Identification of Candidate Genes and Pathways Linked to the Temperament Trait in Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic and Pedigree Data
2.2. Genotypic Data and Quality Control
2.3. Model and Estimation of Genetic Parameters
2.4. Single Step Genome-Wide Association Studies (ssGWAS)
2.5. Identification of Positional Candidate Genes
3. Gene Enrichment Analysis
4. Results
4.1. Estimation of Genetic Parameters
4.2. Single Step Genome-Wide Association Studies (ssGWAS)
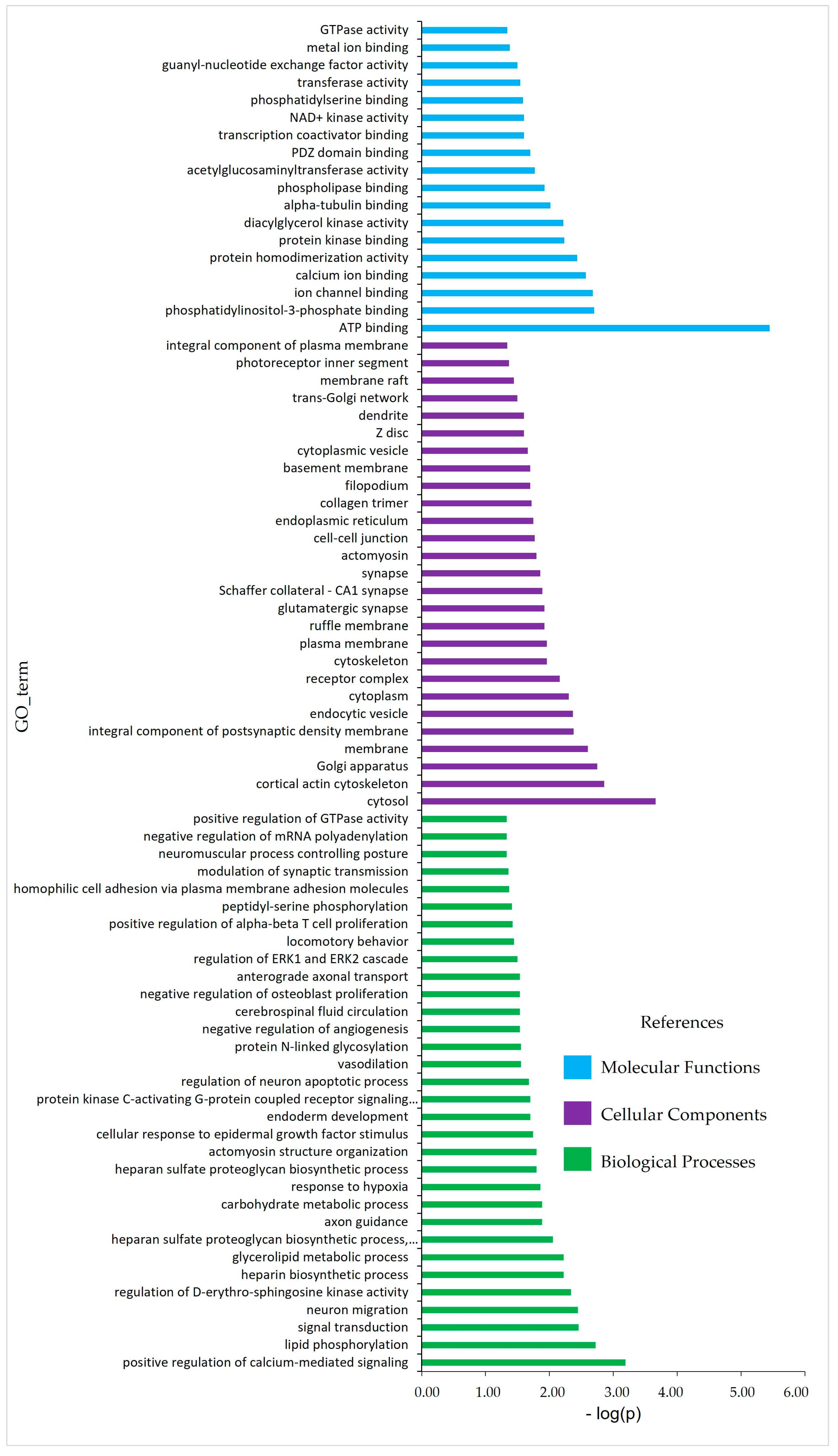
4.3. Enrichment Analysis
5. Discussion
5.1. Heritability (h2)
5.2. Chromosomes and Candidate Genes
5.3. Functional Enrichment Analysis
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blache, D.; Bickell, S.L. Temperament and Reproductive Biology: Emotional Reactivity and Reproduction in Sheep. Rev. Bras. Zootec. 2010, 39, 401–408. [Google Scholar] [CrossRef]
- Désiré, L.; Boissy, A.; Veissier, I. Emotions in Farm Animals: A New Approach to Animal Welfare in Applied Ethology. Behav. Process. 2002, 60, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, M.A.; Rogers, A.R.; Verkerk, G.A. The Effect of Temperament and Responsiveness towards Humans on the Behavior, Physiology and Milk Production of Multi-Parous Dairy Cows in a Familiar and Novel Milking Environment. Physiol. Behav. 2012, 107, 329–337. [Google Scholar] [CrossRef]
- Veissier, I.; Boissy, A.; Désiré, L.; Greiveldinger, L. Animals’ Emotions: Studies in Sheep Using Appraisal Theories. Anim. Welf. 2009, 18, 347–354. [Google Scholar] [CrossRef]
- Bickell, S.; Durmic, Z.; Blache, D.; Vercoe, P.E.; Martin, G.B. Rethinking the Management of Health and Reproduction in Small Ruminants. In Updates on Ruminant Production and Medicine, Proceedings of the 26th World Buiatrics Congress, 14–17 November 2010, Santiago, Chile; Wittwer, F., Chihuailaf, R., Contreras, H., Gallo, C., Kruze, J., Lanuza, F., Letelier, C., Monti, G., Noro, M., Eds.; World Buiatrics Congress: Santiago, Chile, 2010; pp. 317–325. [Google Scholar]
- Van Lier, E.; Hart, K.W.; Viñoles, C.; Paganoni, B.; Blache, D. Calm Merino Ewes Have a Higher Ovulation Rate and More Multiple Pregnancies than Nervous Ewes. Animal 2017, 11, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Wulf, D.M.; Emnett, R.S.; Leheska, J.M.; Moeller, S.J. Relationships among Glycolytic Potential, Dark Cutting (Dark, Firm, and Dry) Beef, and Cooked Beef Palatability. J. Anim. Sci. 2002, 80, 1895–1903. [Google Scholar] [CrossRef]
- Voisinet, B.D.; Grandin, T.; Tatum, J.D.; O’Connor, S.F.; Struthers, J.J. Feedlot Cattle with Calm Temperaments Have Higher Average Daily Gains Than Cattle with Excitable Temperaments. J. Anim. Sci. 1997, 75, 892–896. [Google Scholar] [CrossRef]
- Sart, S.; Bencini, R.; Blache, D.; Martin, G.B. Calm Ewes Produce Milk With More Protein Than Nervous Ewes. Anim. Prod. Aust. 2004, 25, 307. [Google Scholar]
