A Pilot Study of Multiplex Ligation-Dependent Probe Amplification Evaluation of Copy Number Variations in Romanian Children with Congenital Heart Defects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. DNA Extraction and MLPA Analysis
3. Results
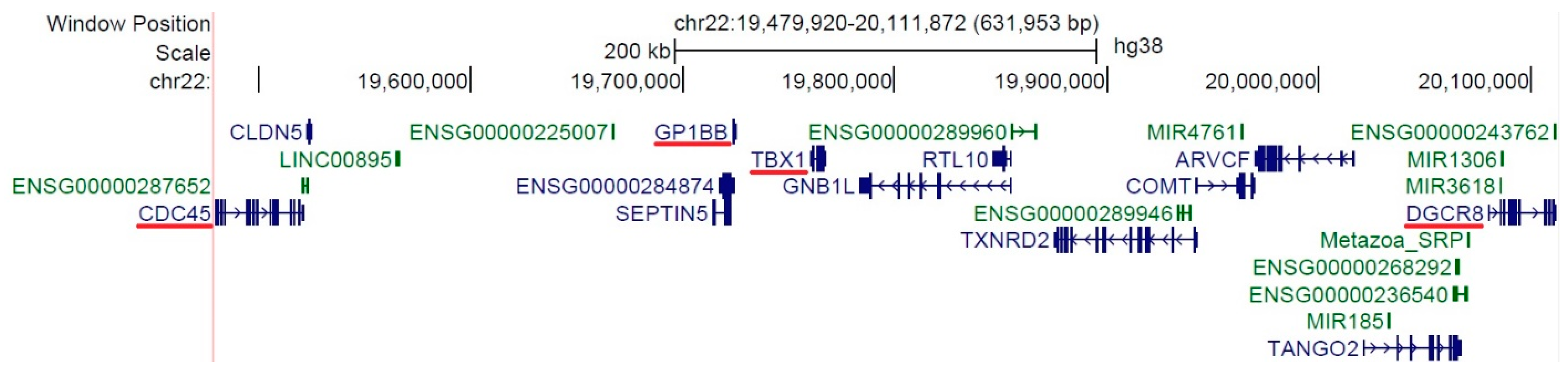
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Chen, S.; Zühlke, L.; Black, G.C.; Choy, M.K.; Li, N.; Keavney, B.D. Global Birth Prevalence of Congenital Heart Defects 1970–2017: Updated Systematic Review and Meta-Analysis of 260 Studies. Int. J. Epidemiol. 2019, 48, 455–463. [Google Scholar] [CrossRef]
- Diab, N.S.; Barish, S.; Dong, W.; Zhao, S.; Allington, G.; Yu, X.; Kahle, K.T.; Brueckner, M.; Jin, S.C. Molecular Genetics and Complex Inheritance of Congenital Heart Disease. Genes 2021, 12, 1020. [Google Scholar] [CrossRef]
- Øyen, N.; Poulsen, G.; Boyd, H.A.; Wohlfahrt, J.; Jensen, P.K.A.; Melbye, M. Recurrence of Congenital Heart Defects in Families. Circulation 2009, 120, 295–301. [Google Scholar] [CrossRef]
- Fahed, A.C.; Gelb, B.D.; Seidman, J.G.; Seidman, C.E. Genetics of Congenital Heart Disease: The Glass Half Empty. Circ Res. 2013, 112, 707–720. [Google Scholar] [CrossRef]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; George, H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global Variation in Copy Number in the Human Genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef]
- Cowan, J.R.; Ware, S.M. Genetics and Genetic Testing in Congenital Heart Disease. Clin Perinatol. 2015, 42, 373–393. [Google Scholar] [CrossRef]
- Breckpot, J.; Thienpont, B.; Arens, Y.; Tranchevent, L.C.; Vermeesch, J.R.; Moreau, Y.; Gewillig, M.; Devriendt, K. Challenges of Interpreting Copy Number Variation in Syndromic and Non-Syndromic Congenital Heart Defects. Cytogenet. Genome Res. 2011, 135, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Hussein, I.R.; Bader, R.S.; Chaudhary, A.G.; Bassiouni, R.; Alquaiti, M.; Ashgan, F.; Schulten, H.J.; Al Qahtani, M.H. Identification of De Novo and Rare Inherited Copy Number Variants in Children with Syndromic Congenital Heart Defects. Pediatr. Cardiol. 2018, 39, 924–940. [Google Scholar] [CrossRef] [PubMed]
- Ison, H.E.; Griffin, E.L.; Parrott, A.; Shikany, A.R.; Meyers, L.; Thomas, M.J.; Syverson, E.; Demo, E.M.; Fitzgerald, K.K.; Fitzgerald-Butt, S.; et al. Genetic Counseling for Congenital Heart Disease—Practice Resource of the National Society of Genetic Counselors. J. Genet. Couns. 2022, 31, 9–33. [Google Scholar] [CrossRef]
