Validation and Analysis of COIL, a Gene Associated with Multiple Lambing Traits in Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sampling
2.2. Determination of Multi-Lambing Performance and Extraction of Tissue DNA in Sheep
2.3. Polymorphism Detection of the COIL Gene in Sheep
2.4. RNA Extraction, Reverse Transcription, and Real-Time PCR
2.5. Construction and Western Blot Validation of the Eukaryotic Expression Vector of the Sheep COIL Gene
2.6. SiRNA Interference
2.7. The Cell Proliferation Activity Assay
2.8. Detection of the Effect of the Sheep COIL Gene on the Biogenesis Pathway of Spliceosomal U snRNPs
2.9. Statistical Methods
3. Results
3.1. Polymorphism Analysis of the COIL Gene in Sheep
3.2. Examining the Relationships between Sheep Lambing Features and Polymorphic Genotypes of the COIL Gene
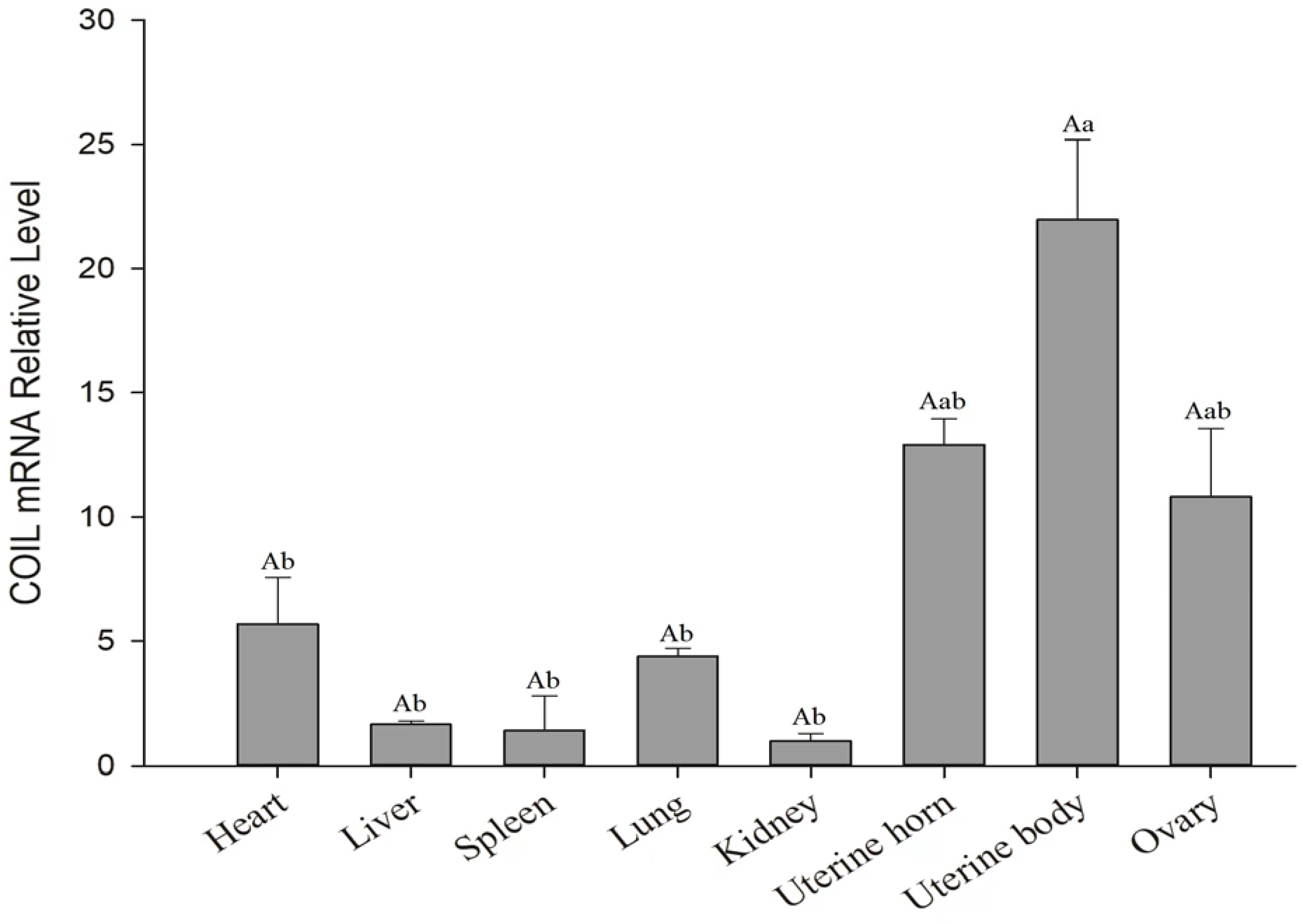
3.3. Tissue Expression Profile Analysis of the COIL Gene in Sheep
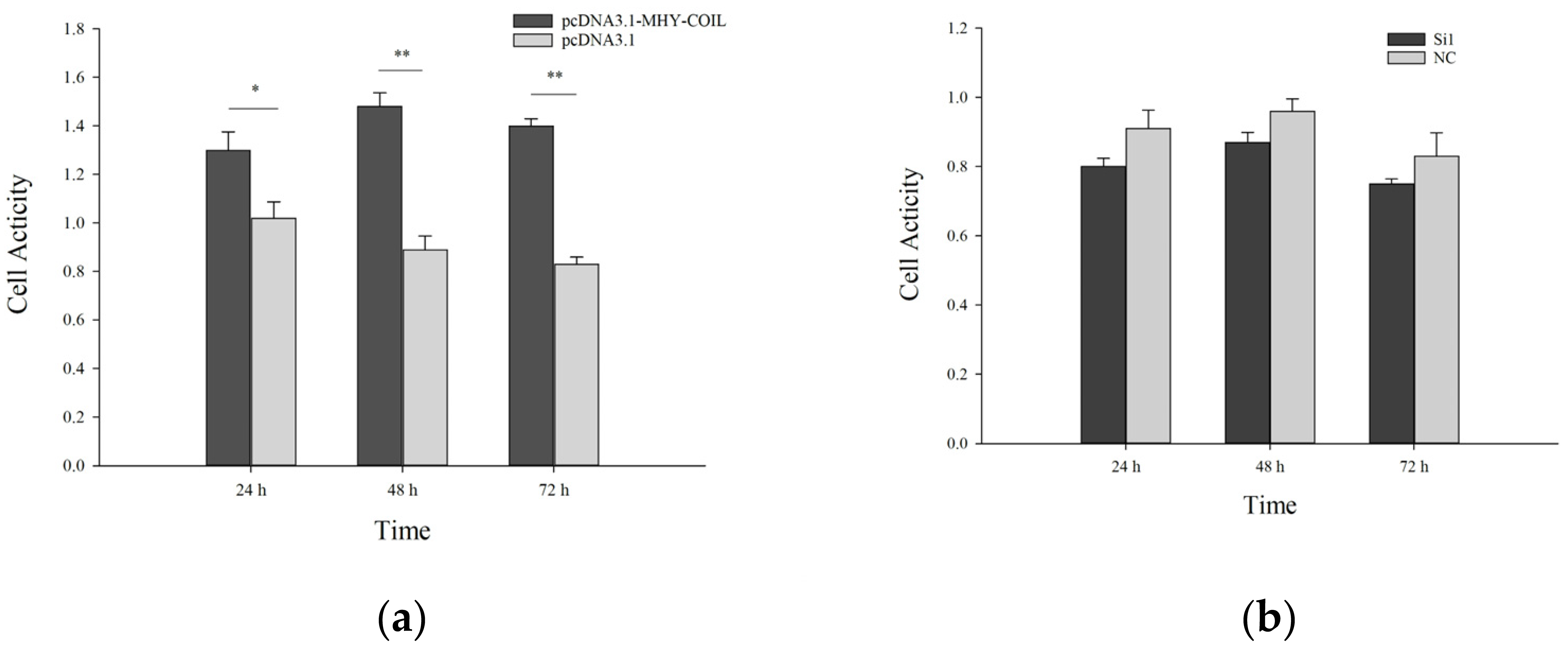
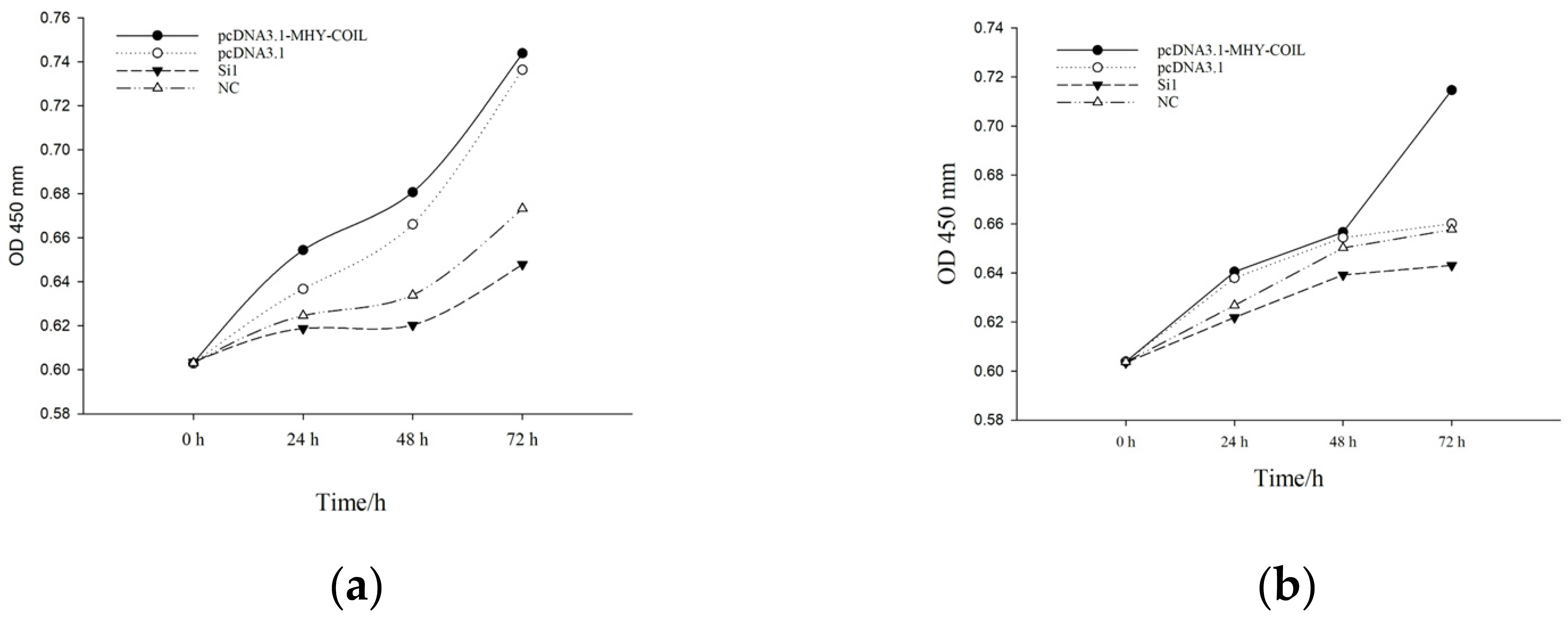
3.4. Effect of Sheep COIL Genes on Cell Proliferation and Activity
3.4.1. The MTT Cell Survival Assay
3.4.2. The CCK-8 Cell Proliferation Assay
3.5. Detection of the Effect of the Sheep COIL Gene on the Biogenesis Pathway of Spliceosomal U snRNPs
4. Discussion
4.1. Mechanism of Action of the COIL Gene
4.2. Study of the Mechanism of the COIL Gene
4.3. Impact of COIL Genes on Spliceosomal U snRNPs’ Biogenesis Pathway
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henchion, M.; Zimmermann, J. Animal food products: Policy, market and social issues and their influence on demand and supply of meat. Proc. Nutr. Soc. 2021, 80, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Tursun, A. Effect of Different Treatments on the Effect of Simultaneous Estrus Lambing in Dorang and Bayinbruck Sheep. Ph.D. Thesis, Xinjiang Agricultural University, Ürümqi, China, 2016. [Google Scholar]