- Hawken, P.A.R.; Williman, M.; Milton, J.; Kelly, R.; Nowak, R.; Blache, D. Nutritional Supplementation during the Last Week of Gestation Increased the Volume and Reduced the Viscosity of Colostrum Produced by Twin Bearing Ewes Selected for Nervous Temperament. Small Rumin. Res. 2012, 105, 308–314. [Google Scholar] [CrossRef]
- Del Campo, M.; Manteca, X.; Soares de Lima, J.M.; Brito, G.; Hernández, P.; Sañudo, C.; Montossi, F. Temperamento, bienestar animal, calidad del producto. Rev. INIA 2011, 24, 11–17. [Google Scholar]
- Haskell, M.J.; Simm, G.; Turner, S.P. Genetic Selection for Temperament Traits in Dairy and Beef Cattle. Front. Genet. 2014, 5, 368. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Ledger, J.; Zheng, C.; Martin, G.B.; Blache, D. Associations between Temperament and Gene Polymorphisms in the Brain Dopaminergic System and the Adrenal Gland of Sheep. Physiol. Behav. 2016, 153, 19–27. [Google Scholar] [CrossRef]
- Qiu, X.; Martin, G.B.; Blache, D. Gene Polymorphisms Associated with Temperament. J. Neurogenet. 2017, 31, 1324857. [Google Scholar] [CrossRef] [PubMed]
- Brookes, A.J. The essence of SNPs. Gene 1999, 234, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Maloney, S.K.; Wang, M.; Rodger, J.; Chen, L.; Blache, D. Association between Temperament Related Traits and Single Nucleotide Polymorphisms in the Serotonin and Oxytocin Systems in Merino Sheep. Genes Brain Behav. 2020, 20, e12714. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, A.B.; Oliveira, H.R.; Chen, S.Y.; Miller, S.P.; Marchant-Forde, J.N.; Grigoletto, L.; Brito, L.F. A Systematic Review of Genomic Regions and Candidate Genes Underlying Behavioral Traits in Farmed Mammals and Their Link with Human Disorders. Animals 2021, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Poissant, J.; Réale, D.; Martin, J.G.A.; Festa-Bianchet, M.; Coltman, D.W. A Quantitative Trait Locus Analysis of Personality in Wild Bighorn Sheep. Ecol. Evol. 2013, 3, 474–481. [Google Scholar] [CrossRef]
- Pant, S.D.; You, Q.; Schenkel, L.C.; Voort, G.V.; Schenkel, F.S.; Wilton, J.; Cain, L.; Karrow, N.A. A Genome-Wide Association Study to Identify Chromosomal Regions Influencing Ovine Cortisol Response. Livest. Sci. 2016, 187, 40–47. [Google Scholar] [CrossRef]
- Blache, D.; Ferguson, D. Genetic Estimates for Temperament Traits in Sheep Breeds. Reproduction 2005, 364, 19. [Google Scholar]
- Blache, D.; Ferguson, D. Increasing Sheep Meat Production Efficiency and Animal Welfare by Selection for Temperament Meat. Statistics 2005, 364, 22. [Google Scholar]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Fernando, R.L.; Vitezica, Z.; Okimoto, R.; Wing, T.; Hawken, R.; Muir, W.M. Genome-Wide Association Mapping Including Phenotypes from Relatives without Genotypes in a Single-Step (SsGWAS) for 6-Week Body Weight in Broiler Chickens. Front. Genet. 2014, 5, 134. [Google Scholar] [CrossRef] [PubMed]
- Medrano, J.F.; Aasen, E.; Sharrow, L. DNA Extraction from Nucleated Red Blood Cells. Biotechniques 1990, 8, 43. [Google Scholar]
