Economy of Effort or Sophisticated Programming? The Prevalence of Bidirectional Promoter Complexes in the Human Genome
Abstract
:1. The Prevalence of Antisense Transcription in Protein-Coding Genes Indicates a Role in Gene Regulation
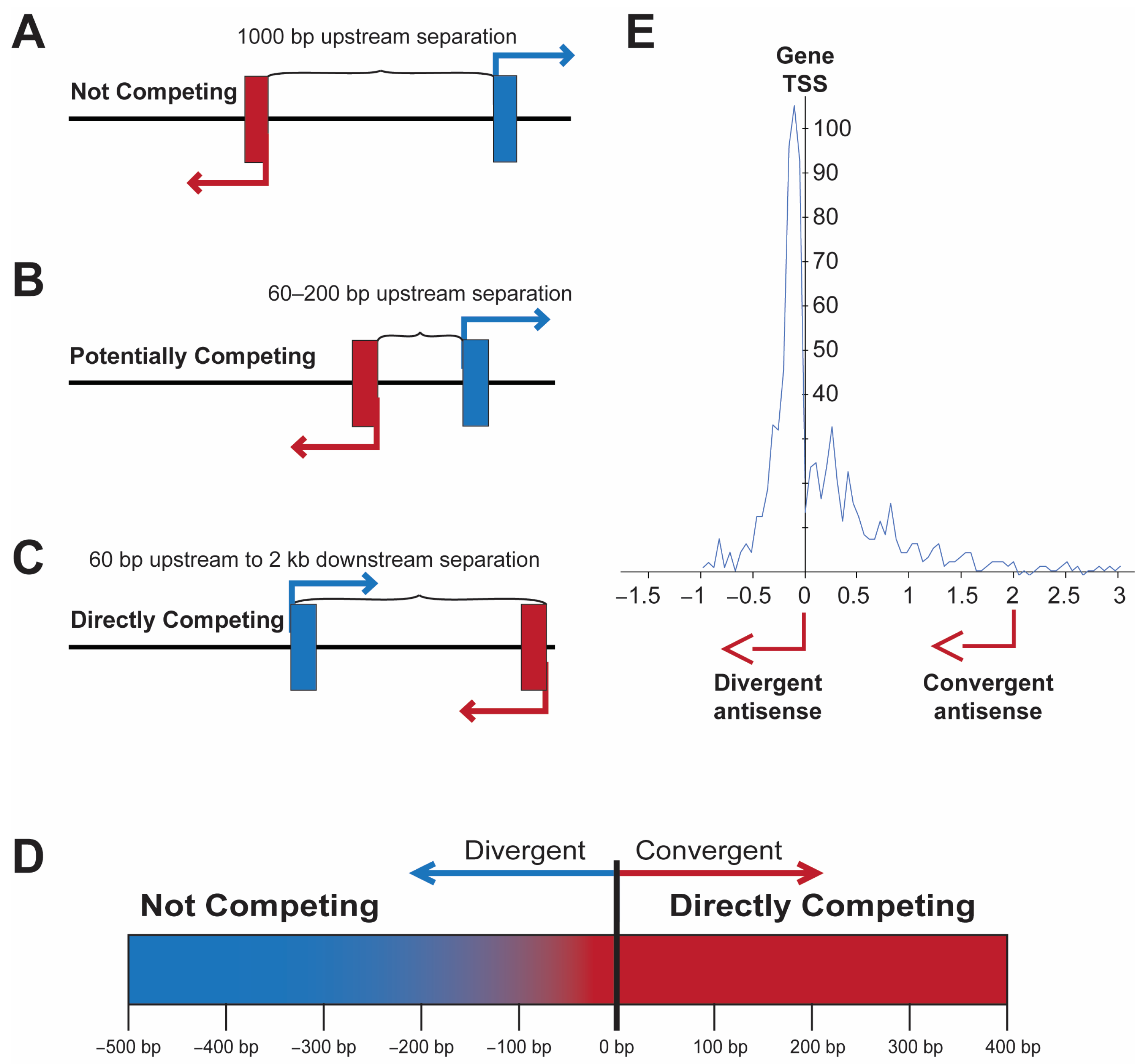
2. Structural Features of Bidirectional Promoters
3. Variegation of MHC Class I Receptor Expression by Bidirectional Promoters
4. GATA Switches and the Role of Noncoding RNA in Differentiation
5. Functional Activities of Noncoding Antisense RNAs
5.1. Diverse Functions of Antisense Transcripts in the GATA Gene Family
5.2. SPI1 Antisense Reduces Gene Expression
5.3. Complex Regulation of the Human E-Cadherin Gene
5.4. Regulation of Gene Expression by lincRNAs
6. Control of Viral Replication by Bidirectional Promoters
6.1. Polyomavirus
6.2. mCMV
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Engström, P.G.; Suzuki, H.; Ninomiya, N.; Akalin, A.; Sessa, L.; Lavorgna, G.; Brozzi, A.; Luzi, L.; Tan, S.L.; Yang, L.; et al. Complex Loci in Human and Mouse Genomes. PLoS Genet. 2006, 2, e47. [Google Scholar] [CrossRef]
- Cheng, J.; Kapranov, P.; Drenkow, J.; Dike, S.; Brubaker, S.; Patel, S.; Long, J.; Stern, D.; Tammana, H.; Helt, G.; et al. Transcriptional Maps of 10 Human Chromosomes at 5-Nucleotide Resolution. Science 2005, 308, 1149–1154. [Google Scholar] [CrossRef]
- He, Y.; Vogelstein, B.; Velculescu, V.E.; Papadopoulos, N.; Kinzler, K.W. The Antisense Transcriptomes of Human Cells. Science 2008, 322, 1855–1857. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Trinklein, N.D.; Aldred, S.F.; Hartman, S.J.; Schroeder, D.I.; Otillar, R.P.; Myers, R.M. An Abundance of Bidirectional Promoters in the Human Genome. Genome Res. 2004, 14, 62–66. [Google Scholar] [CrossRef]
- Rosikiewicz, W.; Sikora, J.; Skrzypczak, T.; Kubiak, M.R.; Makałowska, I. Promoter switching in response to changing environment and elevated expression of protein-coding genes overlapping at their 5′ ends. Sci. Rep. 2021, 11, 8984. [Google Scholar] [CrossRef]
- Li, H.; Rahman, M.A.; Ruesch, M.; Eisele, C.D.; Anderson, E.M.; Wright, P.W.; Cao, J.; Ratnayake, S.; Chen, Q.; Yan, C.; et al. Abundant binary promoter switches in lineage-determining transcription factors indicate a digital component of cell fate determination. Cell Rep. 2023, 42, 113454. [Google Scholar] [CrossRef]
- Lepoivre, C.; Belhocine, M.; Bergon, A.; Griffon, A.; Yammine, M.; Vanhille, L.; Zacarias-Cabeza, J.; Garibal, M.-A.; Koch, F.; Maqbool, M.A.; et al. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genom. 2013, 14, 914. [Google Scholar] [CrossRef]
- Hume, D.A. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood 2000, 96, 2323–2328. [Google Scholar] [CrossRef]
- Olsson, A.; Venkatasubramanian, M.; Chaudhri, V.K.; Aronow, B.J.; Salomonis, N.; Singh, H.; Grimes, H.L. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 2016, 537, 698–702. [Google Scholar] [CrossRef]