- Abdilahi, A.; Abdimahad, K.; Mahamed, A.; Ali, A. Study on breeding practices and reproductive performance of black-head somali sheep under traditional management system: The case of awbarre district, eastern Ethiopia. Open J. Anim. Sci. 2023, 13, 14. [Google Scholar] [CrossRef]
- Li, H.; Xu, H.; Akhatayeva, Z.; Liu, H.; Pan, C. Novel indel variations of the sheep fecb gene and their effects on litter size. Gene 2021, 767, 145176. [Google Scholar] [CrossRef] [PubMed]
- Gall, J.G.; Bellini, M.; Wu, Z.; Murphy, C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 1999, 10, 4385. [Google Scholar] [CrossRef] [PubMed]
- Neuenkirchen, N.; Chari, A.; Fischer, U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Letters 2008, 582, 1997–2003. [Google Scholar] [CrossRef]
- Ma, H.Y.; Fang, C.; Liu, L.L.; Wang, Q.; Aniwashi, J.; Sulaiman, Y.; Abudilaheman, K.; Liu, W.J. Identification of novel genes associated with litter size of indigenous sheep population in Xinjiang, China using specific-locus amplified fragment sequencing technology. PeerJ 2019, 7, e8079. [Google Scholar] [CrossRef]
- Köchl, S.; Niederstätter, H.; Parson, W. DNA Extraction and Quantitation of Forensic Samples Using the Phenol-Chloroform Method and Real-Time PCR. Methods Mol. Biol. 2005, 297, 13. [Google Scholar] [CrossRef]
- Xiao, D.; Li, J.; Wen, J. Improvement of Western blot method for protein expression detection. J. Clin. Exp. Pathol. 2003, 19, 210–211. [Google Scholar]
- Machyna, M.; Heyn, P.; Neugebauer, K.M. Cajal Bodies: Where Form Meets Function. Wiley Interdiscip. Rev. RNA 2013, 4, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Tucker, K.E.; Massello, L.K.; Gao, L.; Barber, T.J.; Hebert, M.D.; Chan, E.K.; Matera, A.G. Structure and characterization of the murine p80 coilin gene, Coil. J. Struct. Biol. 2000, 129, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Y. Production of Adult Skin Fibroblast Cloned Sheep. Ph.D. Thesis, Inner Mongolia Agricultural University, Hohhot, China, 2009. [Google Scholar]
