Genomic Prediction from Multi-Environment Trials of Wheat Breeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phenotypic Data
2.2. Statistical Models
2.3. Cross-Validation Schemes
3. Results
3.1. Descriptive Yield Statistics in the TPEs
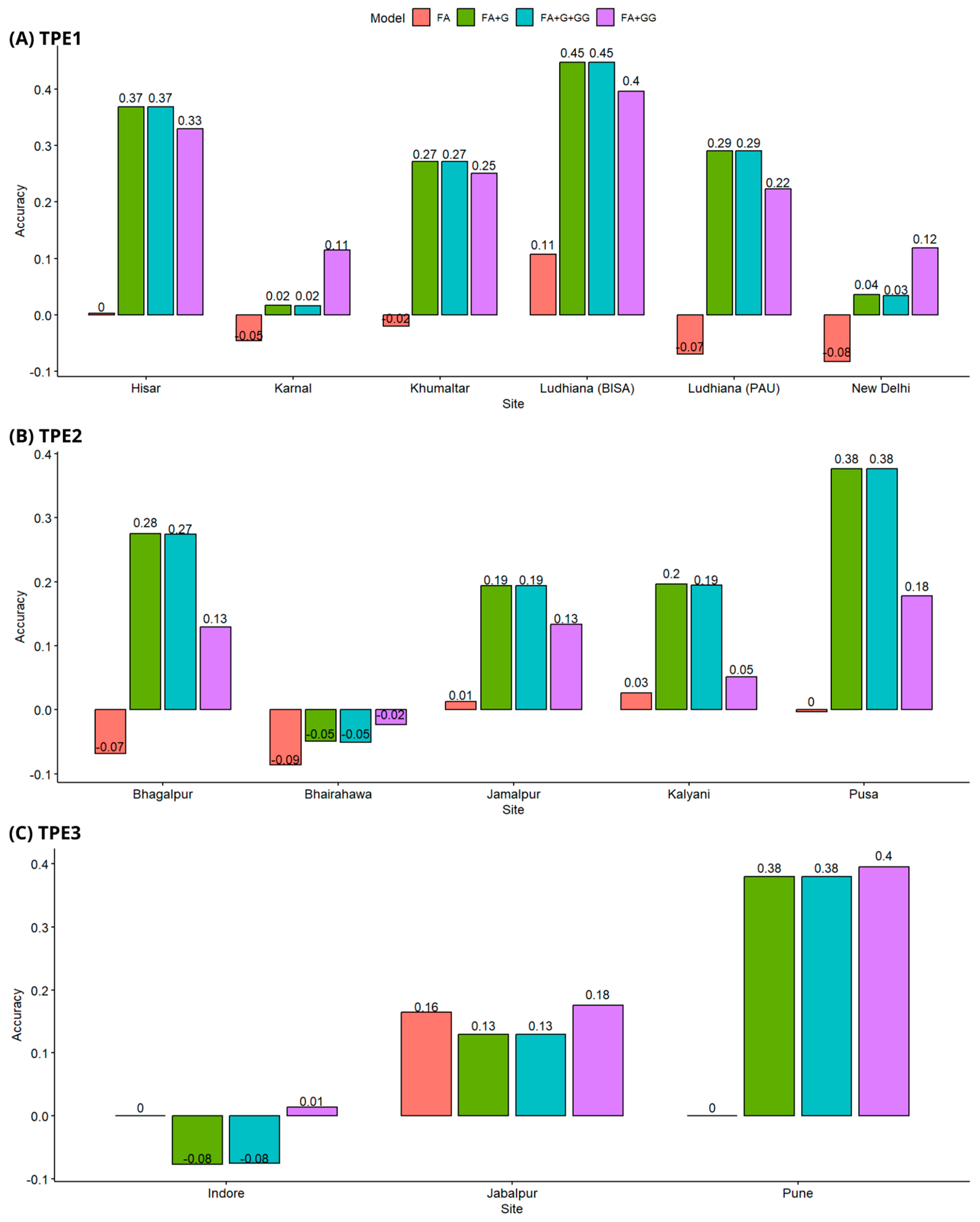
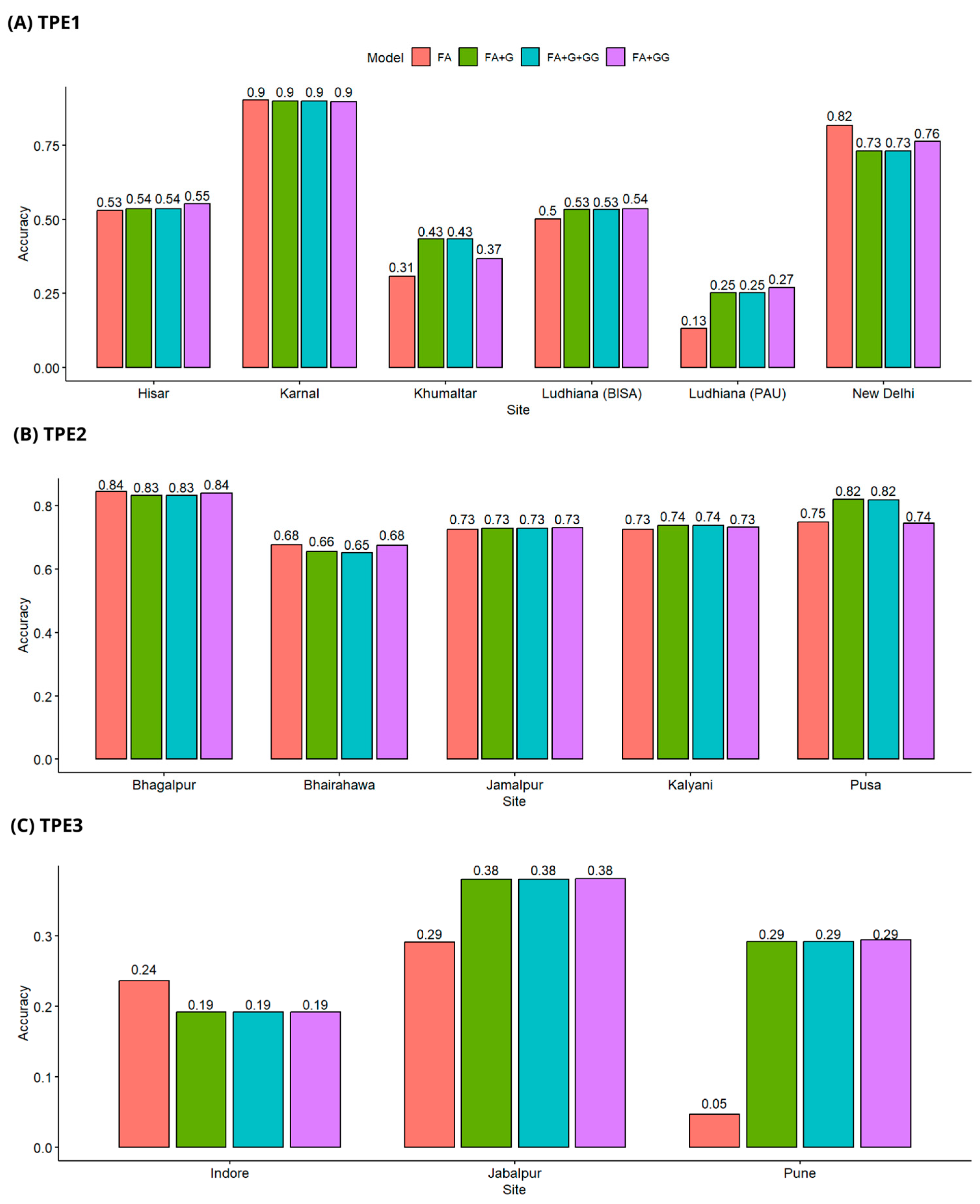
3.2. Prediction of the Breeding Value of Non-Evaluated Genotypes (Cross-Validation CV1)
3.3. Prediction of Breeding Value in Genotypes Evaluated in Some Environments, but Not in All (Cross-Validation CV2)
3.4. Predicting the Breeding Value Using 90% of Genotypes for Training (Cross-Validation CV3)
4. Discussion
4.1. Including Epistasis in the Genomic Prediction Models of Bread Wheat Does Not Increase Prediction Accuracy
4.2. Cross-Validation Schemes
4.3. Advantages of the Factor Analytic Model for Studying Genotype × Environment Interaction in the Context of Genomic Prediction
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Alves, K.; Brito, L.F.; Baes, C.F.; Sargolzaei, M.; Robinson, J.A.B.; Schenkel, F.S. Estimation of Additive and Non-Additive Genetic Effects for Fertility and Reproduction Traits in North American Holstein Cattle Using Genomic Information. J. Anim. Breed. Genet. 2020, 137, 316–330. [Google Scholar] [CrossRef]
- Su, G.; Christensen, O.F.; Ostersen, T.; Henryon, M.; Lund, M.S. Estimating Additive and Non-Additive Genetic Variances and Predicting Genetic Merits Using Genome-Wide Dense Single Nucleotide Polymorphism Markers. PLoS ONE 2012, 7, e45293. [Google Scholar] [CrossRef]
- Mackay, T.F.C. Epistasis and Quantitative Traits: Using Model Organisms to Study Gene–Gene Interactions. Nat. Rev. Genet. 2014, 15, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S.; Mackay, T. Introduction to Quantitative Genetics, 4th ed.; Pearson, Prentice Hall: Harlow, UK, 1996. [Google Scholar]
- Raffo, M.A.; Sarup, P.; Guo, X.; Liu, H.; Andersen, J.R.; Orabi, J.; Jahoor, A.; Jensen, J. Improvement of Genomic Prediction in Advanced Wheat Breeding Lines by Including Additive-by-Additive Epistasis. Theor. Appl. Genet. 2022, 135, 965–978. [Google Scholar] [CrossRef] [PubMed]
