Novel Synergistic Probiotic Intervention: Transcriptomic and Metabolomic Analysis Reveals Ameliorative Effects on Immunity, Gut Barrier, and Metabolism of Mice during Salmonella typhimurium Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Experimental Design, and Treatment
2.2. Sample Collection and Transcriptome Data Analysis
2.2.1. cDNA Library Construction of Transcriptome Samples
2.2.2. Raw Data Processing and Screening of Differentially Expressed Genes
2.2.3. Analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways in Genes Exhibiting Differential Expression
2.3. DEGs’ Validation by qRT-PCR
2.4. Metabolome Data Processing and Analysis
2.4.1. LC-MS Detection of Differential Metabolites and KEGG Pathways’ Analysis
2.4.2. Orthogonal Projection to Latent Structures Discriminant Analysis (OPLS-DA) and Principal Component Analysis
3. Results
3.1. Transcriptomic Data Analysis
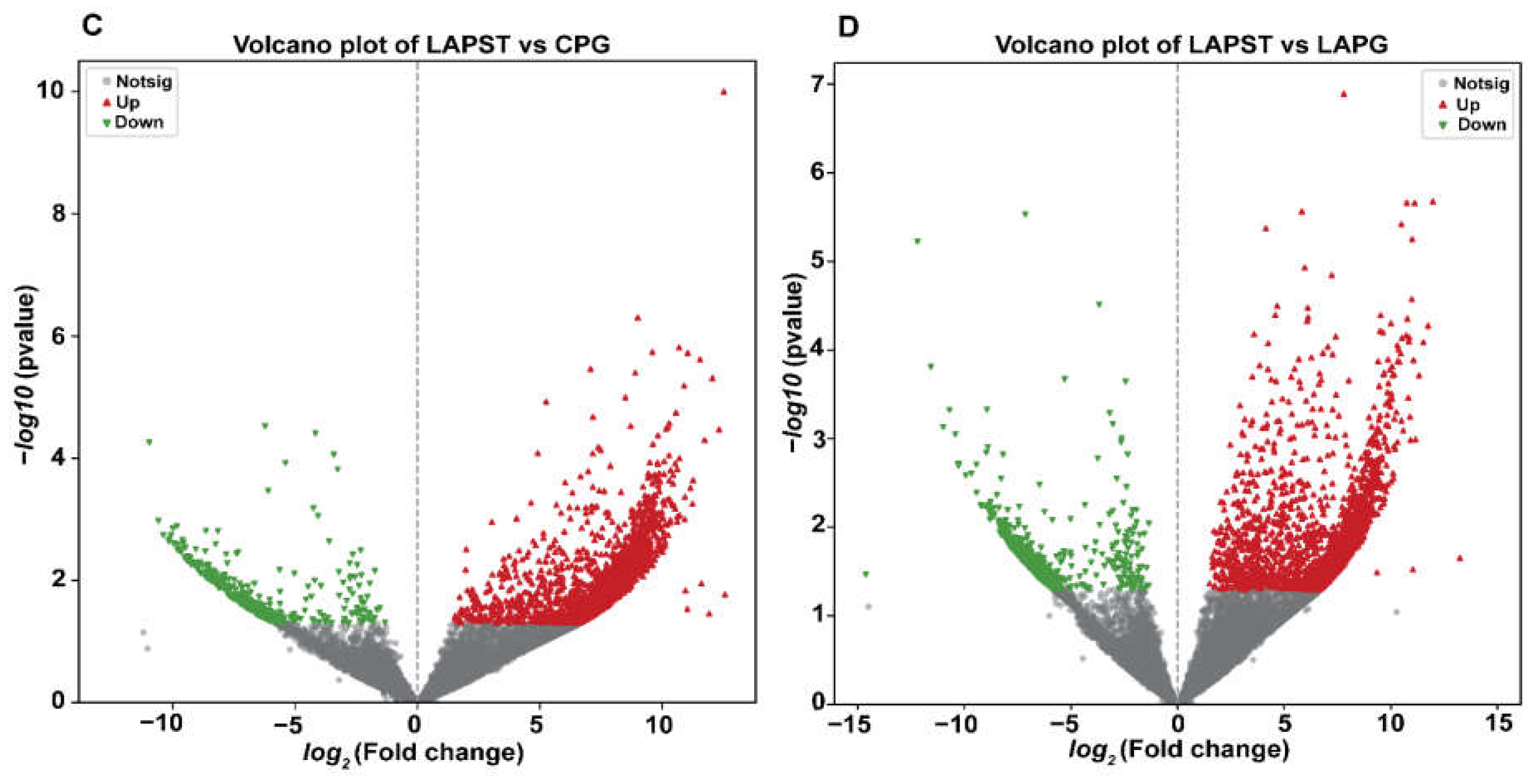
3.1.1. Differential Regulation of Genes Induced by Exposure to S. typhimurium and Probiotics
3.1.2. Genes Regulated by Probiotic Treatment Involved in Immunity, Gut Barrier Integrity, and Metabolism
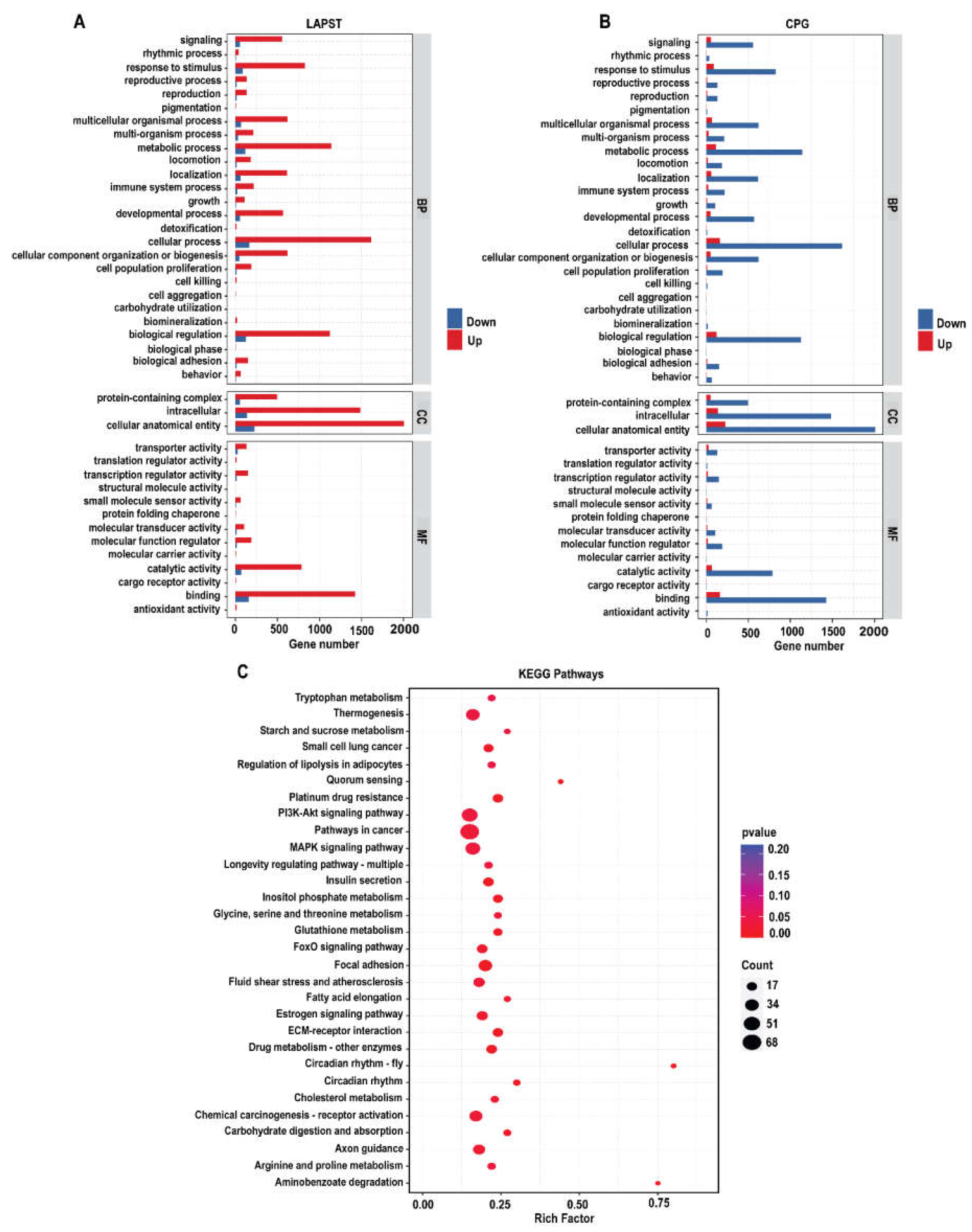
3.1.3. Gene Ontology Annotation and Enrichment Analysis
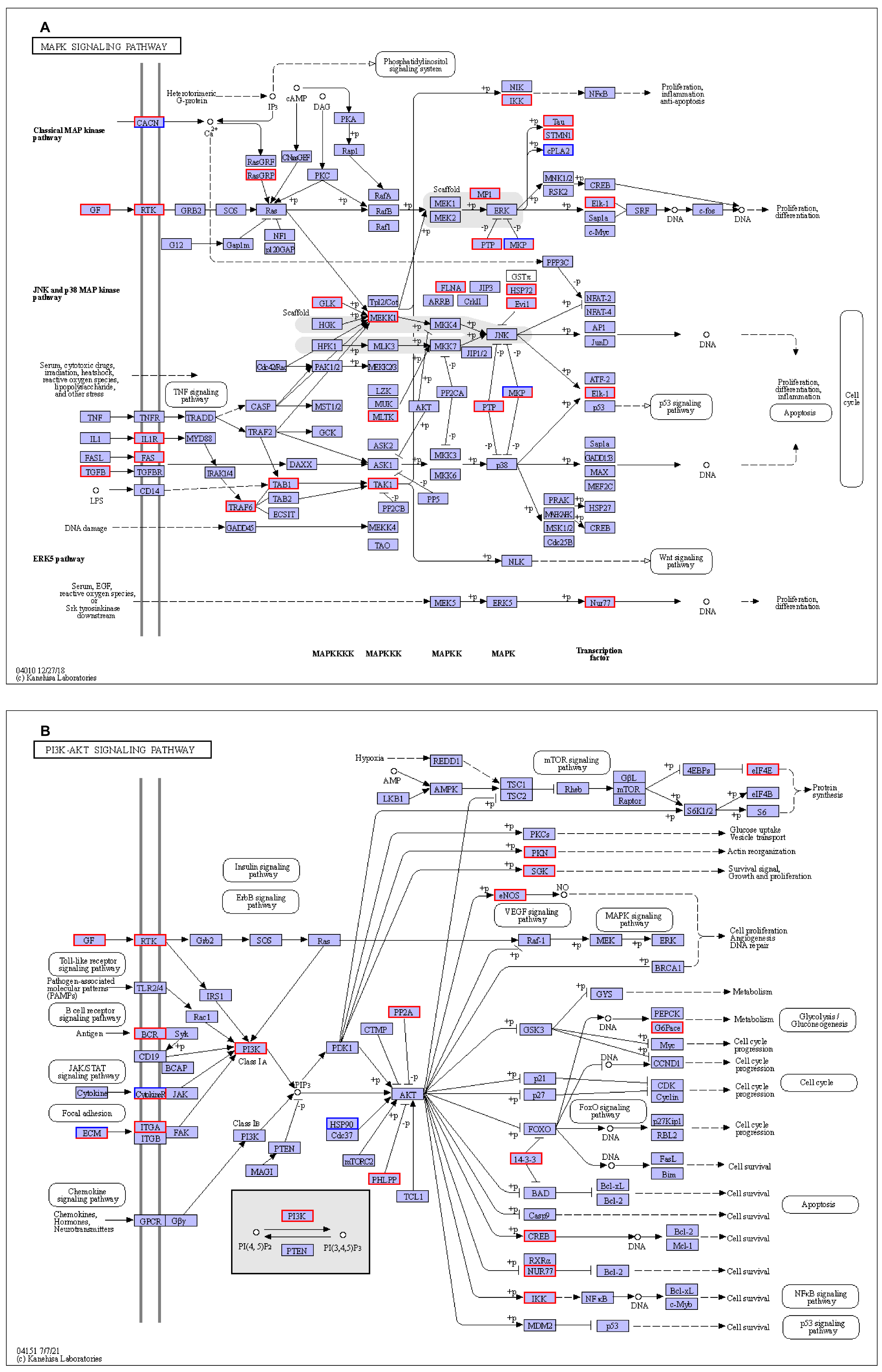
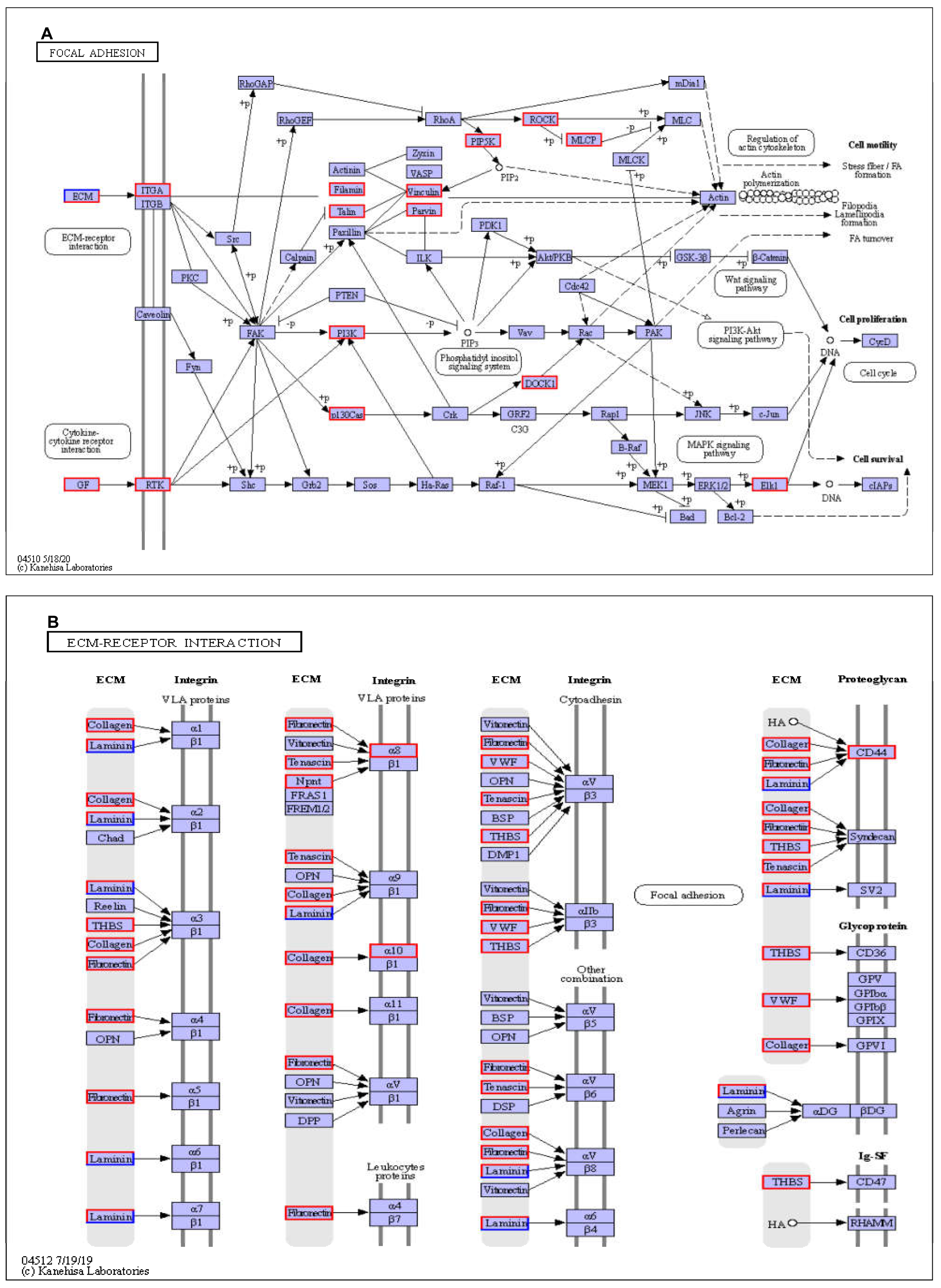
3.1.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways Analysis