- Nees, S.N.; Chung, W.K. The Genetics of Isolated Congenital Heart Disease. Am. J. Med. Genet. Part C Semin. Med. Genet. 2020, 184, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Soemedi, R.; Wilson, I.J.; Bentham, J.; Darlay, R.; Töpf, A.; Zelenika, D.; Cosgrove, C.; Setchfield, K.; Thornborough, C.; Granados-Riveron, J.; et al. Contribution of Global Rare Copy-Number Variants to the Risk of Sporadic Congenital Heart Disease. Am. J. Hum. Genet. 2012, 91, 489–501. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, J.H.; Burt, A.A.; Crosslin, D.R.; Burnham, N.; Kim, C.E.; McDonald-McGinn, D.M.; Zackai, E.H.; Nicolson, S.C.; Spray, T.L.; et al. Burden of Potentially Pathologic Copy Number Variants Is Higher in Children with Isolated Congenital Heart Disease and Significantly Impairs Covariate-Adjusted Transplant-Free Survival. J. Thorac. Cardiovasc. Surg. 2016, 151, 1147–1151. [Google Scholar] [CrossRef]
- Lim, T.B.; Foo, S.Y.R.; Chen, C.K. The Role of Epigenetics in Congenital Heart Disease. Genes 2021, 12, 390. [Google Scholar] [CrossRef] [PubMed]
- Sylva, M.; Van den Hoff, M.J.B.; Moorman, A.F.M. Development of the Human Heart. Am. J. Med. Genet. Part A 2014, 164, 1347–1371. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- George, V.; Colombo, S.; Targoff, K.L. An Early Requirement for Nkx2.5 Ensures the First and Second Heart Field Ventricular Identity and Cardiac Function into Adulthood. Dev. Biol. 2015, 400, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Bolunduț, A.C.; Lazea, C.; Mihu, C.M. Genetic Alterations of Transcription Factors and Signaling Molecules Involved in the Development of Congenital Heart Defects—A Narrative Review. Children 2023, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Mo, R.; Da, M.; Peng, W. Common Variations in BMP4 Confer Genetic Susceptibility to Sporadic Congenital Heart Disease in a Han Chinese Population. Pediatr. Cardiol. 2014, 35, 1442–1447. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Bhaumik, P.; Ghosh, S.; Ozbek, U.; Feingold, E.; Maslen, C.; Sarkar, B.; Pramanik, V.; Biswas, P.; Bandyopadhyay, B.; et al. Polymorphic Haplotypes of CRELD1 Differentially Predispose Down Syndrome and Euploids Individuals to Atrioventricular Septal Defect. Am. J. Med. Genet. Part A 2012, 158, 2843–2848. [Google Scholar] [CrossRef] [PubMed]
- Costain, G.; Silversides, C.K.; Bassett, A.S. The Importance of Copy Number Variation in Congenital Heart Disease. NPJ Genomic Med. 2016, 1, 16031. [Google Scholar] [CrossRef]
- McDonald-McGinn, D.M.; Sullivan, K.E.; Marino, B.; Philip, N.; Swillen, A.; Vorstman, J.A.S.; Zackai, E.H.; Emanuel, B.S.; Vermeesch, J.R.; Morrow, B.E.; et al. 22q11.2 Deletion Syndrome. Nat Rev Dis Prim. 2015, 1, 15071. [Google Scholar] [CrossRef]
- Botto, L.D.; May, K.; Fernhoff, P.M.; Correa, A.; Coleman, K.; Rasmussen, S.A.; Merritt, R.K.; O’Leary, L.A.; Wong, L.; Elixson, E.M.; et al. A Population-Based Study of the 22q11.2 Deletion: Phenotype, Incidence, and Contribution to Major Birth Defects in the Population. Pediatrics 2003, 112, 101–108. [Google Scholar] [CrossRef]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy Number Variation in Human Health, Disease, and Evolution. Annu Rev Genomics Hum Genet. 2009, 10, 451–481. [Google Scholar] [CrossRef]
