Genome-Wide Characterization of the Phenylalanine Ammonia-Lyase Gene Family and Their Potential Roles in Response to Aspergillus flavus L. Infection in Cultivated Peanut (Arachis hypogaea L.)
Abstract
:1. Introduction
2. Material and Methods
2.1. Database Searching and Identification of PAL Genes in Cultivated Peanut Genome
2.2. Phylogenetic Analysis of PAL Proteins
2.3. Chromosome Localization, Gene Structure, Conserved Motif, and Duplication Event Analysis of AhPALs
2.4. Cis-Acting Element Analysis of AhPALs
2.5. In Silico Expression Analysis of AhPAL Genes Using RNA-seq Data
2.6. Plant Materials and Quantitative RT-PCR-Based Expression Assays of AhPALs
3. Results
3.1. Genome-Wide Identification of PAL Genes in the Cultivated Peanut Genome
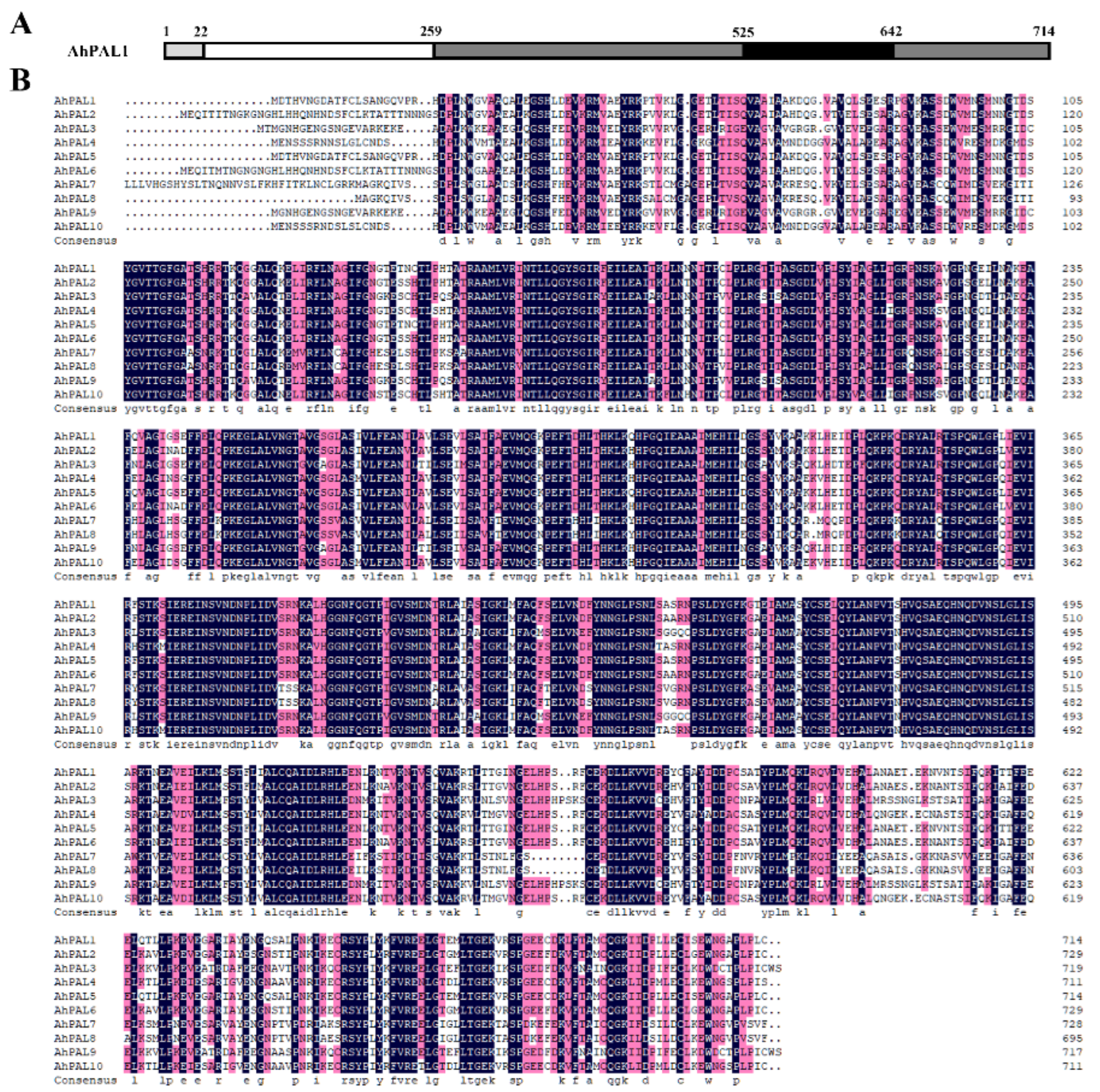
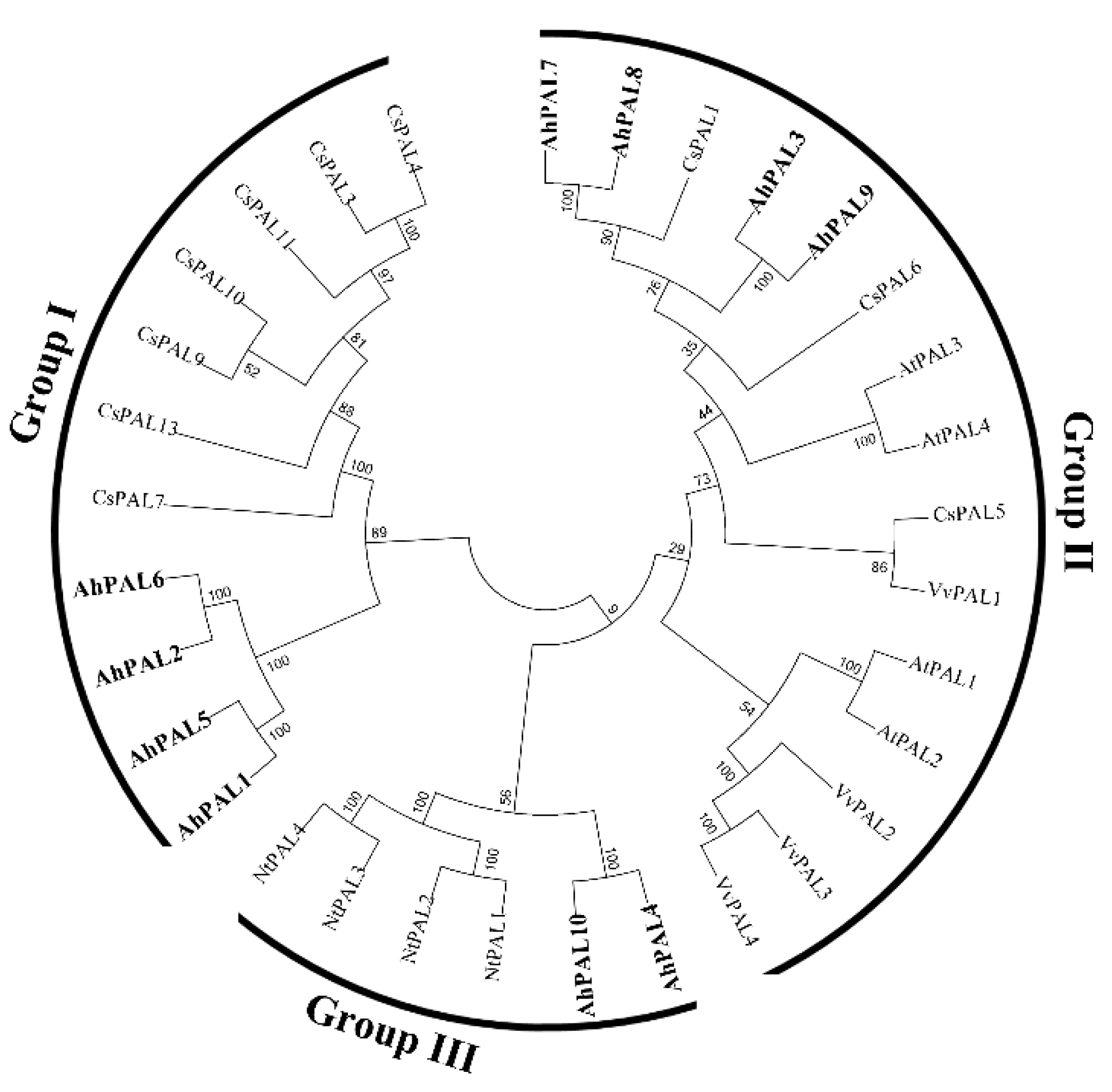
3.2. Sequence Characterization and Phylogenetic Analysis of PAL Family Proteins in Cultivated Peanut
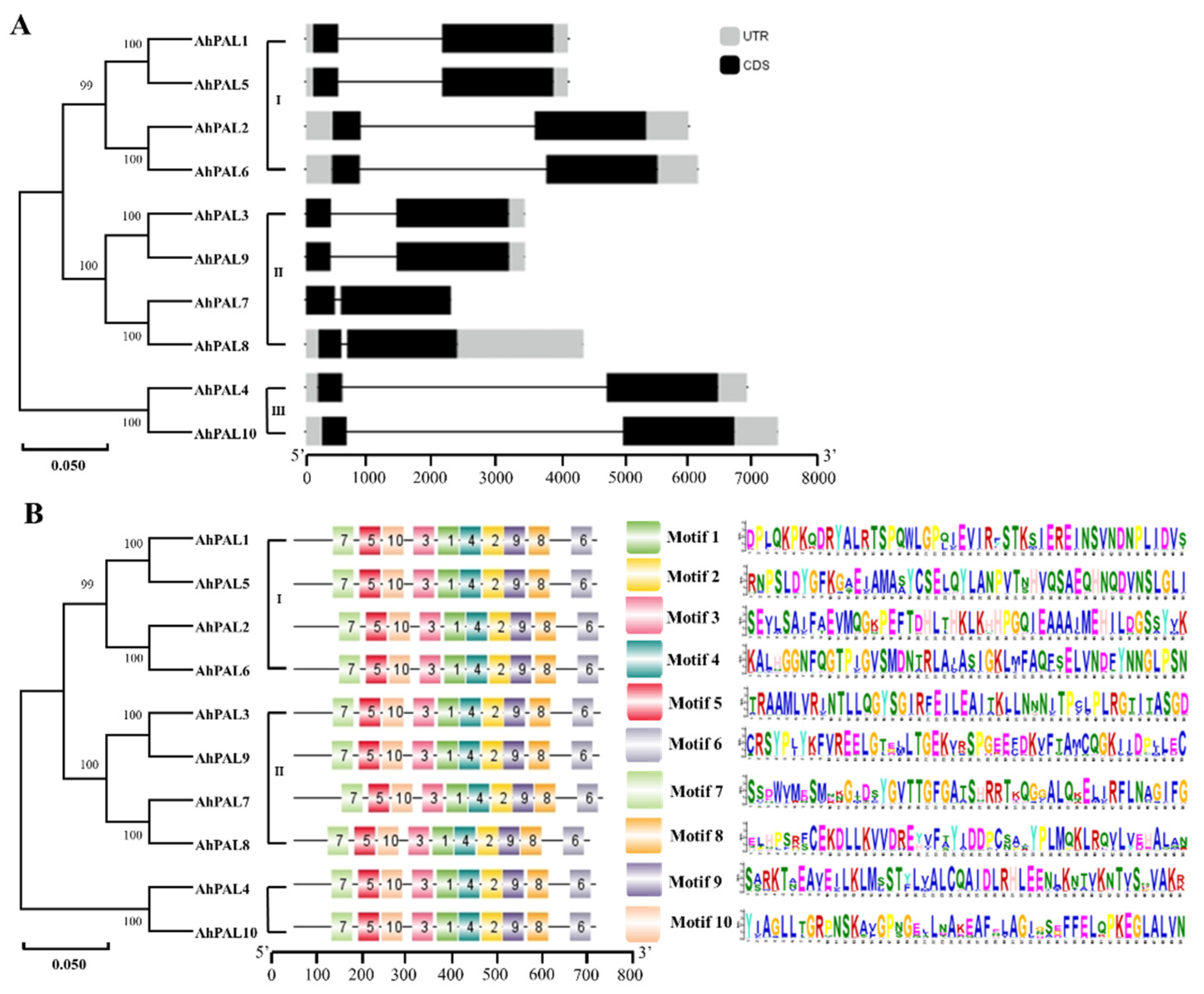
3.3. Exon–Intron and Conserved Motif Analysis of AhPALs
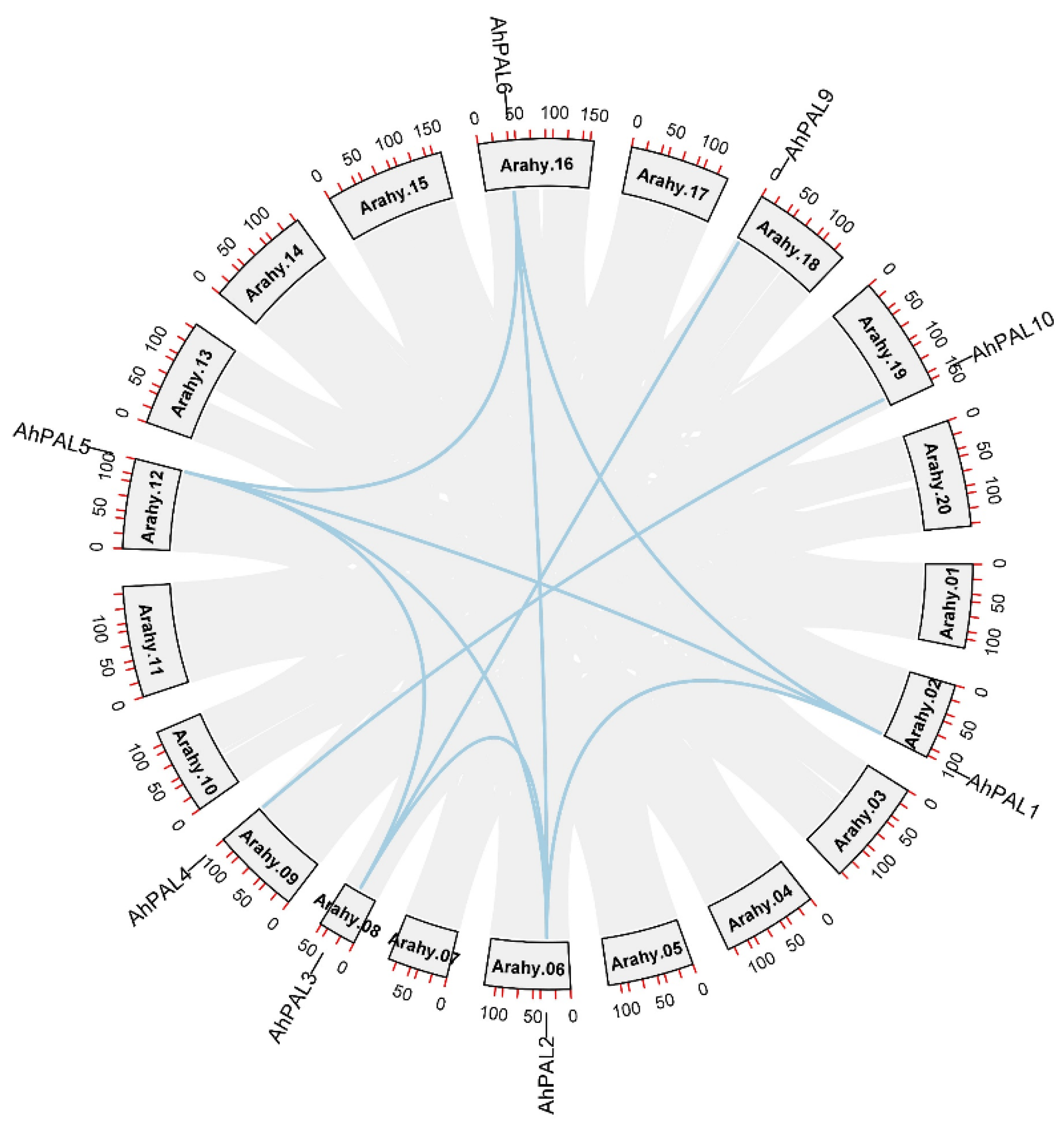
3.4. Duplication Events in AhPAL Genes and Synteny Analysis
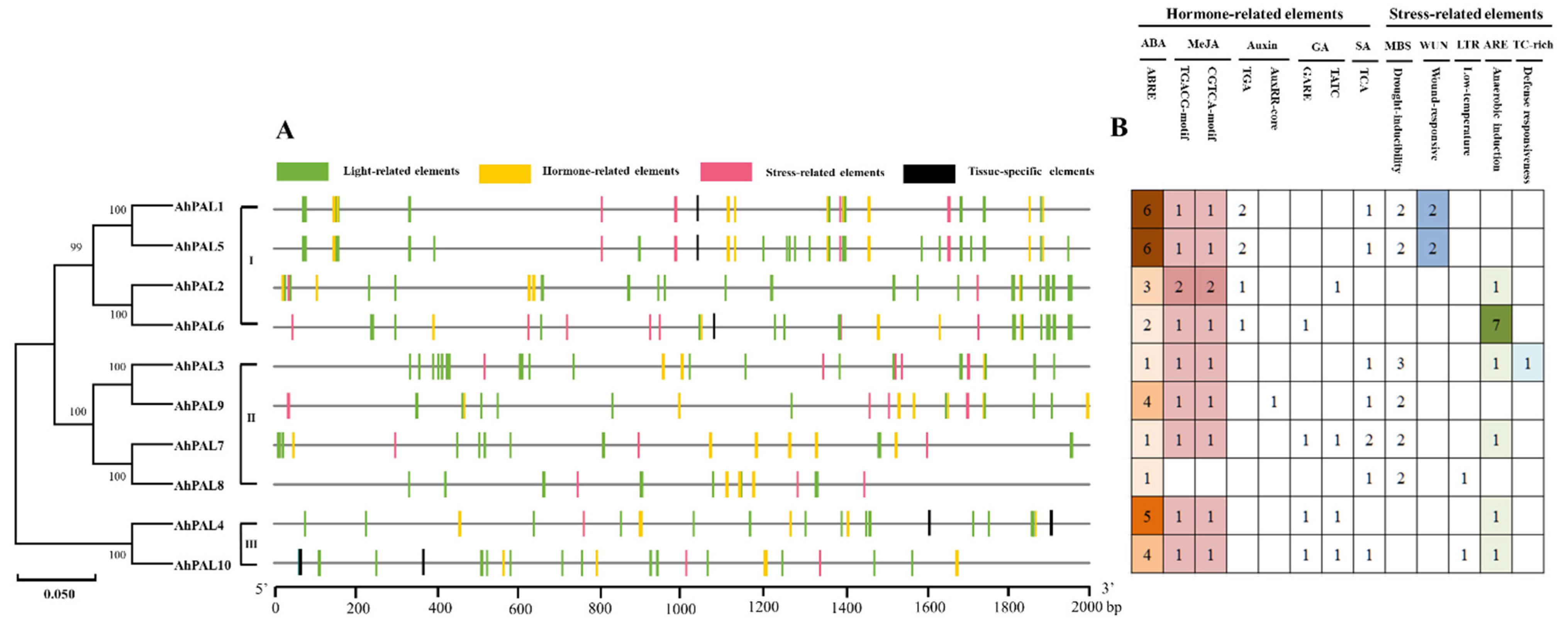
3.5. Cis-Acting Elements of AhPALs Promoter Region
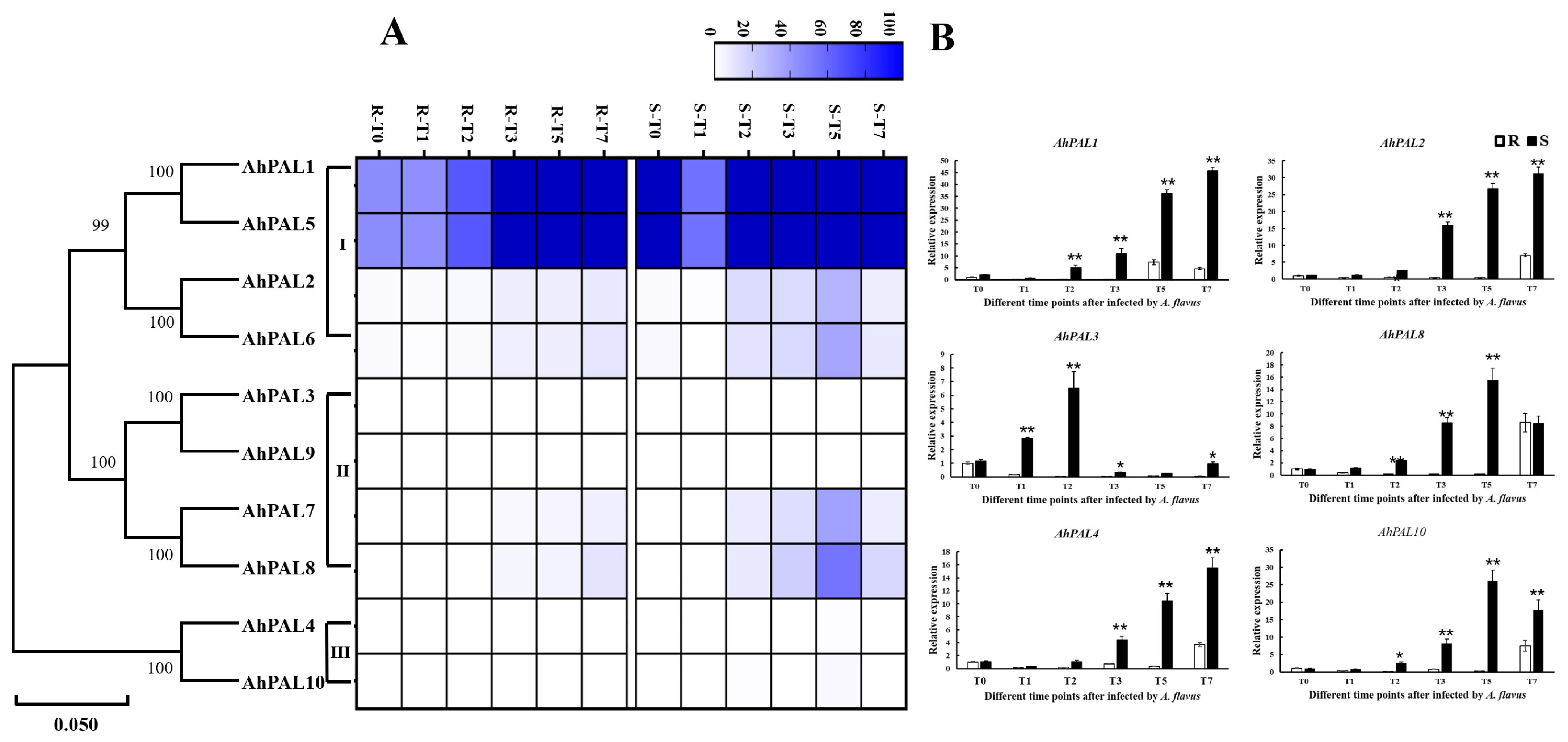
3.6. Expression Patterns of AhPAL Genes in Response to A. flavus Stress in Peanut Seeds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- MacDonald, M.J.; Godwin, B.D. A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 2007, 85, 273–282. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Huang, J.L.; Gu, M.; Lai, Z.B.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signaling of the defense response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef]