- Zambra, N.; Gimeno, D.; Blache, D.; Van Lier, E. Temperament and Its Heritability in Corriedale and Merino Lambs. Animal 2015, 9, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Masuda, Y.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs; University of Georgia: Athens, GA, USA, 2018. [Google Scholar]
- Aguilar, I.; Misztal, I.; Tsuruta, S.; Legarra, A.; Wang, H. PREGSF90—POSTGSF90: Computational Tools for the Implementation of Single-Step Genomic Selection and Genome-Wide Association with Ungenotyped Individuals in BLUPF90 Programs. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot Topic: A Unified Approach to Utilize Phenotypic, Full Pedigree, and Genomic Information for Genetic Evaluation of Holstein Final Score. J. Dairy. Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy. Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Strandén, I.; Garrick, D.J. Technical Note: Derivation of Equivalent Computing Algorithms for Genomic Predictions and Reliabilities of Animal Merit. J. Dairy. Sci. 2009, 92, 2971–2975. [Google Scholar] [CrossRef] [PubMed]
- Brunes, L.C.; Baldi, F.; Lopes, F.B.; Lôbo, R.B.; Espigolan, R.; Costa, M.F.O.; Stafuzza, N.B.; Magnabosco, C.U. Weighted Single-Step Genome-Wide Association Study and Pathway Analyses for Feed Efficiency Traits in Nellore Cattle. J. Anim. Breed. Genet. 2020, 138, 23–44. [Google Scholar] [CrossRef]
- Sigdel, A.; Bisinotto, R.S.; Peñagaricano, F. Genes and Pathways Associated with Pregnancy Loss in Dairy Cattle. Sci. Rep. 2021, 11, 13329. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A Powerful Link between Biological Databases and Microarray Data Analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package BiomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Hazard, D.; Moreno, C.; Foulquié, D.; Delval, E.; François, D.; Bouix, J.; Sallé, G.; Boissy, A. Identification of QTLs for Behavioral Reactivity to Social Separation and Humans in Sheep Using the OvineSNP50 BeadChip. BMC Genom. 2014, 15, 778. [Google Scholar] [CrossRef]
- Cardelino, R.; Rovira, J. (Eds.) Heredabilidad. In Mejoramiento Genético Animal; Hemisferio Sur: Montevideo, Uruguay, 1989; pp. 111–151. [Google Scholar]
- Plush, K.J.; Hebart, M.L.; Brien, F.D.; Hynd, P.I. The Genetics of Temperament in Merino Sheep and Relationships with Lamb Survival. Appl. Anim. Behav. Sci. 2011, 134, 130–135. [Google Scholar] [CrossRef]
- Le Neindre, P.; Trillat, G.; Sapa, J.; Ménissier, F.; Bonnet, J.N.; Chupin, J.M. Individual Differences in Docility in Limousin Cattle. J. Anim. Sci. 1995, 73, 2249–2253. [Google Scholar] [CrossRef] [PubMed]
- Burrow, H.M. Measurements of Temperament and Their Relationships with Performance Traits of Beef Cattle. In Animal Breeding Abstracts; CABI: Wallingford, UK, 1997; Volume 65, pp. 477–495. [Google Scholar]
- Oki, H.; Kusunose, R.; Nakaoka, H.; Nishiura, A.; Miyake, T.; Sasaki, Y. Estimation of Heritability and Genetic Correlation for Behavioural Responses by Gibbs Sampling in the Thoroughbred Racehorse. J. Anim. Breed. Genet. 2007, 124, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Allen, J.G.; Wilton, S.D.; Akkari, P.A.; Huxtable, C.R.; Laing, N.G. A Splice-Site Mutation Causing Ovine McArdle’s Disease. Neuromuscul. Disord. 1997, 7, 336–342. [Google Scholar] [CrossRef]