- Adachi, N.; Lieber, M.R. Bidirectional gene organization: A common architectural feature of the human genome. Cell 2002, 109, 807–809. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.M.; Collins, P.J.; Trinklein, N.D.; Fu, Y.; Xi, H.; Myers, R.M.; Weng, Z. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res. 2007, 17, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.J.; Kobayashi, Y.; Nguyen, L.; Trinklein, N.D.; Myers, R.M. The Ets-Related Transcription Factor GABP Directs Bidirectional Transcription. PLoS Genet. 2007, 3, e208. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pascal, V.; Martin, M.P.; Carrington, M.; Anderson, S.K. Genetic Control of Variegated KIR Gene Expression: Polymorphisms of the Bi-Directional KIR3DL1 Promoter Are Associated with Distinct Frequencies of Gene Expression. PLoS Genet. 2008, 4, e1000254. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Davies, G.E.; Pascal, V.; Wright, P.W.; Hodge, D.L.; Cho, E.H.; Lockett, S.J.; Abshari, M.; Anderson, S.K. Identification of Probabilistic Transcriptional Switches in the Ly49 Gene Cluster. Immunity 2004, 21, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Tsai, K.-L.; Kalisman, N.; Bushnell, D.A.; Asturias, F.J.; Kornberg, R.D. Structure of an RNA polymerase II preinitiation complex. Proc. Natl. Acad. Sci. USA 2015, 112, 13543–13548. [Google Scholar] [CrossRef] [PubMed]
- Schier, A.C.; Taatjes, D.J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef]
- Rahim, M.M.A.; Makrigiannis, A.P. Ly49 receptors: Evolution, genetic diversity, and impact on immunity. Immunol. Rev. 2015, 267, 137–147. [Google Scholar] [CrossRef]
- Anderson, S.K. Probabilistic bidirectional promoter switches: Noncoding RNA takes control. Mol. Ther. Nucleic Acids 2014, 3, e191. [Google Scholar] [CrossRef]
- Wright, P.W.; Huehn, A.; Cichocki, F.; Li, H.; Sharma, N.; Dang, H.; Lenvik, T.R.; Woll, P.; Kaufman, D.; Miller, J.S.; et al. Identification of a KIR antisense lncRNA expressed by progenitor cells. Genes Immun. 2013, 14, 427–433. [Google Scholar] [CrossRef]
- Cichocki, F.; Lenvik, T.; Sharma, N.; Yun, G.; Anderson, S.K.; Miller, J.S. Cutting Edge: KIR Antisense Transcripts Are Processed into a 28-Base PIWI-Like RNA in Human NK Cells. J. Immunol. 2010, 185, 2009–2012. [Google Scholar] [CrossRef]
- Keenan, C.R. Heterochromatin and Polycomb as regulators of haematopoiesis. Biochem. Soc. Trans. 2021, 49, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Onder, T.T.; Kara, N.; Cherry, A.; Sinha, A.U.; Zhu, N.; Bernt, K.M.; Cahan, P.; Mancarci, B.O.; Unternaehrer, J.; Gupta, P.B.; et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature 2012, 483, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Sakai, S.; Ota, K.; Bando, Y.; Uchida, C.; Niida, H.; Kitagawa, M.; Ohhata, T. CCIVR2 facilitates comprehensive identification of both overlapping and non-overlapping antisense transcripts within specified regions. Sci. Rep. 2023, 13, 14807. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Tanasa, B.; Micheletti, R.; Ohgi, K.A.; Aggarwal, A.K.; Rosenfeld, M.G. Shape of promoter antisense RNAs regulates ligand-induced transcription activation. Nature 2021, 595, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, J.-M.; Roméo, P.-H. T-cell Expression of the Human GATA-3 Gene Is Regulated by a Non-Lineage-Specific Silencer. J. Biol. Chem. 1999, 274, 6567–6578. [Google Scholar] [CrossRef] [PubMed]