- Ferrer, J.L.; Austin, M.B.; Stewart, C.; Noel, J. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef]
- Xu, H.; Park, N.I.; Li, X.; Kim, Y.; Lee, S.; Park, S. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 2010, 101, 9715–9722. [Google Scholar] [CrossRef]
- Lepelley, M.; Mahesh, V.; McCarthy, J.; Rigoreau, M.; Crouzillat, D.; Chabrillange, N.; Kochko, A.D.; Campa, C. Characterization, high-resolution mapping and differential expression of three homologous PAL genes in Coffea canephora Pierre (Rubiaceae). Planta 2012, 236, 313–326. [Google Scholar] [CrossRef]
- Hamberger, B.; Ellis, M.; Friedmann, M.; Clarice, D.; Barbazuk, B.; Douglas, C. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can. J. Bot. 2007, 85, 1182–1201. [Google Scholar] [CrossRef]
- Shang, Q.M.; Li, L.; Dong, C.J. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 2012, 236, 1093–1105. [Google Scholar] [CrossRef]
- Dong, C.J.; Cao, N.; Zhang, Z.; Shang, Q. Phenylalanine ammonia-lyase gene families in cucurbit species: Structure, evolution, and expression. J. Integr. Agric. 2016, 15, 1239–1255. [Google Scholar] [CrossRef]
- Joos, H.J.; Hahlbrock, K. Phenylalanine ammonia-lyase in potato (Solanum tuberosum L.). Genomic complexity, structural comparison of two selected genes and modes of expression. Eur. J. Biochem. 1992, 204, 621–629. [Google Scholar] [CrossRef]
- Chang, A.; Lim, M.H.; Lee, S.W.; Robb, J.; Nazar, R. Tomato phenylalanine ammonia-lyase gene family, highly redundant but strongly underutilized. J. Biol. Chem. 2008, 283, 33591–33601. [Google Scholar] [CrossRef]
- Dong, C.J.; Shang, Q.M. Genome-wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta 2013, 238, 35–49. [Google Scholar] [CrossRef]
