Network Pharmacology Prediction and Molecular Docking-Based Strategy to Explore the Potential Mechanism of Gualou Xiebai Banxia Decoction against Myocardial Infarction
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the Active Components of Gualou Xiebai Banxia Decoction and Their Target Genes
2.2. Identification of Potential MI-Related Targets
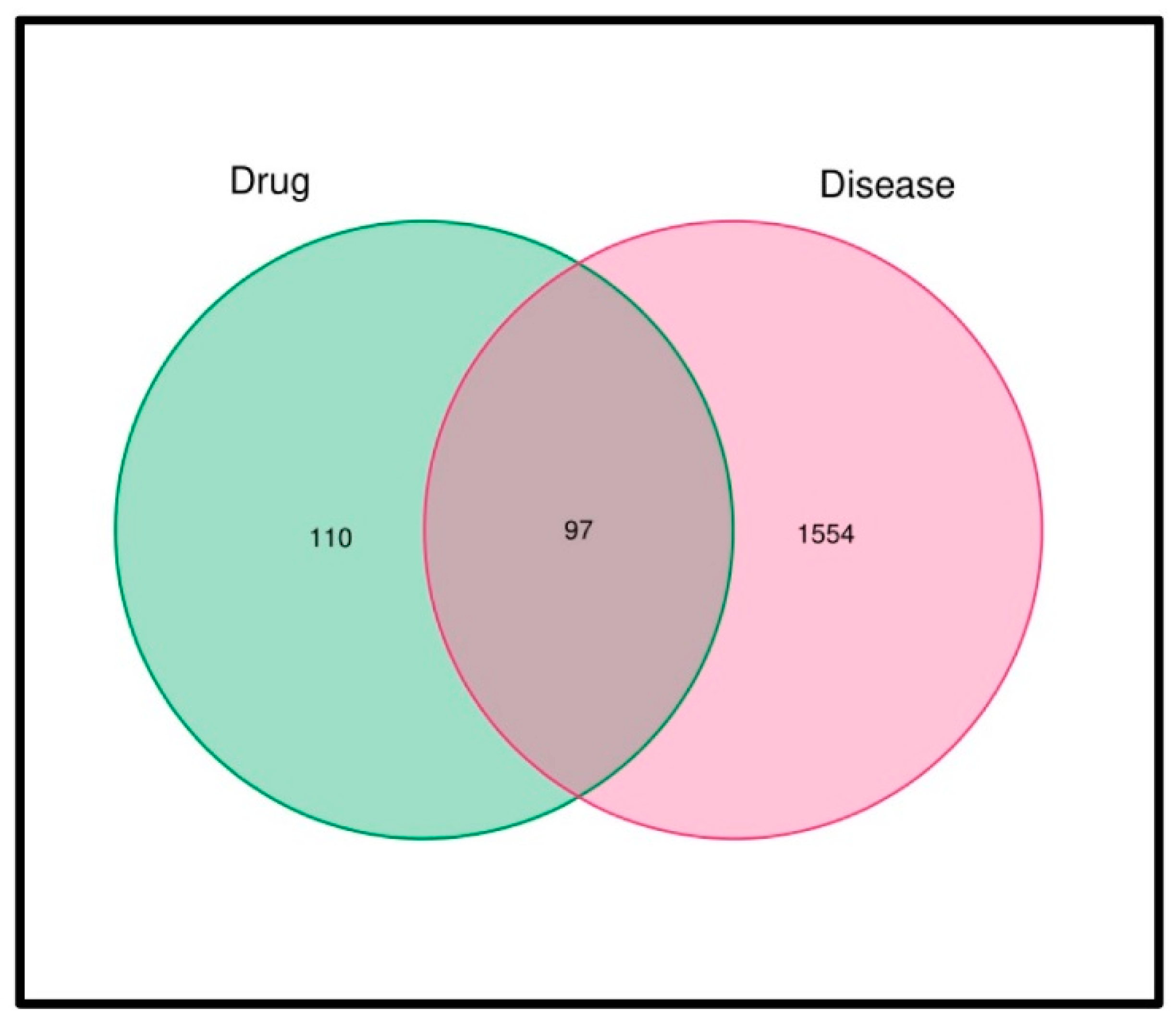
2.3. Identification of Intersections between Drug Targets and Disease-Related Genes
2.4. Construction of the Regulatory Network
2.5. Protein–Protein Interaction (PPI) Network Construction and Core Gene Screening
2.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analyses
2.7. Molecular Docking
3. Results
3.1. Identification of Active Components of Gualou Xiebai Banxia Decoction
3.2. Construction of an Active Component Library and MI Targets
3.3. Interactive Network of Targets
3.4. Regulatory Network of Active Components
3.5. GO and KEGG Pathway Enrichment Analyses
3.6. Target Pathway Analysis
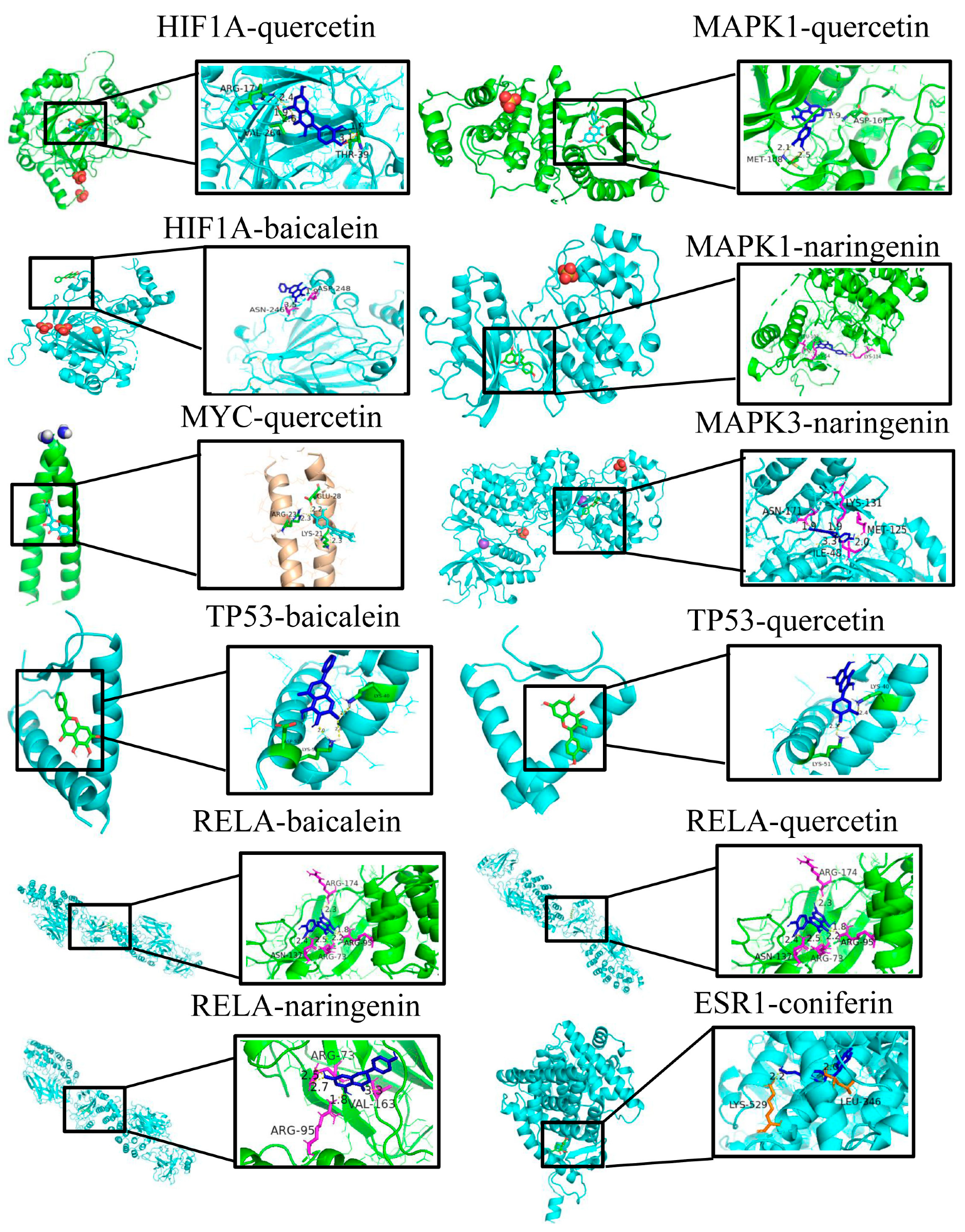
3.7. Molecular Docking
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MI | Myocardial infarction |
| TCM | Traditional Chinese medicine |
| CVDs | Cardiovascular diseases |
| AMI | Acute myocardial ischemia |
| T2DM-AMI | Type II diabetes with acute myocardial ischemia |
| OS | Oxidative stress |
| PI3K | Phosphatidyl inositol 3-kinase |
| AKT | Threonine protein kinase B |
| TCMSP | Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform |
| OB | Oral bioavailability |
| DL | Drug-likeness |
| PPI | Protein–protein interaction |
| BC | Betweenness centrality |
| CC | Closeness centrality |
| DC | Degree centrality |
| EC | Eigenvector centrality |
| LAC | Local average connectivity |
| NC | Network centrality |
| BP | Biological processes |
| CC | Cell components |
| MF | Molecular function |
| AGE-RAGE | Advanced glycation end-RAGE |
| IL-17 | Interleukin (IL)-17 |
| TNF | Tumor necrosis factor |
| HIF-1 | Hypoxia-inducible factor 1 |
| Jak | Janus kinases |
| mTOR | Mammalian/mechanistic target of rapamycin |
| MYC | c-Myc |
| TP53 | Wild-type (WT) human p53 |
| MAK1 | Mitogen-activated protein kinase 1 |
| MAK3 | Mitogen-activated protein kinase 3 |
| ESR1 | Estrogen receptor 1 |
| RELA | NFkappaB-p65 |
| NF-κB | NF-kappaB |
| VEGF | Vascular endothelial-derived growth factor |
References