- Scheinman, E.J.; Avni, O. Transcriptional Regulation of Gata3 in T Helper Cells by the Integrated Activities of Transcription Factors Downstream of the Interleukin-4 Receptor and T Cell Receptor. J. Biol. Chem. 2009, 284, 3037–3048. [Google Scholar] [CrossRef] [PubMed]
- Tindemans, I.; Serafini, N.; Di Santo, J.P.; Hendricks, R.W. GATA-3 Function in Innate and Adaptive Immunity. Immunity 2014, 41, 191–206. [Google Scholar] [CrossRef]
- Gibbons, H.R.; Shaginurova, G.; Kim, L.C.; Chapman, N.; Spurlock, C.F.; Aune, T.M. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Front. Immunol. 2018, 9, 2512. [Google Scholar] [CrossRef]
- Man, H.S.J.; Subramaniam, N.; Downs, T.; Sukumar, A.N.; Saha, A.D.; Nair, R.; Chen, L.; Teitelbaum, D.; Turgeon, P.J.; Ku, K.H.; et al. Long noncoding RNA GATA2-AS1 augments endothelial hypoxia inducible factor 1-α induction and regulates hypoxic signaling. J. Biol. Chem. 2023, 299, 103029. [Google Scholar] [CrossRef]
- Tiyaboonchai, A.; Cardenas-Diaz, F.L.; Ying, L.; Maguire, J.A.; Sim, X.; Jobaliya, C.; Gagne, A.L.; Kishore, S.; Stanescu, D.E.; Hughes, N.; et al. GATA6 Plays an Important Role in the Induction of Human Definitive Endoderm, Development of the Pancreas, and Functionality of Pancreatic β Cells. Stem Cell Rep. 2017, 8, 589–604. [Google Scholar] [CrossRef]
- Jha, R.; Li, D.; Wu, Q.; Ferguson, K.E.; Forghani, P.; Gibson, G.C.; Xu, C. A long non-coding RNA GATA6-AS1 adjacent to GATA6 is required for cardiomyocyte differentiation from human pluripotent stem cells. FASEB J. 2020, 34, 14336–14352. [Google Scholar] [CrossRef]
- Yang, J.; Lu, P.; Li, M.; Yan, C.; Zhang, T.; Jiang, W. GATA6-AS1 Regulates GATA6 Expression to Modulate Human Endoderm Differentiation. Stem Cell Rep. 2020, 15, 694–705. [Google Scholar] [CrossRef]
- Heuts, B.M.H.; Arza-Apalategi, S.; Frölich, S.; Bergevoet, S.M.; Van Den Oever, S.N.; Van Heeringen, S.J.; Van Der Reijden, B.A.; Martens, J.H.A. Identification of transcription factors dictating blood cell development using a bidirectional transcription network-based computational framework. Sci. Rep. 2022, 12, 18656. [Google Scholar] [CrossRef]
- Trinh, B.Q.; Ummarino, S.; Zhang, Y.; Ebralidze, A.K.; Bassal, M.A.; Nguyen, T.M.; Heller, G.; Coffey, R.; Tenen, D.E.; Van Der Kouwe, E.; et al. Myeloid lncRNA LOUP mediates opposing regulatory effects of RUNX1 and RUNX1-ETO in t(8;21) AML. Blood 2021, 138, 1331–1344. [Google Scholar] [CrossRef]
- Van Der Kouwe, E.; Heller, G.; Czibere, A.; Pulikkan, J.A.; Agreiter, C.; Castilla, L.H.; Delwel, R.; Di Ruscio, A.; Ebralidze, A.K.; Forte, M.; et al. Core-binding factor leukemia hijacks the T-cell–prone PU.1 antisense promoter. Blood 2021, 138, 1345–1358. [Google Scholar] [CrossRef]
- Magnani, E.; Macchi, F.; Mancini, M.; Lomazzi, V.; Sara, C.; Pistore, C.; Mandruzzato, M.; Dock-Bregeon, A.-C.; Bonapace, I.M. UHRF1 regulates CDH1 via promoter associated non-coding RNAs in prostate cancer cells. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2018, 1861, 258–270. [Google Scholar] [CrossRef]