- Rawal, H.C.; Singh, N.K.; Sharma, T.R. Conservation, divergence, and genome-wide distribution of PAL and POX A gene families in plants. Int. J. Genom. 2013, 2013, 678969–678974. [Google Scholar]
- Kumar, A.; Ellis, B.E. The phenylalanine ammonia-lyase gene family in raspberry. Structure, expression, and evolution. Plant Physiol. 2001, 127, 230–239. [Google Scholar] [CrossRef]
- Hou, X.M.; Shao, F.J.; Ma, Y.M.; Lu, S. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: Genome-wide characterization, molecular cloning and expression analysis. Mol. Biol. Rep. 2013, 40, 4301–4310. [Google Scholar] [CrossRef]
- Olsen, K.M.; Lea, U.S.; Slimestad, R.; Verheul, M.; Lillo, C. Differential expression of four Arabidopsis PAL genes: PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008, 165, 1491–4199. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.W.; Dron, M.; Cramer, C.L.; Dixon, R.A.; Lamb, C. Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J. Biol. Chem. 1989, 264, 14486–14492. [Google Scholar] [CrossRef]
- Nandini, D.; Bariya, H. Induction of systemic acquired resistance in Arachis hypogaea L. by aspergillus flavus derived elicitors. J. Cell Tissue Res. 2014, 14, 4395–4403. [Google Scholar]
- Firozi, P.; Bhattacharya, R. Effects of natural polyphenols on aflatoxin B1 activation in a reconstituted microsomal monooxygenase system. J. Biochem. Toxicol. 1995, 10, 25–31. [Google Scholar]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal, B.; Soraya, C.M.; Ren, L.; Farmer, A.D.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Zhuang, W.J.