- Vojgani, E.; Hölker, A.C.; Mayer, M.; Schön, C.-C.; Simianer, H.; Pook, T. Genomic Prediction Using Information across Years with Epistatic Models and Dimension Reduction via Haplotype Blocks. PLoS ONE 2023, 18, e0282288. [Google Scholar] [CrossRef] [PubMed]
- Martini, J.W.R.; Wimmer, V.; Erbe, M.; Simianer, H. Epistasis and Covariance: How Gene Interaction Translates into Genomic Relationship. Theor. Appl. Genet. 2016, 129, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, P.S.; Sarup, P.; Fé, D.; Orabi, J.; Snell, P.; Ripa, L.; Mohlfeld, M.; Chu, T.T.; Herrström, J.; Jahoor, A.; et al. Prediction of Additive, Epistatic, and Dominance Effects Using Models Accounting for Incomplete Inbreeding in Parental Lines of Hybrid Rye and Sugar Beet. Front. Plant. Sci. 2023, 14, 1193433. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B.; Carley, C.A.S.; Bethke, P.C.; Coombs, J.J.; Clough, M.E.; da Silva, W.L.; De Jong, W.S.; Douches, D.S.; Frederick, C.M.; Haynes, K.G.; et al. Genetic Variance Partitioning and Genome-Wide Prediction with Allele Dosage Information in Autotetraploid Potato. Genetics 2018, 209, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, P.R.; Resende, M.F.R.; Gezan, S.A.; Resende, M.D.V.; de los Campos, G.; Kirst, M.; Huber, D.; Peter, G.F. Unraveling Additive from Nonadditive Effects Using Genomic Relationship Matrices. Genetics 2014, 198, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Xie, Y.; Zhang, Y.; Wang, R.; Sun, X. Genomic Dissection of Additive and Non-Additive Genetic Effects and Genomic Prediction in an Open-Pollinated Family Test of Japanese Larch. BMC Genom. 2024, 25, 11. [Google Scholar] [CrossRef] [PubMed]
- Flury, C.; Täubert, H.; Simianer, H. Extension of the Concept of Kinship, Relationship, and Inbreeding to Account for Linked Epistatic Complexes. Livest. Sci. 2006, 103, 131–140. [Google Scholar] [CrossRef]
- Abed, A.; Pérez-Rodríguez, P.; Crossa, J.; Belzile, F. When Less Can Be Better: How Can We Make Genomic Selection More Cost-Effective and Accurate in Barley? Theor. Appl. Genet. 2018, 131, 1873–1890. [Google Scholar] [CrossRef] [PubMed]