- Peyvandi, S.; Lupo, P.J.; Garbarini, J.; Woyciechowski, S.; Edman, S.; Emanuel, B.S.; Mitchell, L.E.; Goldmuntz, E. 22q11.2 Deletions in Patients with Conotruncal Defects: Data from 1,610 Consecutive Cases. Pediatr. Cardiol. 2013, 34, 1687–1694. [Google Scholar] [CrossRef]
- Papangeli, I.; Scambler, P. The 22q11 Deletion: DiGeorge and Velocardiofacial Syndromes and the Role of TBX1. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 393–403. [Google Scholar] [CrossRef]
- Sellier, C.; Hwang, V.J.; Dandekar, R.; Durbin-Johnson, B.; Charlet-Berguerand, N.; Ander, B.P.; Sharp, F.R.; Angkustsiri, K.; Simon, T.J.; Tassone, F. Decreased DGCR8 Expression and MiRNA Dysregulation in Individuals with 22q11.2 Deletion Syndrome. PLoS ONE 2014, 9, 13–15. [Google Scholar] [CrossRef]
- Guo, Z.; Li, B.; Tian, P.; Li, D.; Zhang, Y.; Li, Q.; Fan, T.; Yue, J.; Guo, Y. DGCR8 Expression Is Altered in Children with Congenital Heart Defects. Clin. Chim. Acta 2019, 495, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.; Zamore, P.D. Rethinking the Microprocessor. Cell 2006, 125, 827–829. [Google Scholar] [CrossRef] [PubMed]
- Racedo, S.E.; Liu, Y.; Shi, L.; Zheng, D.; Morrow, B.E. Dgcr8 Functions in the Secondary Heart Field for Outflow Tract and Right Ventricle Development in Mammals. Dev. Biol. 2024, 506, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Nagy, O.; Szakszon, K.; Biró, B.O.; Mogyorósy, G.; Nagy, D.; Nagy, B.; Balogh, I.; Ujfalusi, A. Copy Number Variants Detection by Microarray and Multiplex Ligation-Dependent Probe Amplification in Congenital Heart Diseases. J. Biotechnol. 2019, 299, 86–95. [Google Scholar] [CrossRef]
- Russell, M.W.; Chung, W.K.; Kaltman, J.R.; Miller, T.A. Advances in the Understanding of the Genetic Determinants of Congenital Heart Disease and Their Impact on Clinical Outcomes. J. Am. Heart Assoc. 2018, 7, e006906. [Google Scholar] [CrossRef]
- Schouten, J.P.; McElgunn, C.J.; Waaijer, R.; Zwijnenburg, D.; Diepvens, F.; Pals, G. Relative Quantification of 40 Nucleic Acid Sequences by Multiplex Ligation-Dependent Probe Amplification. Nucleic Acids Res. 2002, 30, e57. [Google Scholar] [CrossRef]
- Monteiro, R.A.C.; de Freitas, M.L.; Vianna, G.S.; de Oliveira, V.T.; Pietra, R.X.; Ferreira, L.C.A.; Rocha, P.P.O.; da, S. Gonçalves, M.; da C. César, G.; de S. Lima, J.; et al. Major Contribution of Genomic Copy Number Variation in Syndromic Congenital Heart Disease: The Use of MLPA as the First Genetic Test. Mol. Syndr. 2017, 8, 227–235. [Google Scholar] [CrossRef]
- Mutlu, E.T.; Aykan, H.; Karagöz, T. Analysis of Gene Copy Number Variations in Patients with Congenital Heart Disease Using Multiplex Ligation-Dependent Probe Amplification. Anatol. J. Cardiol. 2018, 20, 9–15. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, J.; Liu, S.; Deng, Y.; Liu, H.; Li, N.; Li, S.; Chen, X.; Lin, Y.; Wang, H.; et al. Copy Number Variation of GATA4 and NKX2-5 in Chinese Fetuses with Congenital Heart Disease. Pediatr. Int. 2015, 57, 234–238. [Google Scholar] [CrossRef]