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Holger, R.; Georg, E.S. Structural basis for the entrance into the phenylpropanoid metabolism catalyzed by phenylalanine ammonia-lyase. Plant Cell 2004, 16, 3426–3436. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protoc Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Zhu, X.; Leng, X.; Sun, X.; Mu, Q.; Wang, B.; Li, X.; Wang, C.; Fang, J. Discovery of conservation and diversification of miR171 genes by phylogenetic analysis based on global genomes. Plant Genome 2015, 8, plantgenome2014-10. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Cui, M.; Haider, M.S.; Chai, P.; Guo, J.; Du, P.; Li, H.; Dong, W.; Huang, B.; Zheng, Z.; Shi, L.; et al. Genome-wide identification and expression analysis of AP2/ERF transcription factor related to drought stress in cultivated peanut (Arachis hypogaea L.). Front. Genet. 2021, 12, 750761. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Magali, L.; Patrice, D.; Gert, T.; Kathleen, M.; Yves, M.; Yves, V.; Pierre, R.; Stephane, R. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar]
- Cui, M.; Han, S.; Wang, D.; Haider, M.S.; Guo, J.; Zhao, Q.; Du, P.; Sun, Z.; Qi, F.; Zheng, Z.; et al. Gene co-expression network analysis of the comparative transcriptome identifies hub genes associated with resistance to Aspergillus flavus L. in cultivated peanut (Arachis hypogaea L.). Front. Plant Sci. 2022, 13, 899177. [Google Scholar] [CrossRef] [PubMed]
- Cramer, C.L.; Edwards, K.; Dron, M.; Liang, X.; Dildine, S.L.; Bolwell, G.P.; Dixon, R.A.; Lamb, C.J.; Schuch, W. Phenylalanine ammonialyase gene organization and structure. Plant Mol. Biol. 1989, 12, 367–383. [Google Scholar] [CrossRef]