- Lorenzana, R.E.; Bernardo, R. Accuracy of Genotypic Value Predictions for Marker-Based Selection in Biparental Plant Populations. Theor. Appl. Genet. 2009, 120, 151–161. [Google Scholar] [CrossRef]
- Wang, D.; Salah El-Basyoni, I.; Stephen Baenziger, P.; Crossa, J.; Eskridge, K.M.; Dweikat, I. Prediction of Genetic Values of Quantitative Traits with Epistatic Effects in Plant Breeding Populations. Heredity 2012, 109, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Montesinos-López, O.A.; Pérez-Rodríguez, P.; Costa-Neto, G.; Fritsche-Neto, R.; Ortiz, R.; Martini, J.W.R.; Lillemo, M.; Montesinos-López, A.; Jarquin, D.; et al. Genome and Environment Based Prediction Models and Methods of Complex Traits Incorporating Genotype × Environment Interaction. In Genomic Prediction of Complex Traits: Methods and Protocols; Ahmadi, N., Bartholomé, J., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; pp. 245–283. [Google Scholar] [CrossRef]
- Schrauf, M.F.; Martini, J.W.R.; Simianer, H.; de los Campos, G.; Cantet, R.; Freudenthal, J.; Korte, A.; Munilla, S. Phantom Epistasis in Genomic Selection: On the Predictive Ability of Epistatic Models. G3 Genes Genomes Genet. 2020, 10, 3137–3145. [Google Scholar] [CrossRef] [PubMed]
- Braun, H.J.; Atlin, G.; Payne, T. Multi-Location Testing as a Tool to Identify Plant Response to Global Climate Change. In Climate Change and Crop Production; Reynolds, P.M., Ed.; CABI Climate Change Series; CAB International: Wallingford, UK, 2010; pp. 115–138. [Google Scholar]
- Crespo-Herrera, L.A.; Crossa, J.; Huerta-Espino, J.; Mondal, S.; Velu, G.; Juliana, P.; Vargas, M.; Pérez-Rodríguez, P.; Joshi, A.K.; Braun, H.J.; et al. Target Population of Environments for Wheat Breeding in India: Definition, Prediction and Genetic Gains. Front. Plant Sci. 2021, 12, 638520. [Google Scholar] [CrossRef] [PubMed]