- Li, Z.; Huang, J.; Liang, B.; Zeng, D.; Luo, S.; Yan, T.; Liao, F.; Huang, J.; Li, J.; Cai, R.; et al. Copy Number Variations in the GATA4, NKX2-5, TBX5, BMP4 CRELD1, and 22q11.2 Gene Regions in Chinese Children with Sporadic Congenital Heart Disease. J. Clin. Lab. Anal. 2019, 33, e22660. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, K.M.; El-Segaier, M.; Fernlund, E.; Errami, A.; Bouvagnet, P.; Nehme, N.; Steensberg, J.; Hjortdal, V.; Soller, M.; Behjati, M.; et al. Screening of Congenital Heart Disease Patients Using Multiplex Ligation-Dependent Probe Amplification: Early Diagnosis of Syndromic Patients. Am. J. Med. Genet. Part A 2012, 158, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Floriani, M.A.; Glaeser, A.B.; Dorfman, L.E.; Agnes, G.; Rosa, R.F.M.; Zen, P.R.G. GATA4 Deletions Associated with Congenital Heart Diseases in South Brazil. J. Pediatr. Genet. 2021, 10, 092–097. [Google Scholar] [CrossRef] [PubMed]
- Aguayo-Gómez, A.; Arteaga-Vázquez, J.; Svyryd, Y.; Calderón-Colmenero, J.; Zamora-González, C.; Vargas-Alarcón, G.; Mutchinick, O.M. Identification of Copy Number Variations in Isolated Tetralogy of Fallot. Pediatr. Cardiol. 2015, 36, 1642–1646. [Google Scholar] [CrossRef] [PubMed]
- Guida, V.; Lepri, F.; Vijzelaar, R.; De Zorzi, A.; Versacci, P.; Digilio, M.C.; Marino, B.; De Luca, A.; Dallapiccola, B. Multiplex Ligation-Dependent Probe Amplification Analysis of GATA4 Gene Copy Number Variations in Patients with Isolated Congenital Heart Disease. Dis. Markers 2010, 28, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Bruneau, B.G. Directed Transdifferentiation of Mouse Mesoderm to Heart Tissue by Defined Factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef]
- Pikkarainen, S.; Tokola, H.; Kerkelä, R.; Ruskoaho, H. GATA Transcription Factors in the Developing and Adult Heart. Cardiovasc. Res. 2004, 63, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Feliciano, J.; Lee, K.H.; Kong, S.W.; Rajagopal, S.; Ma, Q.; Springer, Z.; Izumo, S.; Tabin, C.J.; Pu, W.T. Development of Heart Valves Requires Gata4 Expression in Endothelial-Derived Cells. Development 2006, 133, 3607–3618. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, S.; Barnett, P.; Van Duijvenboden, K.; Weber, D.; Gessler, M.; Christoffels, V.M. GATA-Dependent Regulatory Switches Establish Atrioventricular Canal Specificity during Heart Development. Nat. Commun. 2014, 5, 3680. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Morisaki, H.; Nakaji, M.; Kitano, M.; Kim, K.S.; Sagawa, K.; Ishikawa, S.; Satokata, I.; Mitani, Y.; Kato, H.; et al. Genetic Mutation Analysis in Japanese Patients with Non-Syndromic Congenital Heart Disease. J. Hum. Genet. 2016, 61, 157–162. [Google Scholar] [CrossRef]