- De, L.; Newby, L.; Mills, N. A Proposal for Modest Revision of the Definition of Type 1 and Type 2 Myocardial Infarction. Circulation 2019, 140, 1773–1775. [Google Scholar]
- Lu, L.; Liu, M.; Sun, R.; Zheng, Y.; Zhang, P. Myocardial Infarction: Symptoms and Treatments. Cell Biochem. Biophys. 2015, 72, 865–867. [Google Scholar] [CrossRef]
- Lisowska, A.; Makarewicz-Wujec, M.; Filipiak, K. Risk factors, prognosis, and secondary prevention of myocardial infarction in young adults in Poland. Kardiol. Pol. 2016, 74, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Blakeman, J.; Booker, K. Prodromal myocardial infarction symptoms experienced by women. Heart Lung 2016, 45, 327–335. [Google Scholar] [CrossRef]
- Chatterjee, N.; Levy, W. Sudden cardiac death after myocardial infarction. Eur. J. Heart Fail. 2020, 22, 856–858. [Google Scholar] [CrossRef]
- Parkash, R.; MacIntyre, C.; Dorian, P. Predicting Sudden Cardiac Death After Myocardial Infarction: A Great Unsolved Challenge. Circ. Arrhythm. Electrophysiol. 2021, 14, e009422. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Wan, S.; Gong, S.; Liu, J.; Li, F.; Kou, J. Recent advances of traditional Chinese medicine on the prevention and treatment of COVID-19. Chin. J. Nat. Med. 2020, 18, 881–889. [Google Scholar] [CrossRef]
- Luo, M.; Fan, R.; Wang, X.; Lu, J.; Li, P.; Chu, W.; Hu, Y.; Chen, X. Gualou Xiebai Banxia decoction ameliorates Poloxamer 407-induced hyperlipidemia. Biosci. Rep. 2021, 41, BSR20204216. [Google Scholar] [CrossRef]
- Lin, P.; Wang, Q.; Liu, Y.; Jiang, H.; Lv, W.; Lan, T.; Qin, Z.; Yao, X.; Yao, Z. Qualitative and quantitative analysis of the chemical profile for Gualou-Xiebai-Banxia decoction, a classical traditional Chinese medicine formula for the treatment of coronary heart disease, by UPLC-Q/TOF-MS combined with chemometric analysis. J. Pharm. Biomed. Anal. 2021, 197, 113950. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Peng, Y.; Shen, H.; Shu, L.; Wei, Y.J. Gualou Xiebai decoction inhibits cardiac dysfunction and inflammation in cardiac fibrosis rats. BMC Complement. Altern. Med. 2016, 16, 49. [Google Scholar] [CrossRef]
- Li, C.; Zhang, W.; Yu, Y.; Cheng, C.S.; Han, J.Y.; Yao, X.S.; Zhou, H. Discovery of the mechanisms and major bioactive compounds responsible for the protective effects of Gualou Xiebai Decoction on coronary heart disease by network pharmacology analysis. Phytomedicine 2019, 56, 261–268. [Google Scholar] [CrossRef]