- Laville, E.; Sayd, T.; Morzel, M.; Blinet, S.; Chambon, C.; Lepetit, J.; Renand, G.; Hocquette, J.F. Proteome Changes during Meat Aging in Tough and Tender Beef Suggest the Importance of Apoptosis and Protein Solubility for Beef Aging and Tenderization. J. Agric. Food Chem. 2009, 57, 10755–10764. [Google Scholar] [CrossRef]
- Lametsch, R.; Karlsson, A.H. Electrical Stimulation Affects Metabolic Enzyme Phosphorylation, Protease Activation, and Meat Tenderization in Beef. J. Anim. Sci. 2012, 90, 1638–1649. [Google Scholar] [CrossRef]
- Chen, L.; Li, X.; Ni, N.; Liu, Y.; Chen, L.; Wang, Z.; Shen, Q.W.; Zhang, D. Phosphorylation of Myofibrillar Proteins in Post-mortem Ovine Muscle with Different Tenderness. J. Sci. Food Agric. 2016, 96, 1474–1483. [Google Scholar] [CrossRef]
- Lucia, A.; Nogales-Gadea, G.; Pérez, M.; Martín, M.A.; Andreu, A.L.; Arenas, J. McArdle Disease: What Do Neurologists Need to Know? Nat. Clin. Pract. Neurol. 2008, 4, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Howell, J.M.C.; Walker, K.R.; Creed, K.E.; Dunton, E.; Davies, L.; Quinlivan, R.; Karpati, G. Phosphorylase Re-Expression, Increase in the Force of Contraction and Decreased Fatigue Following Notexin-Induced Muscle Damage and Regeneration in the Ovine Model of McArdle Disease. Neuromuscul. Disord. 2014, 24, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Li, X.; Zhang, D.; Chen, L.; Hou, C.; Zheng, X.; Ren, C.; Ijaz, M. Role of Phosphorylation on Characteristics of Glycogen Phosphorylase in Lamb with Different Glycolytic Rates Post-Mortem. Meat Sci. 2020, 164, 108096. [Google Scholar] [CrossRef]
- Bai, Y.; Li, X.; Zhang, D.; Chen, L.; Hou, C.; Zheng, X.; Ren, C. Effects of Phosphorylation on the Activity of Glycogen Phosphorylase in Mutton during Incubation at 4 °C In Vitro. Food Chem. 2020, 313, 126162. [Google Scholar] [CrossRef]
- Speck, P.A.; Collingwood, K.M.; Bardsley, R.G.; Tucker, G.A.; Gilmour, R.S.; Buttery, P.J. Transient Changes in Growth and in Calpain and Calpastatin Expression in Ovine Skeletal Muscle after Short-Term Dietary Inclusion of Cimaterol. Biochimie 1993, 75, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Page, B.T.; Casas, E.; Heaton, M.P.; Cullen, N.G.; Hyndman, D.L.; Morris, C.A.; Crawford, A.M.; Wheeler, T.L.; Koohmaraie, M.; Keele, J.W.; et al. Evaluation of Single-Nucleotide Polymorphisms in CAPN1 for Association with Meat Tenderness in Cattle. J. Anim. Sci. 2002, 80, 3077–3085. [Google Scholar] [CrossRef] [PubMed]
- Geesink, G.H.; Kuchay, S.; Chishti, A.H.; Koohmaraie, M. μ-Calpain Is Essential for Postmortem Proteolysis of Muscle Proteins. J. Anim. Sci. 2006, 84, 2834–2840. [Google Scholar] [CrossRef] [PubMed]
- MacHado, A.L.; Meira, A.N.; Muniz, E.N.; Azevedo, H.C.; Coutinho, L.L.; Mouraõ, G.B.; Pedrosa, V.B.; Pinto, L.F.B. Single Loci and Haplotypes in CAPN1 and CAST Genes Are Associated with Growth, Biometrics, and In Vivo Carcass Traits in Santa Inês Sheep. Ann. Anim. Sci. 2020, 20, 465–483. [Google Scholar] [CrossRef]