- Dong, Y.J.; Li, Q.W.; Cao, W.G.; Nimal, J.; Wan-Li, L. Construction of IGF2 eukaryotic expression vector and transgenic study of sheep skeletal muscle cells. Heilongjiang Anim. Husb. Vet. Med. 2013, 07, 6–8. [Google Scholar]
- Du, W.J.; Hao, L.; Chao, X.; Piao, S.; Wang, S.; An, T. Transplantation and identification of reconstructed embryos with MSTN gene knockdown from sheep. J. Hunan Agric. Univ. Nat. Sci. Ed. 2018, 44, 539–543. [Google Scholar]
- Yu, M. Construction of Skin-Specific Expression Vector and Interference Vector of Tibetan Sheep Smo Gene and Preliminary Analysis of Their Functions. Ph.D. Thesis, Southwest University for Nationalities, Sichuan, China, 2016. [Google Scholar]
- Li, Y.; Wu, K.F.; Guo, X.D.; Guo, J.T.; Bou, S.G. Optimization of conditions for liposome-mediated in vitro transfection of exogenous genes into bovine fetal fibroblasts. Genetics 2002, 24, 653–655. [Google Scholar]
- Lu, J.; Sun, D.; Xu, L.; Lu, G.; Zhao, F.; Wei, C.; Zhang, L.; Ding, J.; Li, B.; Du, L. Selection of an effective small interference RNA to silence myostatin gene expression in sheep fibroblast cells. Biochem. Genet. 2012, 50, 838–847. [Google Scholar] [CrossRef]
- Dunlap, K.A.; Kwak, H.I.; Burghardt, R.C.; Bazer, F.W.; Magness, R.R.; Johnson, G.A. The Sphingosine 1-Phosphate (S1P) Signaling Pathway Is Regulated During Pregnancy in Sheep. Biol. Reprod. 2010, 82, 876–887. [Google Scholar] [CrossRef]
- Che, H.; Wang, L.; Ren, Y. CCK-8 cell viability assay. Bio-Protocol, 2021; 1481, preprint. [Google Scholar]
- Zhang, Z.; Xu, J.; Lyu, S.; Wang, E. Whole-Genome DNA Methylation Dynamics of Sheep Preimplantation Embryo Investigated by Single-Cell DNA Methylome Sequencing. Front. Genet. 2021, 12, 753144. [Google Scholar] [CrossRef]