- Ding, Y.; Peng, Y.; Li, J.; Shen, H.; Shen, M.Q.; Fang, T.H. Gualou Xiebai Decoction prevents myocardial fibrosis by blocking TGF-β/Smad signalling. J. Pharm. Pharmacol. 2013, 65, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zou, C.; Wei, M.; Gao, Z.Z.; Yang, Y. Pharmacodynamics Study on Gualou Xiebai Dropping Pills and Its Medicinal Ingredients in Prescription. Zhong Yao Cai 2015, 38, 567–571. [Google Scholar] [PubMed]
- Sun, F.; Huang, Y.; Li, L.; Yang, C.; Zhuang, P.; Zhang, Y. Fuzi and Banxia Combination, Eighteen Antagonisms in Chinese Medicine, Aggravates Adriamycin-Induced Cardiomyopathy Associated with PKA/beta2AR-Gs Signaling. Evid. Based Complement. Altern. Med. 2018, 2018, 2875873. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhou, J.; Sang, X.; Zhao, Q. Gualou-Xiebai-Banxia decoction protects against type II diabetes with acute myocardial ischemia by attenuating oxidative stress and apoptosis via PI3K/Akt/eNOS signaling. Chin. J. Nat. Med. 2021, 19, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Athanasios, A.; Charalampos, V.; Vasileios, T.; Md Ashraf, G. Protein-Protein Interaction (PPI) Network: Recent Advances in Drug Discovery. Curr. Drug Metab. 2017, 18, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, Z.; Zhang, L. Triptolide: Progress on research in pharmacodynamics and toxicology. J. Ethnopharmacol. 2014, 155, 67–79. [Google Scholar] [CrossRef]
- Ma, D.; Ma, G. Mechanism prediction of Simiao Yongan Decoction in treatment of psoriasis arthritis based on network pharmacology. Zhongguo Zhong Yao Za Zhi 2020, 45, 2611–2618. [Google Scholar]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Xu, L.; Liu, Z.; Yang, M.; Jiang, X.; Zhang, Q.; Huang, J. Utilising network pharmacology to explore the underlying mechanism of Wumei Pill in treating pancreatic neoplasms. BMC Complement. Altern. Med. 2019, 19, 158. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, W.; Huang, C.; Li, Y.; Yu, H.; Wang, Y.; Duan, J.; Ling, Y. A novel chemometric method for the prediction of human oral bioavailability. Int. J. Mol. Sci. 2012, 13, 6964–6982. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, W.; Hu, E.; Sun, Z.; Li, P.; Yu, Z.; Zhu, X.; Zheng, F.; Xing, Z.; Xia, Z.; et al. Integrated metabolomics and network pharmacology to reveal the mechanisms of hydroxysafflor yellow A against acute traumatic brain injury. Comput. Struct. Biotechnol. J. 2021, 19, 1002–1013. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shi, X.; Li, X.; Zhang, L.; Wu, P.; Mao, J.; Xing, R.; Zhang, N.; Wang, P. Network Pharmacology Approach to Uncover the Mechanism Governing the Effect of Simiao Powder on Knee Osteoarthritis. BioMed Res. Int. 2020, 2020, 6971503. [Google Scholar] [CrossRef] [PubMed]