- Pisignano, G.; Napoli, S.; Magistri, M.; Mapelli, S.N.; Pastori, C.; Di Marco, S.; Civenni, G.; Albino, D.; Enriquez, C.; Allegrini, S.; et al. A promoter-proximal transcript targeted by genetic polymorphism controls E-cadherin silencing in human cancers. Nat. Commun. 2017, 8, 15622. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Deniz, E.; Erman, B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct. Integr. Genom. 2017, 17, 135–143. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in Bacteriophage Lambda Development. Annu. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef]
- Callen, B.P.; Shearwin, K.E.; Egan, J.B. Transcriptional Interference between Convergent Promoters Caused by Elongation over the Promoter. Mol. Cell 2004, 14, 647–656. [Google Scholar] [CrossRef]
- O’Grady, T.; Cao, S.; Strong, M.J.; Concha, M.; Wang, X.; Splinter Bondurant, S.; Adams, M.; Baddoo, M.; Srivastav, S.K.; Lin, Z.; et al. Global Bidirectional Transcription of the Epstein-Barr Virus Genome during Reactivation. J. Virol. 2014, 88, 1604–1616. [Google Scholar] [CrossRef]
- Hirsch, H.H.; Steiger, J. Polyomavirus BK. Lancet Infect. Dis. 2003, 3, 611–623. [Google Scholar] [CrossRef]
- Moens, U.; Johansen, T.; Johnsen, J.I.; Seternes, O.M.; Traavik, T. Noncoding control region of naturally occurring BK virus variants: Sequence comparison and functional analysis. Virus Genes 1995, 10, 261–275. [Google Scholar] [CrossRef]
- Bethge, T.; Ajuh, E.; Hirsch, H.H. Imperfect Symmetry of Sp1 and Core Promoter Sequences Regulates Early and Late Virus Gene Expression of the Bidirectional BK Polyomavirus Noncoding Control Region. J. Virol. 2016, 90, 10083–10101. [Google Scholar] [CrossRef]
- Stinski, M.F.; Isomura, H. Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency. Med. Microbiol. Immunol. 2008, 197, 223–231. [Google Scholar] [CrossRef]
- Simon, C.O.; Kühnapfel, B.; Reddehase, M.J.; Grzimek, N.K.A. Murine Cytomegalovirus Major Immediate-Early Enhancer Region Operating as a Genetic Switch in Bidirectional Gene Pair Transcription. J. Virol. 2007, 81, 7805–7810. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anderson, E.M.; Anderson, S.K. Economy of Effort or Sophisticated Programming? The Prevalence of Bidirectional Promoter Complexes in the Human Genome. Genes 2024, 15, 252. https://doi.org/10.3390/genes15020252
Anderson EM, Anderson SK. Economy of Effort or Sophisticated Programming? The Prevalence of Bidirectional Promoter Complexes in the Human Genome. Genes. 2024; 15(2):252. https://doi.org/10.3390/genes15020252
Chicago/Turabian StyleAnderson, Erik M., and Stephen K. Anderson. 2024. "Economy of Effort or Sophisticated Programming? The Prevalence of Bidirectional Promoter Complexes in the Human Genome" Genes 15, no. 2: 252. https://doi.org/10.3390/genes15020252
APA StyleAnderson, E. M., & Anderson, S. K. (2024). Economy of Effort or Sophisticated Programming? The Prevalence of Bidirectional Promoter Complexes in the Human Genome. Genes, 15(2), 252. https://doi.org/10.3390/genes15020252