- Meira, A.N.; De Farias Jucá, A.; De Souza, T.C.; De Souza, T.C.; Coutinho, L.L.; Mourao, G.B.; Azevedo, H.C.; Muniz, E.N.; Pedrosa, V.B.; Batista Pinto, L.F. Post-Mortem Carcass Traits Are Associated with μ-Calpain and Calpastatin Variants in Santa Inês Sheep. Anim. Sci. Pap. Rep. 2020, 38, 369–380. [Google Scholar]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- van der Laan, S.W.; Harshfield, E.L.; Hemerich, D.; Stacey, D.; Wood, A.M.; Asselbergs, F.W. From Lipid Locus to Drug Target through Human Genomics. Cardiovasc. Res. 2018, 114, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Jackman, S.L.; Turecek, J.; Belinsky, J.E.; Regehr, W.G. The Calcium Sensor Synaptotagmin 7 Is Required for Synaptic Facilitation. Nature 2016, 529, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Satterfield, R.; Young, S.M.; Jonas, P.; Chen, C.; Satterfield, R.; Young, S.M.; Jonas, P. Triple Function of Synaptotagmin 7 Ensures Efficiency of High-Frequency Transmission at Central GABAergic Synapses. Cell Rep. 2017, 21, 2082–2089. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Wang, Q.; Liu, Y.; Marchetto, M.C.; Linker, S.; Lu, S.; Chen, Y.; Liu, C. Synaptotagmin-7 Is a Key Factor for Bipolar-like Behavioral Abnormalities in Mice. Proc. Natl. Acad. Sci. USA 2020, 117, 4392–4399. [Google Scholar] [CrossRef] [PubMed]
- Koohmaraie, M. Effect of PH, Temperature, and Inhibitors on Autolysis and Catalytic Activity of Bovine Skeletal Muscle μ-Calpain. J. Anim. Sci. 1992, 70, 3071–3080. [Google Scholar] [CrossRef] [PubMed]
- Tanda, G.; Ebbs, A.L.; Kopajtic, T.A.; Elias, L.M.; Campbell, B.L.; Newman, A.H.; Katz, J.L. Effects of Muscarinic M1 Receptor Blockade on Cocaine-Induced Elevations of Brain Dopamine Levels and Locomotor Behavior in Rats. J. Pharmacol. Exp. Ther. 2007, 321, 334–344. [Google Scholar] [CrossRef]
- Megdiche, S.; Mastrangelo, S.; Ben Hamouda, M.; Lenstra, J.A.; Ciani, E. A Combined Multi-Cohort Approach Reveals Novel and Known Genome-Wide Selection Signatures for Wool Traits in Merino and Merino-Derived Sheep Breeds. Front. Genet. 2019, 10, 1025. [Google Scholar] [CrossRef]
- Moberg, G.P. Biological Response to Stress: Implications for Animal Welfare. In The Biology of Animal Stress: Basic Principles and Implications for Animal Welfare; CABI Publishing: Wallingford, UK, 2000; pp. 1–21. [Google Scholar] [CrossRef]
- Karadurmus, D.; Rial, D.; De Backer, J.-F.; Communi, D.; de Kerchove d’Exaerde, A.; Schiffmann, S.N. GPRIN3 Controls Neuronal Excitability, Morphology, and Striatal-Dependent Behaviors in the Indirect Pathway of the Striatum. J. Neurosci. 2019, 39, 7513–7528. [Google Scholar] [CrossRef]
- Selye, H. The Effect of Adaptation to Various Damaging Agents in the Female Sex Organs in the Rat. Endocrinology 1939, 25, 615–624. [Google Scholar] [CrossRef]
- Giménez Martín, C.; Zafra Gómez, F.; Aragón Rueda, C. Fisiopatología de Los Transportadores de Glutamato y de Glicina: Nuevas Dianas Terapéuticas. Rev. Neurol. 2018, 67, 491. [Google Scholar] [CrossRef]