- Vella, D.; Marini, S.; Vitali, F.; Di Silvestre, D.; Mauri, G.; Bellazzi, R. MTGO: PPI Network Analysis Via Topological and Functional Module Identification. Sci. Rep. 2018, 8, 5499. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.; Nastou, K.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Zhao, Q.; Jiang, C.; Gao, Q.; Zhang, Y.; Wang, G.; Chen, X.; Wu, S.; Tang, J. Gene expression and methylation profiles identified CXCL3 and CXCL8 as key genes for diagnosis and prognosis of colon adenocarcinoma. J. Cell Physiol. 2020, 235, 4902–4912. [Google Scholar] [CrossRef]
- Yu, H.; Hu, K.; Zhang, T.; Ren, H. Identification of Target Genes Related to Sulfasalazine in Triple-Negative Breast Cancer through Network Pharmacology. Med. Sci. Monit. 2020, 26, e926550. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; Huang, Y.; Ni, Y.-H.; Xu, Z.-M. Systematic elucidation of the mechanism of geraniol via network pharmacology. Drug Des. Dev. Ther. 2019, 13, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Chen, G.; Seukep, A.; Guo, M. Recent Advances in Molecular Docking for the Research and Discovery of Potential Marine Drugs. Mar. Drugs 2020, 18, 545. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, Y.; Lu, A.; Zhang, G. Systems Pharmacology in Small Molecular Drug Discovery. Int. J. Mol. Sci. 2016, 17, 246. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, X.; Li, S. An Integrative Platform of TCM Network Pharmacology and Its Application on a Herbal Formula, Qing-Luo-Yin. Evid. Based Complement. Altern. Med. 2013, 2013, 456747. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wang, J.; Liu, J.; Pei, M.; Huang, C.; Wang, Y. System-level multi-target drug discovery from natural products with applications to cardiovascular diseases. Mol. Divers. 