- Bu, H.; Sun, G.; Zhu, Y.; Yang, Y.; Tan, Z.; Zhao, T.; Hu, S. The M310T Mutation in the GATA4 Gene Is a Novel Pathogenic Target of the Familial Atrial Septal Defect. BMC Cardiovasc. Disord. 2021, 21, 12. [Google Scholar] [CrossRef]
- Sarwar, S.; Ehsan, F.; Shabana; Tahir, A.; Jamil, M.; Shahid, S.U.; Khan, A.; Hasnain, S. First Report of Polymorphisms in MTRR, GATA4, VEGF, and ISL1 Genes in Pakistani Children with Isolated Ventricular Septal Defects (VSD). Ital. J. Pediatr. 2021, 47, 4–9. [Google Scholar] [CrossRef]
- Behiry, E.G.; Al-Azzouny, M.A.; Sabry, D.; Behairy, O.G.; Salem, N.E. Association of NKX2-5, GATA4, and TBX5 Polymorphisms with Congenital Heart Disease in Egyptian Children. Mol. Genet. Genomic Med. 2019, 7, e612. [Google Scholar] [CrossRef]
- Fan, D.; Pang, S.; Chen, J.; Shan, J.; Cheng, Q.; Yan, B. Identification and Functional Study of GATA4 Gene Regulatory Variants in Atrial Septal Defects. BMC Cardiovasc. Disord. 2021, 21, 321. [Google Scholar] [CrossRef]
- Reamon-Buettner, S.M.; Borlak, J. NKX2-5: An Update on This Hypermutable Homeodomain Protein and Its Role in Human Congenital Heart Disease (CHD). Hum. Mutat. 2010, 31, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Nomura-Kitabayashi, A.; Sultana, N.; Cai, W.; Cai, X.; Moon, A.M.; Cai, C.L. Mesodermal Nkx2.5 Is Necessary and Sufficient for Early Second Heart Field Development. Dev. Biol. 2014, 390, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.D.; Zhang, B.; Lee, B.; Evans, S.I.; Lassar, A.B.; Lee, K.H. Evolutionary Conservation of Nkx2.5 Autoregulation in the Second Heart Field. Dev. Biol. 2013, 374, 198–209. [Google Scholar] [CrossRef]
- Jay, P.Y.; Harris, B.S.; Maguire, C.T.; Buerger, A.; Wakimoto, H.; Tanaka, M.; Kupershmidt, S.; Roden, D.M.; Schultheiss, T.M.; O’Brien, T.X.; et al. Nkx2-5 Mutation Causes Anatomic Hypoplasia of the Cardiac Conduction System. J. Clin. Investig. 2004, 113, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Narasimhan, C.; Balekundri, V.I.; Agrawal, D.; Kumar, A.; Mohapatra, B. Functional Analysis of Novel Genetic Variants of NKX2-5 Associated with Nonsyndromic Congenital Heart Disease. Am. J. Med. Genet. Part A 2021, 185, 3644–3663. [Google Scholar] [CrossRef]
- Rozqie, R.; Satwiko, M.G.; Anggrahini, D.W.; Sadewa, A.H.; Gunadi; Hartopo, A.B.; Mumpuni, H.; Dinarti, L.K. NKX2-5 Variants Screening in Patients with Atrial Septal Defect in Indonesia. BMC Med. Genomics 2022, 15, 91. [Google Scholar] [CrossRef]
- Takeuchi, J.K.; Ohgi, M.; Koshiba-Takeuchi, K.; Shiratori, H.; Sakaki, I.; Ogura, K.; Saijoh, Y.; Ogura, T. Tbx5 Specifies the Left/Right Ventricles and Ventricular Septum Position during Cardiogenesis. Development 2003, 130, 5953–5964. [Google Scholar] [CrossRef]