- Iremonger, K.J.; Constantin, S.; Liu, X.; Herbison, A.E. Glutamate Regulation of GnRH Neuron Excitability. Brain Res. 2010, 1364, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, G. Functional Amino Acids in Growth, Reproduction, and Health. Adv. Nutr. 2010, 1, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Crane, A.R.; Redden, R.R.; Van Emon, M.L.; Neville, T.L.; Reynolds, L.P.; Caton, J.S.; Schauer, C.S. Impacts of Supplemental Arginine on the Reproductive Performance of Fall Lambing Ewes. J. Anim. Sci. 2016, 94, 3540–3549. [Google Scholar] [CrossRef] [PubMed]
- Damián, J.P.; de Soto, L.; Espindola, D.; Gil, J.; Van Lier, E. Intranasal Oxytocin Affects the Stress Response to Social Isolation in Sheep. Physiol. Behav. 2021, 230, 113282. [Google Scholar] [CrossRef] [PubMed]
- Champagne, F.; Diorio, J.; Sharma, S.; Meaney, M.J. Naturally Occurring Variations in Maternal Behavior in the Rat Are Associated with Differences in Estrogen-Inducible Central Oxytocin Receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 12736–12741. [Google Scholar] [CrossRef] [PubMed]
- Feldman, R.; Zagoory-Sharon, O.; Weisman, O.; Schneiderman, I.; Gordon, I.; Maoz, R.; Shalev, I.; Ebstein, R.P. Sensitive Parenting Is Associated with Plasma Oxytocin and Polymorphisms in the OXTR and CD38 Genes. Biol. Psychiatry 2012, 72, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Dobson, H.; Smith, R.F. What Is Stress, and How Does It Affect Reproduction? Anim. Reprod. Sci. 2000, 60, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Damián, J.P.; Bausero, M.; Bielli, A. Acute Stress, Hypothalamic-Hypophyseal-Gonadal Axis and Testicular Function—A Review. Ann. Anim. Sci. 2015, 15, 31–50. [Google Scholar] [CrossRef]
- Reif, A.; Lesch, K.-P. Toward a Molecular Architecture of Personality. Behav. Brain Res. 2003, 139, 1–20. [Google Scholar] [CrossRef]
- Noblett, K.L.; Coccaro, E.F. Molecular Genetics of Personality. Curr. Psychiatry Rep. 2005, 7, 73–80. [Google Scholar] [CrossRef]
- Meyer-Lindenberg, A.; Domes, G.; Kirsch, P.; Heinrichs, M. Oxytocin and Vasopressin in the Human Brain: Social Neuropeptides for Translational Medicine. Nat. Rev. Neurosci. 2011, 12, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Kumsta, R.; Heinrichs, M. Oxytocin, Stress and Social Behavior: Neurogenetics of the Human Oxytocin System. Curr. Opin. Neurobiol. 2013, 23, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Tost, H.; Kolachana, B.; Hakimi, S.; Lemaitre, H.; Verchinski, B.A.; Mattay, V.S.; Weinberger, D.R.; Meyer–Lindenberg, A. A Common Allele in the Oxytocin Receptor Gene (OXTR) Impacts Prosocial Temperament and Human Hypothalamic-Limbic Structure and Function. Proc. Natl. Acad. Sci. USA 2010, 107, 13936–13941. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.I.; Zai, C.C.; Abu, Z.; Nowrouzi, B.; Beitchman, J.H. The Role of Oxytocin and Oxytocin Receptor Gene Variants in Childhood-onset Aggression. Genes Brain Behav. 2012, 11, 545–551. [Google Scholar] [CrossRef]