2014, 18, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.; Chuang, M.; Liu, J.; Liu, X.Q.; Ho, L.K.; Pan, W.H.; Zhang, X.; Liu, C.; Tsai, S.; Kong, C.; et al. Baicalein protects against retinal ischemia by antioxidation, antiapoptosis, downregulation of HIF-1alpha, VEGF, and MMP-9 and upregulation of HO-1. J. Ocul. Pharmacol. Ther. 2013, 29, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, G.; Le, Y.; Ju, J.; Zhang, P.; Wan, D.; Zhao, Q.; Jin, G.; Su, H.; Liu, J.; et al. Quercetin promotes human epidermal stem cell proliferation through the estrogen receptor/β-catenin/c-Myc/cyclin A2 signaling pathway. Acta Biochim. Biophys. Sin. 2020, 52, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wei, F.; Liu, H.; Zhao, S.; Du, G.; Qin, X. Integrating hippocampal metabolomics and network pharmacology deciphers the antidepressant mechanisms of Xiaoyaosan. J. Ethnopharmacol. 2021, 268, 113549. [Google Scholar] [CrossRef] [PubMed]
- Kramarz, B.; Lovering, R. Gene Ontology: A Resource for Analysis and Interpretation of Alzheimer’s Disease Data. In Alzheimer’s Disease; Wisniewski, T., Ed.; Exon Publications: Brisbane, Australia, 2019. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, Y. Analysis of key GO terms and KEGG pathways associated with carcinogenic chemicals. Comb. Chem. High. Throughput Screen. 2017, 20, 861–871. [Google Scholar] [CrossRef]
- Lin, C.K.; Lin, Y.H.; Huang, T.C.; Shi, C.S.; Yang, C.T.; Yang, Y.L. VEGF mediates fat embolism-induced acute lung injury via VEGF receptor 2 and the MAPK cascade. Sci. Rep. 2019, 9, 11713. [Google Scholar] [CrossRef] [PubMed]
- Shao, L. Network Pharmacology Evaluation Method Guidance-Draft. World J. Tradit. Chin. Med. 2021, 7, 146–154. [Google Scholar]
- Cotter, K.A.; Rubin, M.A. Sequence of events in prostate cancer. Nature 2018, 560, 557–559. [Google Scholar] [CrossRef] [PubMed]






| Drug Name | Mol ID | Molecule Name | OB (%) | DL |
|---|---|---|---|---|
| Gualou | MoL001494 | Mandenol | 42 | 0.19 |
| Gualou | MoL002881 | Diosmetin | 31.14 | 0.27 |
| Gualou | MoL004355 | Spinasterol | 42.98 | 0.76 |
| Gualou | MoL005530 | Hydroxygenkwanin | 36.47 | 0.27 |
| Gualou | MoL 006756 | Schottenol | 37.42 | 0.75 |
| Gualou | MoL007156 | 10α-cucurbita-5,24-diene-3β-ol | 44.02 | 0.74 |
| Gualou | MoL007171 | 5-dehydrokarounidiol | 30.23 | 0.77 |
| Gualou | MoL007172 | 7-oxo-dihydrokaro-unidiol | 36.85 | 0.75 |
| Gualou | MoL007175 | karounidiol 3-o-benzoate | 43.99 | 0.5 |
| Gualou | MoL007179 | Linolenic acid ethyl ester | 46.1 | 0.2 |
| Gualou | MoL007180 | vitamin-e | 32.29 | 0.7 |
| Xiebai | MoL001973 | Sitosteryl acetate | 40.39 | 0.85 |
| Xiebai | MoL002341 | Hesperetin | 70.31 | 0.27 |
| Xiebai | MoL000332 | n-coumaroyltyramine | 85.63 | 0.2 |
| Xiebai | MoL000358 | β-sitosterol | 36.91 | 0.75 |