3.2. Validation of DEGs’ by qRT-PCR
3.3. Effects of Probiotic Treatment on Gut Metabolites (LC/MS Analysis)
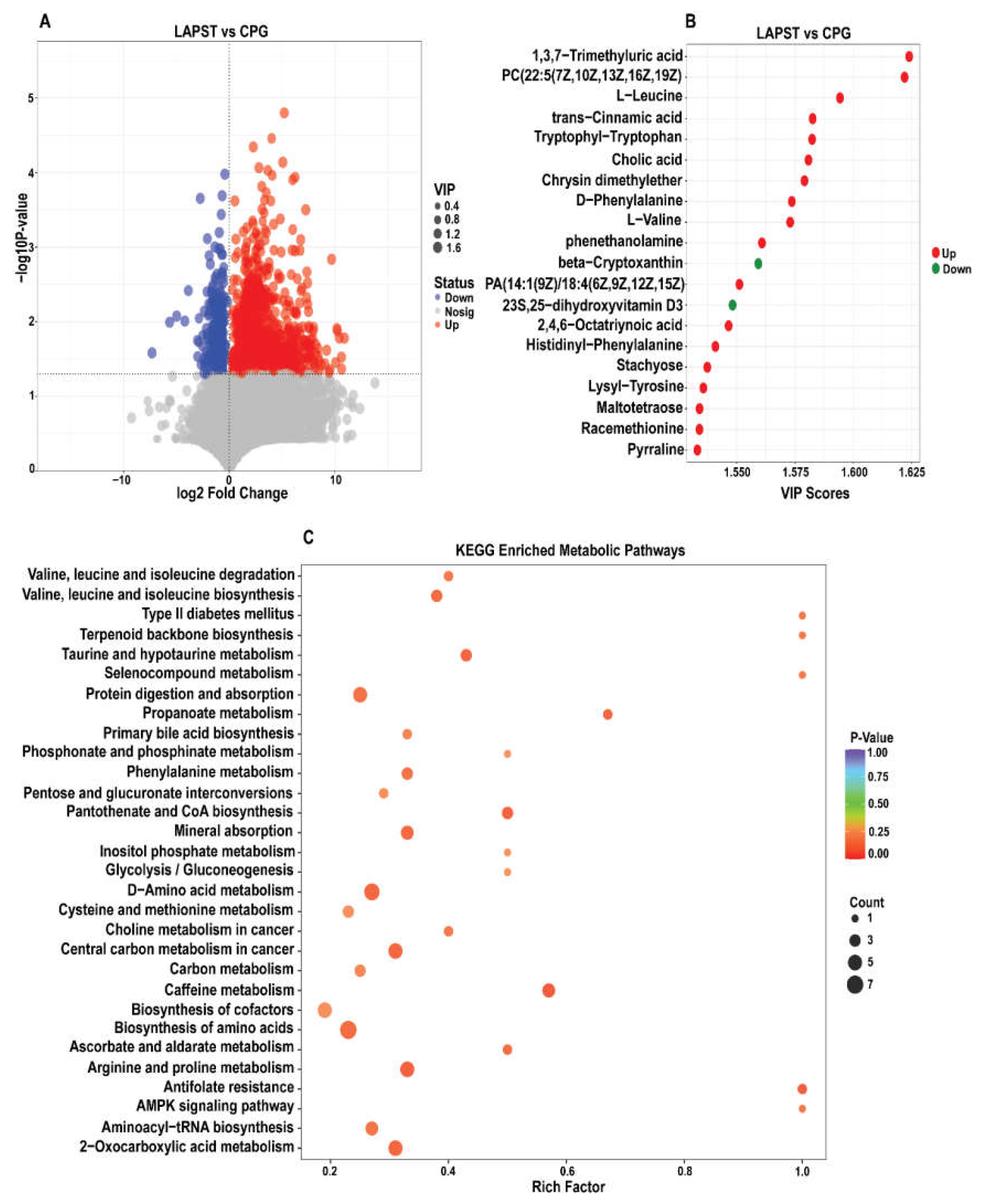
3.3.1. Differential Metabolites Analysis
3.3.2. Differential Metabolites KEGG Metabolic Pathways Analysis
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, food safety and food handling practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef]
- Popa, G.L.; Papa, M. Salmonella spp. infection-a continuous threat worldwide. Germs 2021, 11, 88. [Google Scholar] [CrossRef]
- Thompson, A.; Fulde, M.; Tedin, K. The metabolic pathways utilized by Salmonella typhimurium during infection of host cells. Environ. Microbiol. Rep. 2018, 10, 140–154. [Google Scholar] [CrossRef]