- Coast, O.; Posch, B.C.; Rognoni, B.G.; Bramley, H.; Gaju, O.; Mackenzie, J.; Pickles, C.; Kelly, A.M.; Lu, M.; Ruan, Y.-L.; et al. Wheat Photosystem II Heat Tolerance: Evidence for Genotype-by-Environment Interactions. Plant J. 2022, 111, 1368–1382. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.M.; Smith, A.B.; Eccleston, J.A.; Cullis, B.R. The Accuracy of Varietal Selection Using Factor Analytic Models for Multi-Environment Plant Breeding Trials. Crop. Sci. 2007, 47, 1063–1070. [Google Scholar] [CrossRef]
- Meyer, K. Factor-Analytic Models for Genotype × Environment Type Problems and Structured Covariance Matrices. Genet. Sel. Evol. 2009, 41, 21. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, I.C.M.; Guilhen, J.H.S.; Ribeiro, P.C.d.O.; Gezan, S.A.; Schaffert, R.E.; Simeone, M.L.F.; Damasceno, C.M.B.; Carneiro, J.E.d.S.; Carneiro, P.C.S.; Parrella, R.A.d.C.; et al. Genotype-by-Environment Interaction and Yield Stability Analysis of Biomass Sorghum Hybrids Using Factor Analytic Models and Environmental Covariates. Field Crop. Res. 2020, 257, 107929. [Google Scholar] [CrossRef]
- Pietragalla, J.; Pask, A. Chapter 18. Grain Yield and Yield Components. In Physiological Breeding II: A Field Guide to Wheat Phenotyping; Pask, A., Pietragalla, J., Mullan, D., Chávez, P., Reynolds, M.P., Eds.; CIMMYT: México-Veracruz, México, 2013; pp. 95–103. [Google Scholar]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.-L. Development of High-Density Genetic Maps for Barley and Wheat Using a Novel Two-Enzyme Genotyping-by-Sequencing Approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Crossa, J.; Burgueño, J.; Cornelius, P.L.; McLaren, G.; Trethowan, R.; Krishnamachari, A. Modeling Genotype × Environment Interaction Using Additive Genetic Covariances of Relatives for Predicting Breeding Values of Wheat Genotypes. Crop. Sci. 2006, 46, 1722–1733. [Google Scholar] [CrossRef]
- Smith, A.; Cullis, B.; Thompson, R. Analyzing Variety by Environment Data Using Multiplicative Mixed Models and Adjustments for Spatial Field Trend. Biometrics 2001, 57, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias-Pazaran, G. Genome-Assisted Prediction of Quantitative Traits Using the R Package Sommer. PLoS ONE 2016, 11, e0156744. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. 2023. Available online: https://www.R-project.org/ (accessed on 15 December 2023).