| Xiebai | MoL004328 | naringenin | 59.29 | 0.21 |
| Xiebai | MoL000483 | (Z)-3-(4-hydroxy-3-methoxy-phenyl)-N-[2-(4-hydroxyphenyl)ethyl]acrylamide | 118.35 | 0.26 |
| Xiebai | MoL000631 | coumaroyltyramine | 112.9 | 0.2 |
| Xiebai | MoL007640 | macrostemonoside e_qt | 35.26 | 0.87 |
| Xiebai | MoL007650 | PGA(sup 1) | 43.98 | 0.25 |
| Xiebai | MoL007651 | Prostaglandin B1 | 40.21 | 0.25 |
| Xiebai | MoL000098 | quercetin | 46.43 | 0.28 |
| Banxia | MoL001755 | 24-Ethylcholest-4-en-3-one | 36.08 | 0.76 |
| Banxia | MoL002670 | Cavidine | 35.64 | 0.81 |
| Banxia | MoL002714 | baicalein | 33.52 | 0.21 |
| Banxia | MoL002776 | Baicalin | 40.12 | 0.75 |
| Banxia | MoL000358 | β-sitosterol | 36.91 | 0.75 |
| Banxia | MoL000449 | Stigmasterol | 43.83 | 0.76 |
| Banxia | MoL005030 | gondoic acid | 30.7 | 0.2 |
| Banxia | MoL000519 | coniferin | 31.11 | 0.32 |
| Banxia | MoL006936 | 10,13-eicosadienoic | 39.99 | 0.2 |
| Banxia | MoL006937 | 12,13-epoxy-9-hydroxynonadeca-7,10-dienoic acid | 42.15 | 0.24 |
| Banxia | MoL006957 | (3S,6S)-3-(benzyl)-6-(4-hydroxybenzyl)piperazine-2,5-quinone | 46.89 | 0.27 |
| Banxia | MoL003678 | Cycloartenol | 38.69 | 0.78 |
| Banxia | MoL006967 | β-D-Ribofuranoside, xanthine-9 | 44.72 | 0.21 |
| Targets | Full Name |
|---|---|
| NR3C2 | nuclear receptor subfamily 3 group C member 2 |
| PTGS1 | prostaglandin-endoperoxide synthase 1 |
| CHRM3 | M3 muscarinic acetylcholine receptor |
| KCNH2 | K(+) voltage-gated channel subfamily H member 2 |
| CHRM1 | Epigenetic regulation of cholinergic receptor M1 |
| ADRB1 | adrenergic receptor β 1 |
| SCN5A | cardiac sodium channel |
| PTGS2 | Prostaglandin-Endoperoxide Synthase 2 |
| ADRA2C | α-2C-adrenergic receptor gene |
| CHRM4 | cholinergic receptor, muscarinic 4 |
| RXRA | retinoid X receptor protein |
| OPRD1 | opioid receptor delta 1 |
| ADRA1B | α-adrenergic receptor-1b |
| ADRB2 | beta2-adrenergic receptor |
| ADRA1D | α(1) adrenoreceptor subtype D |
| OPRM1 | opioid Receptor Mu 1 |
| DRD1 | dopamine receptors 1 |
| SLC6A4 | solute carrier family 6 member 4 |
| F7 | genes of factor VII |
| DPP4 | dipeptidyl peptidase-4 |
| RELA | NFkappaB-p65 |
| AKT1 | protein kinase B |
| VEGFA | vascular endothelial growth factor A |
| BCL2 | B-cell lymphoma 2 |
| MMP9 | gelatinase matrix metalloproteinase 9 |
| CASP3 | Caspase-3 |
| TP53 | tumor protein p53 |
| HIF1A | hypoxia-inducible factor 1alpha |