- Sun, H.; Wan, Y.; Du, P.; Bai, L. The epidemiology of monophasic Salmonella typhimurium. Foodborne Pathog. Dis. 2020, 17, 87–97. [Google Scholar] [CrossRef]
- Nealon, N.; Worcester, C.; Ryan, E. Lactobacillus paracasei metabolism of rice bran reveals metabolome associated with Salmonella typhimurium growth reduction. J. Appl. Microbiol. 2017, 122, 1639–1656. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.; Hofacre, C.L.; Lee, M.D.; Maurer, J.J.; Doyle, M.P. Animal sources of salmonellosis in humans. J. Am. Vet. Med. Assoc. 2002, 221, 492–497. [Google Scholar] [CrossRef]
- Sannathimmappa, M.B.; Nambiar, V.; Aravindakshan, R.J.; Promotion, H. Antibiotics at the crossroads–Do we have any therapeutic alternatives to control the emergence and spread of antimicrobial resistance? J. Educ. Health Promot. 2021, 10, 438. [Google Scholar]
- Bäumler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef]
- Liu, Y.; Mian, M.F.; Neufeld, K.-A.M.; Forsythe, P. CD4+ CD25+ T Cells are Essential for Behavioral Effects of Lactobacillus rhamnosus JB-1 in Male BALB/c mice. Brain Behav. Immun. 2020, 88, 451–460. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, X.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Qian, L.; Wang, Q.; Xue, Z.; Zhai, Q. Lactobacillus plantarum CCFM8610 alleviates irritable bowel syndrome and prevents gut microbiota dysbiosis: A randomized, double-blind, placebo-controlled, pilot clinical trial. Engineering 2021, 7, 376–385. [Google Scholar] [CrossRef]
- Panigrahi, P.; Parida, S.; Nanda, N.C.; Satpathy, R.; Pradhan, L.; Chandel, D.S.; Baccaglini, L.; Mohapatra, A.; Mohapatra, S.S.; Misra, P.R. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 2017, 548, 407–412. [Google Scholar] [CrossRef]
- Michael, P.; Shivshetty, N. Chapter-5 Probiotics. Probiotics. 2023, p. 87. Available online: https://www.researchgate.net/profile/Neelesh-Maurya/publication/374750375 (accessed on 26 March 2024).