- Reichert, A.I.; He, X.Z.; Dixon, R.A. Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): Characterization of the four tobacco PAL genes and active heterotetrameric enzymes. Biochem. J. 2009, 424, 233–242. [Google Scholar] [CrossRef]
- Baloglu, M.C. Genome-wide in silico identification and comparison of growth regulating factor (GRF) genes in Cucurbitaceae family. Plant Omics 2014, 7, 260–270. [Google Scholar]
- Song, H.; Wang, P.; Lin, J.Y.; Zhao, C.; Bi, Y.; Wang, X. Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 2016, 26, 534. [Google Scholar] [CrossRef]
- Zhao, K.; Li, K.; Ning, L.; He, J.; Yin, D. Genome-wide analysis of the growth-regulating factor family in peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 2019, 2, 4120. [Google Scholar] [CrossRef]
- Wan, L.; Ren, W.; Miao, H.; Zhang, J.; Fang, J. Genome-wide identification, expression, and association analysis of the monosaccharide transporter (MST) gene family in peanut (Arachis hypogaea L.). 3 Biotech. 2020, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Chao, G.; Sun, J.; Wang, C.; Dong, Y.; Xiao, S.; Wang, X.; Jiao, Z.; Chen, Z.H. Genome-wide analysis of basic/helix-loophelix gene family in peanut and assessment of its roles in pod development. PLoS ONE 2017, 12, e0181843. [Google Scholar]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, C.; Zhang, W.; Pervaiz, T.; Haider, M.S.; Tang, W.; Fang, J. Characterization of Vv-miR156: Vv-SPL pairs involved in the modulation of grape berry development and ripening. Mol. Genet. Genom. 2018, 293, 1333–1354. [Google Scholar] [CrossRef]
- Meng, J.; Yang, J.; Peng, M.; Liu, X.; He, H. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Medicago truncatula. Life 2020, 10, 176–200. [Google Scholar] [CrossRef]