- von Borell, E.; Dobson, H.; Prunier, A. Stress, Behaviour and Reproductive Performance in Female Cattle and Pigs. Horm. Behav. 2007, 52, 130–138. [Google Scholar] [CrossRef]
- Ribeiro, E.D.A.; Pinotsis, N.; Ghisleni, A.; Salmazo, A.; Konarev, P.V.; Kostan, J.; Sjöblom, B.; Schreiner, C.; Polyansky, A.A.; Gkougkoulia, E.A.; et al. The Structure and Regulation of Human Muscle α-Actinin. Cell 2014, 159, 1447–1460. [Google Scholar] [CrossRef]
- Murphy, A.C.H.; Young, P.W. The Actinin Family of Actin Cross-Linking Proteins—A Genetic Perspective. Cell Biosci. 2015, 5, 49. [Google Scholar] [CrossRef]
- Arvanitis, M.; Tampakakis, E.; Zhang, Y.; Wang, W.; Auton, A.; Agee, M.; Aslibekyan, S.; Bell, R.K.; Bryc, K.; Clark, S.K.; et al. Genome-Wide Association and Multi-Omic Analyses Reveal ACTN2 as a Gene Linked to Heart Failure. Nat. Commun. 2020, 11, 1122. [Google Scholar] [CrossRef]
- Herrero, A. Especificidad Espacio-Temporal de las Señales Ras-ERK en la Determinación de Respuestas Biológicas; Instituto de Biomedicina y Biotecnología de Cantabria: Cantabria, Spain, 2012. [Google Scholar]
- Sapolsky, R.M.; Romero, L.M.; Munck, A.U. How Do Glucocorticoids Influence Stress Responses? Integrating Permissive, Suppressive, Stimulatory, and Preparative Actions. Endocr. Rev. 2000, 21, 55–89. [Google Scholar] [CrossRef]
- Cannon, W.B. The Emergency Function Medulla in Pain and The Major Emotions. Am. J. Physiol. -Leg. Content 1914, 33, 356–372. [Google Scholar] [CrossRef]
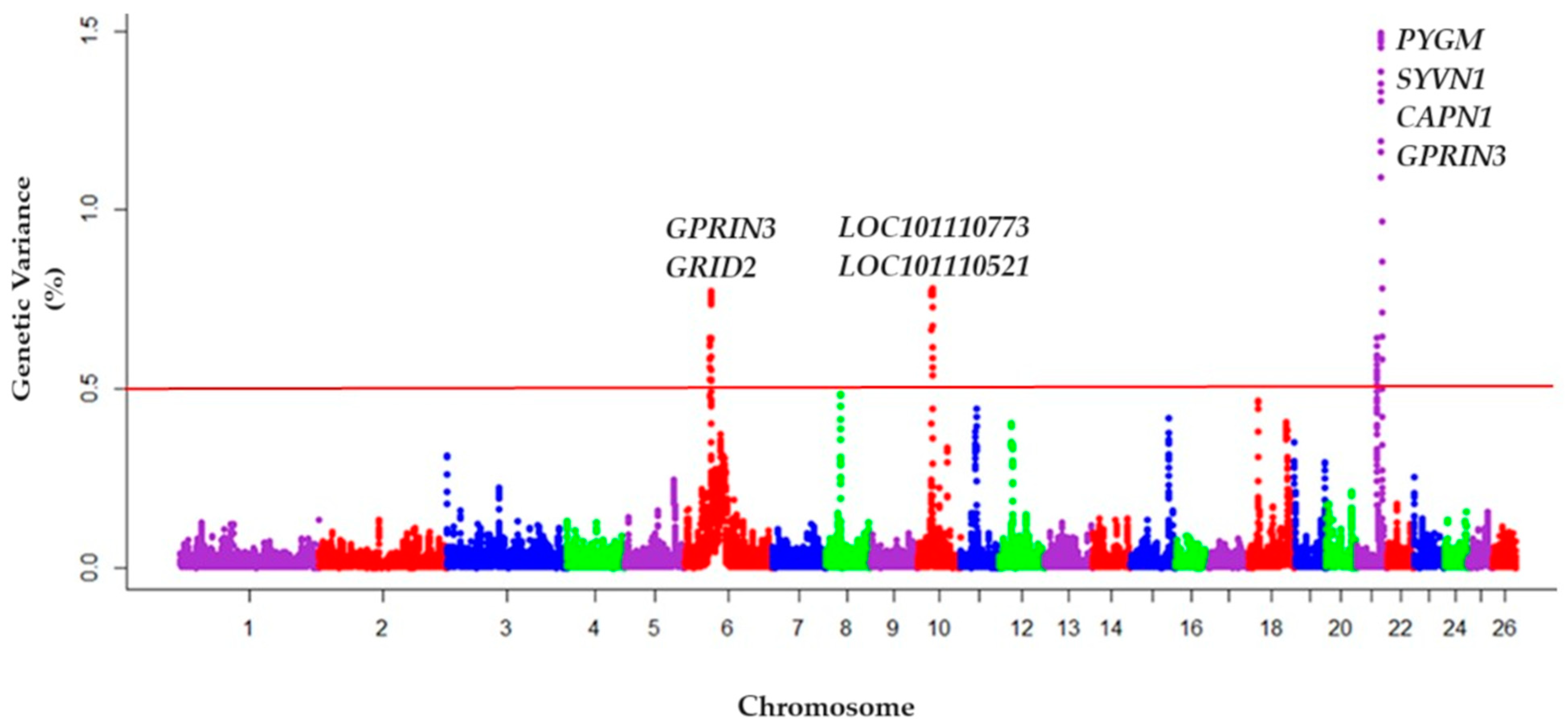
| rs Code | Pos (bp) | CHR | % Var | Variant Type | Candidate Gene |
|---|---|---|---|---|---|
| rs402505013 | 42,295,749 | 21 | 1.4691 | Splice acceptor var | PYGM |
| rs419347404 | 42,601,748 | 21 | 1.4519 | Intron variant | - |
| rs161627521 | 42,654,067 | 21 | 1.3288 | Intron variant | SYVN1 |
| rs413708295 | 42,714,381 | 21 | 1.1617 | Intron variant | CAPN1 |
| rs421553713 | 42,714,613 | 21 | 1.0906 | Intron variant | CAPN1 |
| rs161627624 | 42,715,850 | 21 | 0.8537 | Synonymous variant | CAPN1 |
| rs408317317 | 29,353,089 | 10 | 0.7803 | Intron variant | LOC101110773 |
| rs428995675 | 29,072,930 | 10 | 0.7738 | Intron variant | LOC101110521 |
| rs407693533 | 35,491,698 | 6 | 0,7722 | Intron variant | - |
| rs422603241 | 31,453,177 | 6 | 0.7688 | Intron variant | GRID2 |
| rs422288687 | 29,304,176 | 10 | 0.7678 | Intron variant | LOC101110773 |
| rs427220269 | 29,054,709 | 10 | 0.7670 | Intron variant | LOC101110521 |
| rs421383362 | 29,188,403 | 10 | 0.7636 | Intron variant | LOC101110521 |
| rs409829992 | 29,162,222 | 10 | 0.7624 | Intron variant | LOC101110521 |
| rs399382510 | 29,202,499 | 10 | 0.7619 | Intergenic variant | - |
| rs411759303 | 35,511,497 | 6 | 0.7430 | Missense variant | GPRIN3 |
| rs400430030 | 29,421,760 | 10 | 0.6745 | Intron variant | LOC101110773 |
| rs419116702 | 29,030,595 | 10 | 0.6646 | Intron variant | LOC101110521 |
| rs398879843 | 35,191,867 | 6 | 0.6423 | Intergenic variant | - |
| rs161618576 | 39,638,962 | 21 | 0.6420 | Downstream gene variant | - |
