C | T | 0.048 | 0.305 | 0.373 | 0.428 | 0.208 |
| CASP8 | rs3754934/ Incidence | Intron Variant | C/A/T | C | 0.034 | 0.260 | 0.055 | 0.048 | 0.244 |
| KRAS | rs61764370/ Incidence | 3 Prime UTR Variant | A/C | A | 0.047 | 0.006 | 0.071 | 0.096 | SAS only (0.020) |
| GCKR | rs780092/ Incidence | Intron Variant | A/G/T | A | 0.033 | 0.238 | 0.137 | 0.169 | 0.293 |
| IQGAP1 | rs2290203/ Incidence | 3 Prime UTR Variant | G/A | A | 0.05 | 0.436 | 0.295 | 0.215 | 0.442 |
| ZNF577 | rs10411161/ Incidence | 3 Prime UTR Variant | C/T | C | 0.037 | 0.343 | 0.207 | 0.133 | 0.364 |
| MRTFA | rs6001930/ Incidence | Intron Variant | T/C/G | C | 0.037 | 0.149 | 0.076 | 0.102 | 0.178 |
| RMEL3 | rs16886165/ Incidence | Intergenic Variant | T/C/G | G | 0.036 | 0.362 | 0.147 | 0.150 | 0.343 |
| SLC4AZ | rs4973768/ Incidence | 3 Prime UTR Variant | C/T | T | 0.037 | 0.317 | 0.526 | 0.481 | 0.312 |
| TOX3 | rs8051542/ Incidence | Intron Variant | T/C/G | C | 0.002 | 0.292 | 0.432 | 0.432 | 0.226 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Costa Nunes, G.G.; de Freitas, L.M.; Monte, N.; Gellen, L.P.A.; Santos, A.P.; de Moraes, F.C.A.; da Costa, A.C.A.; de Lima, M.C.; Fernandes, M.R.; dos Santos, S.E.B.; et al. Genomic Variants and Worldwide Epidemiology of Breast Cancer: A Genome-Wide Association Studies Correlation Analysis. Genes 2024, 15, 145. https://doi.org/10.3390/genes15020145
da Costa Nunes GG, de Freitas LM, Monte N, Gellen LPA, Santos AP, de Moraes FCA, da Costa ACA, de Lima MC, Fernandes MR, dos Santos SEB, et al. Genomic Variants and Worldwide Epidemiology of Breast Cancer: A Genome-Wide Association Studies Correlation Analysis. Genes. 2024; 15(2):145. https://doi.org/10.3390/genes15020145
Chicago/Turabian Styleda Costa Nunes, Giovanna Gilioli, Lilian Marques de Freitas, Natasha Monte, Laura Patrícia Albarello Gellen, Aline Pasquini Santos, Francisco Cezar Aquino de Moraes, Ana Caroline Alves da Costa, Milena Cardoso de Lima, Marianne Rodrigues Fernandes, Sidney Emanuel Batista dos Santos, and et al. 2024. "Genomic Variants and Worldwide Epidemiology of Breast Cancer: A Genome-Wide Association Studies Correlation Analysis" Genes 15, no. 2: 145. https://doi.org/10.3390/genes15020145