| CCNB1 | cyclin B1 |
| AHR | aryl hydrocarbon receptor |
| ADRA1A | α-1A-adrenoreceptor |
| CHRM2 | cholinergic muscarinic 2 receptor |
| PRKCA | protein kinase C α |
| PON1 | Paraoxonase-1 |
| ADH1C | Alcohol dehydrogenase 1C |
| IGHG1 | immunoglobulin heavy constant γ 1 |
| ADRA2A | adrenergic receptor α-2A |
| SLC6A2 | solute carrier family 6 member 2 |
| LTA4H | leukotriene A4 hydrolase |
| MAOA | Monoamine oxidase A |
| ESR1 | estrogen receptor 1 |
| PPARG | peroxisome proliferator-activated receptor γ |
| NOS2 | nitric oxide synthase |
| MAPK3 | mitogen-activated protein kinase 3 |
| MAPK1 | mitogen-activated protein kinase 1 |
| LDLR | Low-density lipoprotein receptor |
| SOD1 | superoxide dismutase 1 |
| MTTP | microsomal triglyceride transfer protein |
| APOB | apolipoprotein B |
| HMGCR | 3-Hydroxy-3-methylglutaryl coenzyme A reductase |
| CYP19A1 | cytochrome P4501A1 |
| UGT1A1 | uridine diphosphate glucuronosyltransferase 1A1 |
| GSR | galvanic skin response |
| ABCC1 | multidrug resistance protein 1 |
| SOAT2 | sterol O-acyltransferase 2 |
| AKR1C1 | Aldo-keto-reductases 1C1 |
| GOT1 | glutamate oxaloacetate transaminase 1 |
| CES1 | Carboxylesterase 1 |
| PKIA | Protein Kinase Inhibitor α |
| CCND1 | Cyclin D1 |
| MMP2 | matrix metalloproteinase 2 |
| EGF | epidermal growth factor |
| RB1 | retinoblastoma susceptibility gene |
| TNFAIP6 | TNFalpha-stimulated gene-6 |
| NFKBIA | nuclear factor-kappa-B inhibitor-α |
| TOP1 | topoisomerase I |
| MMP1 | matrix metallopeptidase 1 |
| STAT1 | signal transducer and activator of transcription 1 |
| HSPA5 | Heat shock 70 kDa protein 5 |
| ACACA | acetyl-CoA carboxylase α |
| HMOX1 | heme oxygenase 1 |
| CYP3A4 | cytochrome P450 3A4 |
| CYP1A2 | cytochrome P450 1A2 |
| CAV1 | Caveolin-1 |
| MYC | c-Myc |
| GJA1 | Gap Junction Protein α 1 |
| CYP1A1 | cytochrome P450 1A1 |
| ICAM1 | intercellular adhesion molecule 1 |
| SELE | E-selectin |
| VCAM1 | vascular cell adhesion molecule 1 |
| NOS3 | nitric oxide synthase 3 |
| HSPB1 | HSP β-1 |
| NR1I2 | nuclear receptor subfamily 1 group I member 2 |
| SERPINE1 | serine protease inhibitor clade E member 1 |
| IFNG | interferon γ |
| ALOX5 | arachidonate 5-lipoxygenase |
| NCF1 | neutrophil cytosolic factor 1 |
| ABCG2 | ATP-binding cassette (ABC) superfamily G member 2 |
| SLC2A4 | solute carrier family 2 member 4 |
| COL3A1 | Collagen type III α 1 |
| CXCL2 | CXC motif chemokine ligand 2 |