- Esmaeili-Fard, S.M.; Gholizadeh, M.; Hafezian, S.H.; Abdollahi-Arpanahi, R. Genome-wide association study and pathway analysis identify NTRK2 as a novel candidate gene for litter size in sheep. PLoS ONE 2021, 16, e0244408. [Google Scholar] [CrossRef]
- Jia, J.L.; Ding, Q.; Lai, Y.; Zhang, Y.; Zhang, L.P. Screening of Sheep BMPR-1B Interacting Proteins by CoIP and MS. J. Domest. Anim. Ecol. 2017, 38, 10–13. [Google Scholar]
- Ferreira, J.A. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J. Cell Biol. 1994, 126, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Tucker, K.E. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 2001, 154, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Chari, A.; Golas, M.M.; Klingenh Ger, M.; Euenkirchen, N.; Sander, B.; Englbrecht, C. An Assembly Chaperone Collaborates with the SMN Complex to Generate Spliceosomal SnRNPs. Cell 2008, 135, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Golembe, T.J.; Yong, J.; Dreyfuss, G. Specific Sequence Features, Recognized by the SMN Complex, Identify snRNAs and Determine Their Fate as snRNPs. Mol. Cell. Biol. 2005, 25, 10989–11004. [Google Scholar] [CrossRef] [PubMed]
- Giulia, B.; Roberto, M.; Phuong, C.T.; Magdalena, M.; Maciej, C.; Sowndarya, M. The small Cajal body-specific RNA 15 (SCARNA15) directs p53 and redox homeostasis via selective splicing in cancer cells. NAR Cancer 2021, 3, zcab026. [Google Scholar]
- Mahmoudi, S.; Henriksson, S.; Weibrecht, I.; Smith, S.; Söderberg, O.; Strömblad, S. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 2010, 8, e1000521. [Google Scholar] [CrossRef] [PubMed]
- Bergstrand, S.; O’Brien, E.M.; Farnebo, M. The Cajal Body Protein WRAP53β Prepares the Scene for Repair of DNA Double-Strand Breaks by Regulating Local Ubiquitination. Front. Mol. Biosci. 2019, 6, 51. [Google Scholar] [CrossRef]
- Yuan, F.; Li, G.; Tong, T. Nucleolar and coiled-body phosphoprotein 1 (NOLC1) regulates the nucleolar retention of TRF2. Cell Death Discov. 2017, 3, 17043. [Google Scholar] [CrossRef]