| rs424142667 | 35,511,899 | 6 | 0.6415 | Missense variant | GPRIN3 |
| rs410592527 | 35,184,703 | 6 | 0.6228 | Intergenic variant | - |
| rs408222545 | 39,642,723 | 21 | 0.6204 | Upstream gene variant | - |
| rs419203432 | 29,415,140 | 10 | 0.6173 | Intron variant | LOC101110773 |
| rs161618641 | 39,646,260 | 21 | 0.5915 | Upstream gene variant | - |
| rs416558978 | 35,254,368 | 6 | 0.5873 | Intergenic variant | - |
| rs417392501 | 39,651,749 | 21 | 0.5832 | Downstream gene variant | - |
| rs404318469 | 42,719,476 | 21 | 0.5826 | Intron variant | CAPN1 |
| rs403363266 | 39,653,383 | 21 | 0.5657 | 3 prime UTR variant | FADS1 |
| rs419203432 | 29,415,140 | 10 | 0.5595 | Intron variant | LOC101110773 |
| rs403382566 | 35,512,106 | 6 | 0.5508 | Missense variant | GPRIN3 |
| rs421709693 | 39,432,569 | 21 | 0.5434 | Intergenic variant | SYT7 |
| rs427110197 | 39,654,860 | 21 | 0.5384 | Intron variant | FADS1 |
| rs398157763 | 29,455,959 | 10 | 0.5363 | 3 prime UTR variant | LOC101110773 |
| rs399480023 | 31,217,615 | 6 | 0.5249 | Intron variant | GRID2 |
| rs399060511 | 35,275,766 | 6 | 0.5247 | Intergenic variant | - |
| rs406335698 | 35,276,244 | 6 | 0.5219 | Intergenic variant | - |
| Chromosome | Start Position (bp) | End Position (bp) | Candidate Genes |
|---|---|---|---|
| 6 | 35,138,403 | 35,512,106 | GRID2, GPRIN3 |
| 10 | 29,030,595 | 29,455,959 | LOC101110773, LOC101110521 |
| 21 | 39,432,569 | 42,419,476 | PYGM, SYVN1, CAPN1, FADS1, SYT7 |
| Pathways | p-Value |
|---|---|
| Signaling | |
| Phosphatidylinositol signaling system | 0.0042 |
| Adherens junction | 0.0094 |
| Phoaphopase D signaling pathway | 0.0240 |
| Pathways in cancer | 0.0310 |
| Axon guidance | 0.0370 |
| Calcium signaling pathway | 0.0480 |
| Metabolism | |
| Glycosaminoglycan biosynthesis—heparin sulfate/heparin | 0.0070 |
| ECM—receptor interaction | 0.0270 |
| Metabolic pathway | 0.0340 |
| Steroidogenesis | |
| Ovarian steroidogenesis | 0.0170 |
| Others | |
| Arrhythmogenic right ventricular cardiomyopathy | 0.0370 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romaniuk, E.; Vera, B.; Peraza, P.; Ciappesoni, G.; Damián, J.P.; Van Lier, E. Identification of Candidate Genes and Pathways Linked to the Temperament Trait in Sheep. Genes 2024, 15, 229. https://doi.org/10.3390/genes15020229
Romaniuk E, Vera B, Peraza P, Ciappesoni G, Damián JP, Van Lier E. Identification of Candidate Genes and Pathways Linked to the Temperament Trait in Sheep. Genes. 2024; 15(2):229. https://doi.org/10.3390/genes15020229
Chicago/Turabian StyleRomaniuk, Estefanía, Brenda Vera, Pablo Peraza, Gabriel Ciappesoni, Juan Pablo Damián, and Elize Van Lier. 2024. "Identification of Candidate Genes and Pathways Linked to the Temperament Trait in Sheep" Genes 15, no. 2: 229. https://doi.org/10.3390/genes15020229
APA StyleRomaniuk, E., Vera, B., Peraza, P., Ciappesoni, G., Damián, J. P., & Van Lier, E. (2024). Identification of Candidate Genes and Pathways Linked to the Temperament Trait in Sheep. Genes, 15(2), 229. https://doi.org/10.3390/genes15020229