| NR1I3 | constitutive androstane receptor |
| INSR | insulin receptors |
| PPARD | peroxisome proliferator-activated receptor β/delta |
| CHUK | conserved helix-loop-helix ubiquitous kinase |
| SPP1 | secreted phosphoprotein 1 |
| Target Gene Symbol | BC | CC | DC | EC | LAC | NC |
|---|---|---|---|---|---|---|
| ESR1 | 0 | 1 | 6 | 0.377964467 | 5 | 6 |
| TP53 | 0 | 1 | 6 | 0.377964616 | 5 | 6 |
| MYC | 0 | 1 | 6 | 0.377964497 | 5 | 6 |
| MAPK1 | 0 | 1 | 6 | 0.377964497 | 5 | 6 |
| RELA | 0 | 1 | 6 | 0.377964497 | 5 | 6 |
| HIF1A | 0 | 1 | 6 | 0.377964497 | 5 | 6 |
| MAPK3 | 0 | 1 | 6 | 0.377964497 | 5 | 6 |
| Molecule Name | Molecular Formula | Structure |
|---|---|---|
| Baicalein | C15H10O5 | 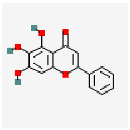 |
| Quercetin | C15H10O7 | 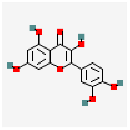 |
| Naringenin | C15H12O5 |  |
| Coniferin | C16H22O8 |  |
| Baicalein | Quercetin | Naringenin | Coniferin | |
|---|---|---|---|---|
| HIF1A docking score (kcal/mol) | −4.7 | −7.4 | \ | \ |
| MYC docking score (kcal/mol) | \ | −5.6 | \ | \ |
| TP53 docking score (kcal/mol) | −6.3 | −6.3 | \ | \ |
| MAK1 docking score (kcal/mol) | \ | −8.5 | −8.4 | \ |
| MAK3 docking score (kcal/mol) | \ | \ | −8.7 | \ |
| RELA docking score (kcal/mol) | −6.9 | −7.7 | −7.4 | \ |
| ESR1 docking score (kcal/mol) | \ | \ | \ | −7.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-L.; Chen, Y. Network Pharmacology Prediction and Molecular Docking-Based Strategy to Explore the Potential Mechanism of Gualou Xiebai Banxia Decoction against Myocardial Infarction. Genes 2024, 15, 392. https://doi.org/10.3390/genes15040392
Wang W-L, Chen Y. Network Pharmacology Prediction and Molecular Docking-Based Strategy to Explore the Potential Mechanism of Gualou Xiebai Banxia Decoction against Myocardial Infarction. Genes. 2024; 15(4):392. https://doi.org/10.3390/genes15040392
Chicago/Turabian StyleWang, Wei-Lu, and Yan Chen. 2024. "Network Pharmacology Prediction and Molecular Docking-Based Strategy to Explore the Potential Mechanism of Gualou Xiebai Banxia Decoction against Myocardial Infarction" Genes 15, no. 4: 392. https://doi.org/10.3390/genes15040392
APA StyleWang, W.-L., & Chen, Y. (2024). Network Pharmacology Prediction and Molecular Docking-Based Strategy to Explore the Potential Mechanism of Gualou Xiebai Banxia Decoction against Myocardial Infarction. Genes, 15(4), 392. https://doi.org/10.3390/genes15040392