- Gilmour, A.R.; Gogel, B.J.; Cullis, B.R.; Welham, S.J.; Thompson, R. ASReml User Guide Release 4.1; VSN International Ltd.: Hemel Hempstead, UK, 2015; Volume Release 4.1. [Google Scholar]
- Kassambara, A. Ggpubr: “ggplot2” Based Publication Ready Plots. 2023. Available online: https://rpkgs.datanovia.com/ggpubr/#ggpubr-ggplot2-based-publication-ready-plots (accessed on 15 December 2023).
- Duenk, P.; Bijma, P.; Calus, M.P.L.; Wientjes, Y.C.J.; van der Werf, J.H.J. The Impact of Non-Additive Effects on the Genetic Correlation between Populations. G3 Genes Genomes Genet. 2020, 10, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Goddard, M.E.; Visscher, P.M. Data and Theory Point to Mainly Additive Genetic Variance for Complex Traits. PLoS Genet. 2008, 4, e1000008. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.B. Epistasis and Plant Breeding. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001; Volume 21, pp. 27–92. [Google Scholar] [CrossRef]
- Wientjes, Y.C.J.; Bijma, P.; Calus, M.P.L.; Zwaan, B.J.; Vitezica, Z.G.; van den Heuvel, J. The Long-Term Effects of Genomic Selection: 1. Response to Selection, Additive Genetic Variance, and Genetic Architecture. Genet. Sel. Evol. 2022, 54, 19. [Google Scholar] [CrossRef] [PubMed]
- McGaugh, S.E.; Lorenz, A.J.; Flagel, L.E. The Utility of Genomic Prediction Models in Evolutionary Genetics. Proc. R. Soc. B 2021, 288, 20210693. [Google Scholar] [CrossRef] [PubMed]

| Cross-Validation Schemes | Set | Ludhiana (BISA) | Ludhiana (PAU) | Khumaltar | Karnal | New Delhi | Hisar |
|---|---|---|---|---|---|---|---|
| CV1 | Training | 130 | 130 | 130 | 130 | 130 | 130 |
| Testing | 32 | 32 | 32 | 32 | 32 | 32 | |
| CV2 | Training | 130 | 162 | 162 | 130 | 162 | 130 |
| Testing | 32 | 0 | 0 | 32 | 0 | 32 | |
| CV3 | Training | 20 | 162 | 162 | 162 | 162 | 162 |
| Testing | 142 | 0 | 0 | 0 | 0 | 0 |