- Moreno-Castro, C.; Prieto-Sánchez, S.; Sánchez-Hernández, N.; Hernández-Munain, C.; Sué, C. Role for the splicing factor TCERG1 in Cajal body integrity and snRNP assembly. J. Cell Sci. 2019, 132, jcs232728. [Google Scholar] [CrossRef]
- Schaffert, N. Role of Cajal bodies in spliceosomal U snRNP biogenesis. Boehringer Ingelh. Fonds Futur. 2005, 20, 276–280. [Google Scholar]



| Gene Name | SNP Site | Position | Primer Sequence |
|---|---|---|---|
| COIL | COILSNP1 | 7321466 | F1: GAAGGTCGGAGTCAACGGATTCCATGAAAGAACCTGGGAAA F2: GAAGGTGACCAAGTTCATGCTCCATGAAAGAACCTGGGAAC R1: CCTCAGCTCCATTTTCGTTG |
| COIL | COILSNP2 | 7314134 | F1: GAAGGTCGGAGTCAACGGATTGACTCCGAGGAGGAATCGC F2: GAAGGTGACCAAGTTCATGCTGACTCCGAGGAGGAATCGG R1: GTGGCATGGTCGTCCGTAC |
| COIL | COILSNP3 | 7321563 | F1:GAAGGTCGGAGTCAACGGATTGCACAGTCTGTGAAAGAGTGGA F2: GAAGGTGACCAAGTTCATGCTGCACAGTCTGTGAAAGAGTGGG R1: TCTAGCAGGAAGAGCTTTAGGG |
| Gene | Sequence (5′-3′) | Length (bp) |
|---|---|---|
| COIL | Si1 | CCAUCAUCACAGGCUCCAATT |
| Si2 | CCAGAAGUGCAGCCCUAAATT | |
| Si3 | GCAAAGAAGCGGGCAUUUATT |
| SNP Site | Breed | Number | Genotype Frequency | Allele Frequency | |||
|---|---|---|---|---|---|---|---|
| CC | CG | GG | C | G | |||
| COILSNP1 | Altay sheep | 65 | 0.450 | 0.483 | 0.067 | 0.692 | 0.308 |
| Turpan black sheep | 240 | 0.570 | 0.338 | 0.092 | 0.739 | 0.261 | |
| Hetian duotai hong sheep | 62 | 0.580 | 0.360 | 0.060 | 0.760 | 0.240 | |
| Cele black sheep | 44 | 0.720 | 0.260 | 0.020 | 0.850 | 0.150 | |
| Duolang sheep | 16 | 0.190 | 0.560 | 0.250 | 0.470 | 0.530 | |
| AA | AC | CC | A | C | |||
| COILSNP2 | Altay sheep | 65 | / | / | / | / | / |
| Turpan black sheep | 240 | / | / | / | / | / | |
| Hetian duotai hong sheep | 62 | 0.990 | / | 0.010 | 0.990 | 0.010 | |
| Cele black sheep | 44 | / | / | / | / | / | |
| Duolang sheep | 16 | / | / | / | / | / | |
| GG | GA | AA | G | A | |||
| COILSNP3 | Altay sheep | 65 | 0.323 | 0.677 | / | 0.662 | 0.338 |