- Achnine, L.; Blancaflor, E.B.; Rasmussen, S.; Dixon, R. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 2004, 16, 3098–3109. [Google Scholar] [CrossRef]
- Jong, F.; Hanley, S.J.; Beale, M.H.; Karp, A. Characterization of the willow phenylalanine ammonia-lyase (PAL) gene family reveals expression differences compared with poplar. Phytochemistry 2015, 117, 90–97. [Google Scholar] [CrossRef]
- Yan, F.; Li, H.; Zhao, P. Genome-wide identification and transcriptional expression of the PAL gene family in common walnut (Juglans regia L.). Genes 2019, 10, 46. [Google Scholar] [CrossRef]
- Shi, R.; Sun, Y.H.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2010, 51, 144–163. [Google Scholar] [CrossRef]
- Wang, Z.B.; Chen, X.; Wang, W.; Cheng, K.; Kong, J. Transcriptome-wide identification and characterization of Ornithogalum saundersiae phenylalanine ammonia lyase gene family. RSC Adv. 2014, 4, 27159–27175. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Li, C.; Han, S.; Lopez-Baltazar, J.; Zhang, X.; Wang, X. Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 2016, 12, 35245. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.N.; Agarwal, G.; Pandey, M.K.; Sudini, H.; Jayale, A.; Purohit, S.; Desai, A.; Wan, L.; Guo, B.; Liao, B.; et al. Aspergillus flavus infection triggered immune responses and host-pathogen cross-talks in groundnut during in-vitro seed colonization. Sci. Rep. 2017, 29, 9659–9672. [Google Scholar] [CrossRef] [PubMed]