| Ludhiana (BISA) | Ludhiana (PAU) | Khumaltar | Karnal | New Delhi | Hisar | |
|---|---|---|---|---|---|---|
| Ludhiana (BISA) | 1 | |||||
| Ludhiana (PAU) | 0.12 | 1 | ||||
| Khumaltar | 0.20 | 0.00 | 1 | |||
| Karnal | 0.59 | 0.14 | 0.21 | 1 | ||
| New Delhi | 0.55 | 0.18 | 0.23 | 0.90 | 1 | |
| Hisar | 0.35 | 0.18 | 0.48 | 0.42 | 0.48 | 1 |
| Bhagalpur | Jamalpur | Pusa | Bhairahawa | Kalyani | |
|---|---|---|---|---|---|
| Bhagalpur | 1 | ||||
| Jamalpur | 0.69 | 1 | |||
| Pusa | 0.63 | 0.64 | 1 | ||
| Bhairahawa | 0.57 | 0.59 | 0.50 | 1 | |
| Kalyani | 0.69 | 0.64 | 0.60 | 0.62 | 1 |
| Pune | Indore | Jabalpur | |
|---|---|---|---|
| Pune | 1 | ||
| Indore | 0.09 | 1 | |
| Jabalpur | −0.04 | 0.24 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Barrios, G.; Crespo-Herrera, L.; Cruz-Izquierdo, S.; Vitale, P.; Sandoval-Islas, J.S.; Gerard, G.S.; Aguilar-Rincón, V.H.; Corona-Torres, T.; Crossa, J.; Pacheco-Gil, R.A. Genomic Prediction from Multi-Environment Trials of Wheat Breeding. Genes 2024, 15, 417. https://doi.org/10.3390/genes15040417
García-Barrios G, Crespo-Herrera L, Cruz-Izquierdo S, Vitale P, Sandoval-Islas JS, Gerard GS, Aguilar-Rincón VH, Corona-Torres T, Crossa J, Pacheco-Gil RA. Genomic Prediction from Multi-Environment Trials of Wheat Breeding. Genes. 2024; 15(4):417. https://doi.org/10.3390/genes15040417
Chicago/Turabian StyleGarcía-Barrios, Guillermo, Leonardo Crespo-Herrera, Serafín Cruz-Izquierdo, Paolo Vitale, José Sergio Sandoval-Islas, Guillermo Sebastián Gerard, Víctor Heber Aguilar-Rincón, Tarsicio Corona-Torres, José Crossa, and Rosa Angela Pacheco-Gil. 2024. "Genomic Prediction from Multi-Environment Trials of Wheat Breeding" Genes 15, no. 4: 417. https://doi.org/10.3390/genes15040417
APA StyleGarcía-Barrios, G., Crespo-Herrera, L., Cruz-Izquierdo, S., Vitale, P., Sandoval-Islas, J. S., Gerard, G. S., Aguilar-Rincón, V. H., Corona-Torres, T., Crossa, J., & Pacheco-Gil, R. A. (2024). Genomic Prediction from Multi-Environment Trials of Wheat Breeding. Genes, 15(4), 417. https://doi.org/10.3390/genes15040417