- Burnicka-Turek, O.; Broman, M.T.; Steimle, J.D.; Boukens, B.J.; Petrenko, N.B.; Ikegami, K.; Nadadur, R.D.; Qiao, Y.; Arnolds, D.E.; Yang, X.H.; et al. Transcriptional Patterning of the Ventricular Cardiac Conduction System. Circ. Res. 2020, 127, E94–E106. [Google Scholar] [CrossRef]
- Vanlerberghe, C.; Jourdain, A.S.; Ghoumid, J.; Frenois, F.; Mezel, A.; Vaksmann, G.; Lenne, B.; Delobel, B.; Porchet, N.; Cormier-Daire, V.; et al. Holt-Oram Syndrome: Clinical and Molecular Description of 78 Patients with TBX5 Variants. Eur. J. Hum. Genet. 2019, 27, 360–368. [Google Scholar] [CrossRef]
- Kimura, M.; Kikuchi, A.; Ichinoi, N.; Kure, S. Novel TBX5 Duplication in a Japanese Family with Holt–Oram Syndrome. Pediatr. Cardiol. 2015, 36, 244–247. [Google Scholar] [CrossRef]
- Patel, C.; Silcock, L.; McMullan, D.; Brueton, L.; Cox, H. TBX5 Intragenic Duplication: A Family with an Atypical Holt-Oram Syndrome Phenotype. Eur. J. Hum. Genet. 2012, 20, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Bogarapu, S.; Bleyl, S.B.; Calhoun, A.; Viskochil, D.; Saarel, E.V.; Everitt, M.D.; Frank, D.U. Phenotype of a Patient with Contiguous Deletion of TBX5 and TBX3: Expanding the Disease Spectrum. Am. J. Med. Genet. Part A 2014, 164, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Cenni, C.; Andres, S.; Hempel, M.; Strom, T.M.; Thomas, E.; Davies, A.; Timoney, N.; Frigiola, A.; Logan, M.; Holder-Espinasse, M. TBX3 and TBX5 Duplication: A Family with an Atypical Overlapping Holt-Oram/Ulnar-Mammary Syndrome Phenotype. Eur. J. Med. Genet. 2021, 64, 104213. [Google Scholar] [CrossRef] [PubMed]


| Characteristic | Syndromic (28 Subjects) | Isolated (28 Subjects) | p-Value |
|---|---|---|---|
| Age 1 (months), median [Q1–Q3] | 28 [5–90.25] | 46 [8.5–122.3] | 0.2584 |
| Female gender, n | 14/28 | 18/28 | 0.4182 |
| Urban, n | 16/28 | 14/28 | 0.7891 |
| Consanguinity, n | 1/28 | 0/28 | NA |
| Family history of CHD, n | 7/28 | 8/28 | >0.9999 |
| Gestational age (weeks), median [Q1–Q3] | 38 [37–40] | 38 [37–40] | 0.9796 |
| Birth weight Z-score, median [Q1–Q3] | −1.12 [−1.54–−0.34] | −0.43 [−1.41–0.33] | 0.0866 |
| Birth length Z-score, median [Q1–Q3] | −0.65 [−1.86–0.12] | 0.41 [−0.65–1.57] | 0.0237 |
| SGA 2, n | 3/28 | 3/28 | >0.9999 |
| Weight1 Z-score, median [Q1–Q3] | −2.18 [−3.6–−1.23] | −0.97 [−2.11–0.35] | 0.0138 |
| Height1 Z-score, median [Q1–Q3] | −1.25 [−2.4–−0.65] | −0.13 [−0.89–0.63] | 0.0014 |
| Short stature 3, n | 6/28 | 3/28 | 0.4688 |
| Type of CHD, n | |||
| Ventricular septal defect | 8/28 | 14/28 | 0.1707 |
| Tetralogy of Fallot | 5/28 | 2/28 | 0.4216 |
| Pulmonary stenosis | 4/28 | 3/28 | >0.9999 |
| Atrial septal defect | 2/28 | 3/28 | >0.9999 |
| Atrioventricular canal defect | 2/28 | 2/28 | >0.9999 |
| Aortic stenosis | 1/28 | 2/28 | >0.9999 |
| Patent ductus arteriosus | 0/28 | 3/28 | NA |
| Truncus arteriosus | 3/28 | 0/28 | NA |
| Coarctation of aorta | 0/28 | 2/28 | NA |
| Double-outlet right ventricle | 1/28 | 1/28 | >0.9999 |
| Transposition of great arteries | 0/28 | 1/28 | NA |