| Turpan black sheep | 240 | / | / | / | / | / | |
| Hetian duotai hong sheep | 62 | / | 0.530 | 0.470 | 0.263 | 0.737 | |
| Cele black sheep | 44 | / | / | / | / | / | |
| Duolang sheep | 16 | / | / | / | / | / | |
| SNP Site | Breed | PIC | Heterozygosity | Ne |
|---|---|---|---|---|
| COILSNP1 | Altay sheep | 0.336 | 0.427 | 1.744 |
| Turpan black sheep | 0.311 | 0.386 | 1.628 | |
| Hetian duotai hong sheep | 0.297 | 0.363 | 1.569 | |
| Cele black sheep | 0.224 | 0.257 | 0.345 | |
| Duolang sheep | 0.374 | 0.498 | 1.992 | |
| COILSNP2 | Hetian duotai hong sheep | 0.020 | 0.020 | 1.020 |
| COILSNP3 | Altay sheep | 0.347 | 0.446 | 1.806 |
| Hetian duotai hong sheep | 0.312 | 0.387 | 1.632 |
| Population | Trait | Genotype | ||
|---|---|---|---|---|
| CC | CG | GG | ||
| Hetian duotai hong sheep | Average number of lambs | 1.60 ± 0.55 a | 1.39 ± 0.54 ab | 1.17 ± 0.40 b |
| Cele black sheep | Average number of lambs | 1.65 ± 0.48 a | 1.39 ± 0.49 b | 1.50 ± 0.71 ab |
| Duolang sheep | Average number of lambs | 1.15 ± 0.37 b | 1.35 ± 0.49 b | 1.80 ± 0.45 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, H.; Wen, Y.; Ma, H.; Liu, W. Validation and Analysis of COIL, a Gene Associated with Multiple Lambing Traits in Sheep. Genes 2024, 15, 235. https://doi.org/10.3390/genes15020235
Cao H, Wen Y, Ma H, Liu W. Validation and Analysis of COIL, a Gene Associated with Multiple Lambing Traits in Sheep. Genes. 2024; 15(2):235. https://doi.org/10.3390/genes15020235
Chicago/Turabian StyleCao, Hang, Yilin Wen, Haiyu Ma, and Wujun Liu. 2024. "Validation and Analysis of COIL, a Gene Associated with Multiple Lambing Traits in Sheep" Genes 15, no. 2: 235. https://doi.org/10.3390/genes15020235
APA StyleCao, H., Wen, Y., Ma, H., & Liu, W. (2024). Validation and Analysis of COIL, a Gene Associated with Multiple Lambing Traits in Sheep. Genes, 15(2), 235. https://doi.org/10.3390/genes15020235