- Wanner, L.A.; Li, G.; Ware, D.; Somssich, I.; Davis, K. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995, 27, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Soni, P.; Nayak, S.N.; Kumar, R.; Pandey, M.; Singh, N.; Sudini, H.K.; Bajaj, P.; Fountain, J.C.; Singam, P.; Hong, Y.; et al. Transcriptome analysis identified coordinated control of key pathways regulating cellular physiology and metabolism upon Aspergillus flavus infection resulting in reduced aflatoxin production in groundnut. J. Fungi 2020, 6, 370–392. [Google Scholar] [CrossRef] [PubMed]
- Kabir, S.M.T.; Hossain, M.S.; Bashar, K.K.; Honi, U.; Ahmed, B.; Emdad, E.M. Genome-wide identification and expression profiling of AP2/ERF superfamily genes under stress conditions in dark jute (Corchorus olitorius L.). Ind. Crops Prod. 2021, 166, 113469–113487. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | Subcellular Location | Amino Acid (AA) | Molecular Weight (kDa) | Theoretical pI | Chromosome | Gene Position | |
|---|---|---|---|---|---|---|---|---|
| AhPAL1 | arahy. FI9C9D.1 | Cytoplasm | 714 | 77.94 | 5.96 | Arahy.02 | 102,530,759 | 102,534,874 |
| AhPAL2 | arahy. V1SQAY.1 | Cytoplasm | 729 | 79.3 | 6.21 | Arahy.06 | 32,131,529 | 32,137,537 |
| AhPAL3 | arahy. 9CK4GV.1 | Cytoplasm | 720 | 78.94 | 6.19 | Arahy.08 | 29,020,167 | 29,023,604 |
| AhPAL4 | arahy. 0B4MFB.1 | Cytoplasm | 711 | 77.56 | 6.21 | Arahy.09 | 118,976,212 | 118,983,138 |
| AhPAL5 | arahy. 5H4H17.1 | Cytoplasm | 714 | 77.94 | 5.96 | Arahy.12 | 120,128,684 | 120,132,799 |
| AhPAL6 | arahy. PUMP6L.1 | Cytoplasm | 729 | 79.29 | 6.15 | Arahy.16 | 42,030,770 | 42,036,927 |
| AhPAL7 | arahy. EEZ4Y8.1 | Cytoplasm | 728 | 79.7 | 6.68 | Arahy.17 | 389,497 | 391,773 |
| AhPAL8 | arahy. G9Z3VW.1 | Cytoplasm | 695 | 75.75 | 6.04 | Arahy.17 | 1,175,458 | 1,179,817 |
| AhPAL9 | arahy. U2YH27.1 | Cytoplasm | 718 | 78.55 | 6.11 | Arahy.18 | 4,683,011 | 4,686,452 |