| Interrupted aortic arch | 1/28 | 0/28 | NA |
| Hypoplastic left-heart syndrome | 1/28 | 0/28 | NA |
| Tricuspid atresia | 1/28 | 0/28 | NA |
| ALCAPA | 1/28 | 0/28 | NA |
| Complex | 1/28 | 1/28 | NA |
| Reference | Population | CNV | Frequency | Congenital Heart Defects |
|---|---|---|---|---|
| Mutlu et al., 2018 [35] | Türkiye | Deletion 22q11.2 | 3/45 | ASD, ASD + VSD, VSD |
| Liu et al., 2015 [36] | China | Deletion 22q11.2 | 3/117 | TOF |
| Duplication 22q11.2 | 2/117 | Endocardial cushion defect, PS + VSD | ||
| Nagy et al., 2019 [31] | Hungary | Deletion 22q11.2 | 2/49 | TOF |
| Duplication GATA4 (8p23.1) | 2/49 | CoA, Patent foramen ovale | ||
| Li et al., 2018 [37] | China | Deletion 22q11.2 | 5/167 | ASD + VSD, TOF, VSD |
| Sorensen et al., 2012 [38] | Denmark, Sweeden, France | Deletion GATA4 (8p23.1) | 1/402 | VSD |
| Deletion 22q11.2 | 2/402 | PA + VSD, TOF | ||
| Duplication 22q11.2 | 2/402 | CoA, PS | ||
| Triplication NKX2-5 (5q35.3) | 1/402 | ASD + PS | ||
| Complex (deletion GATA4 + duplication 22q11.2) | 1/402 | AVSD + hypoplastic right ventricle | ||
| Floriani et al., 2020 [39] | Brazil | Deletion GATA4 (8p23.1) | 2/207 | ASD + PLSVC + VSD, ASD + PLSVC + PS + VSD |
| Deletion 22q11.2 | 4/207 | ASD, TOF, VSD | ||
| Duplication 22q11.2 | 1/207 | Subvalvular aortic ring | ||
| Aguayo-Gomez et al., 2015 [40] | Mexico | No CNV found | 0/52 | - |
| Guida et al., 2010 [41] | Italy | No CNV found | 0/161 | - |
| Our study | Romania | Deletion 22q11.2 | 2/56 | Interrupted aortic arch type B + VSD, VSD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolunduț, A.C.; Nazarie, F.; Lazea, C.; Șufană, C.; Miclea, D.; Lazăr, C.; Mihu, C.M. A Pilot Study of Multiplex Ligation-Dependent Probe Amplification Evaluation of Copy Number Variations in Romanian Children with Congenital Heart Defects. Genes 2024, 15, 207. https://doi.org/10.3390/genes15020207
Bolunduț AC, Nazarie F, Lazea C, Șufană C, Miclea D, Lazăr C, Mihu CM. A Pilot Study of Multiplex Ligation-Dependent Probe Amplification Evaluation of Copy Number Variations in Romanian Children with Congenital Heart Defects. Genes. 2024; 15(2):207. https://doi.org/10.3390/genes15020207
Chicago/Turabian StyleBolunduț, Alexandru Cristian, Florina Nazarie, Cecilia Lazea, Crina Șufană, Diana Miclea, Călin Lazăr, and Carmen Mihaela Mihu. 2024. "A Pilot Study of Multiplex Ligation-Dependent Probe Amplification Evaluation of Copy Number Variations in Romanian Children with Congenital Heart Defects" Genes 15, no. 2: 207. https://doi.org/10.3390/genes15020207
APA StyleBolunduț, A. C., Nazarie, F., Lazea, C., Șufană, C., Miclea, D., Lazăr, C., & Mihu, C. M. (2024). A Pilot Study of Multiplex Ligation-Dependent Probe Amplification Evaluation of Copy Number Variations in Romanian Children with Congenital Heart Defects. Genes, 15(2), 207. https://doi.org/10.3390/genes15020207