| AhPAL10 | arahy. NB2RRK.1 | Cytoplasm | 711 | 77.79 | 5.93 | Arahy.19 | 146,304,646 | 146,312,059 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, P.; Cui, M.; Zhao, Q.; Chen, L.; Guo, T.; Guo, J.; Wu, C.; Du, P.; Liu, H.; Xu, J.; et al. Genome-Wide Characterization of the Phenylalanine Ammonia-Lyase Gene Family and Their Potential Roles in Response to Aspergillus flavus L. Infection in Cultivated Peanut (Arachis hypogaea L.). Genes 2024, 15, 265. https://doi.org/10.3390/genes15030265
Chai P, Cui M, Zhao Q, Chen L, Guo T, Guo J, Wu C, Du P, Liu H, Xu J, et al. Genome-Wide Characterization of the Phenylalanine Ammonia-Lyase Gene Family and Their Potential Roles in Response to Aspergillus flavus L. Infection in Cultivated Peanut (Arachis hypogaea L.). Genes. 2024; 15(3):265. https://doi.org/10.3390/genes15030265
Chicago/Turabian StyleChai, Pengpei, Mengjie Cui, Qi Zhao, Linjie Chen, Tengda Guo, Jingkun Guo, Chendi Wu, Pei Du, Hua Liu, Jing Xu, and et al. 2024. "Genome-Wide Characterization of the Phenylalanine Ammonia-Lyase Gene Family and Their Potential Roles in Response to Aspergillus flavus L. Infection in Cultivated Peanut (Arachis hypogaea L.)" Genes 15, no. 3: 265. https://doi.org/10.3390/genes15030265
APA StyleChai, P., Cui, M., Zhao, Q., Chen, L., Guo, T., Guo, J., Wu, C., Du, P., Liu, H., Xu, J., Zheng, Z., Huang, B., Dong, W., Han, S., & Zhang, X. (2024). Genome-Wide Characterization of the Phenylalanine Ammonia-Lyase Gene Family and Their Potential Roles in Response to Aspergillus flavus L. Infection in Cultivated Peanut (Arachis hypogaea L.). Genes, 15(3), 265. https://doi.org/10.3390/genes15030265