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Factories 2017, 16, 79. [Google Scholar] [CrossRef]
- Marteau, P.; Seksik, P.; Jian, R. Probiotics and intestinal health effects: A clinical perspective. Br. J. Nutr. 2002, 88, s51–s57. [Google Scholar] [CrossRef]
- Elmadfa, I.; Klein, P.; Meyer, A.L. Immune-stimulating effects of lactic acid bacteria in vivo and in vitro. Proc. Nutr. Soc. 2010, 69, 416–420. [Google Scholar] [CrossRef]
- Keereelang, J.; Mangumphan, K.; Chitmanat, C.; Tongsiri, S.; Linh, N.V.; Van Doan, H. Dietary effect of Lactobacillus plantarum (TISTR 912) on digestive enzyme activity, growth performance, immune response, and disease resistance of black sharkminnow (Labeo chrysophekadion) against Aeromonas hydrophila infection. Aquac. Rep. 2022, 27, 101409. [Google Scholar] [CrossRef]
- Kwoji, I.D.; Aiyegoro, O.A.; Okpeku, M.; Adeleke, M.A. Multi-strain probiotics: Synergy among isolates enhances biological activities. Biology 2021, 10, 322. [Google Scholar] [CrossRef]
- Puvanasundram, P.; Chong, C.M.; Sabri, S.; Yusoff, M.S.; Karim, M. Multi-strain probiotics: Functions, effectiveness and formulations for aquaculture applications. Aquac. Rep. 2021, 21, 100905. [Google Scholar] [CrossRef]
- Tremblay, A.; Xu, X.; Colee, J.; Tompkins, T.A. Efficacy of a multi-strain probiotic formulation in pediatric populations: A comprehensive review of clinical studies. Nutrients 2021, 13, 1908. [Google Scholar] [CrossRef]
- Seddik, H.A.; Bendali, F.; Gancel, F.; Fliss, I.; Spano, G.; Drider, D. Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob. Proteins 2017, 9, 111–122. [Google Scholar] [CrossRef]
- María Remes-Troche, J.; Coss-Adame, E.; Ángel Valdovinos-Díaz, M.; Gómez-Escudero, O.; Eugenia Icaza-Chávez, M.; Antonio Chávez-Barrera, J.; Zárate-Mondragón, F.; Antonio Velarde-Ruíz Velasco, J.; Rafael Aceves-Tavares, G.; Antonio Lira-Pedrín, M. Lactobacillus acidophilus LB: A useful pharmabiotic for the treatment of digestive disorders. Ther. Adv. Gastroenterol. 2020, 13, 1756284820971201. [Google Scholar] [CrossRef]
- Schwarzer, M.; Makki, K.; Storelli, G.; Machuca-Gayet, I.; Srutkova, D.; Hermanova, P.; Martino, M.E.; Balmand, S.; Hudcovic, T.; Heddi, A. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 2016, 351, 854–857. [Google Scholar] [CrossRef] [PubMed]
- Klarin, B.; Adolfsson, A.; Torstensson, A.; Larsson, A. Can probiotics be an alternative to chlorhexidine for oral care in the mechanically ventilated patient? A multicentre, prospective, randomised controlled open trial. Crit. Care 2018, 22, 272. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Polk, D.B. Probiotics and probiotic-derived functional factors—Mechanistic insights into applications for intestinal homeostasis. Front. Immunol. 2020, 11, 1428. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, K.; Zhang, A.; Chang, W.; Zheng, A.; Chen, Z.; Cai, H.; Liu, G. Effects of Lactobacillus acidophilus on the growth performance, immune response, and intestinal barrier function of broiler chickens challenged with Escherichia coli O157. Poult. Sci. 2021, 100, 101323. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sheng, Y.; Pan, Q.; Xue, Y.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W.; et al. Identification of the key physiological characteristics of Lactobacillus plantarum strains for ulcerative colitis alleviation. Food Funct. 2020, 11, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Kim, T.-Y.; Kim, Y.; Lee, S.-H.; Kim, S.; Kang, S.W.; Yang, J.-Y.; Baek, I.-J.; Sung, Y.H.; Park, Y.-Y.; et al. Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host 2018, 24, 833–846.e836. [Google Scholar] [CrossRef] [PubMed]
- Kayama, H.; Okumura, R.; Takeda, K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu. Rev. Immunol. 2020, 38, 23–48. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, Y.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Protective effects of different Bacteroides vulgatus strains against lipopolysaccharide-induced acute intestinal injury, and their underlying functional genes. J. Adv. Res. 2022, 36, 27–37. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef]
- Goh, Y.J.; Barrangou, R.; Klaenhammer, T.R. In vivo transcriptome of Lactobacillus acidophilus and colonization impact on murine host intestinal gene expression. mBio 2021, 12, e03399-20. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.L.; De Vries, M.C.; Wels, M.; Molenaar, D.; Mangell, P.; Ahrne, S.; De Vos, W.M.; Vaughan, E.E.; Kleerebezem, M. Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 2010, 4, 1481–1484. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Salonen, A.; Kekkonen, R.A.; Salojärvi, J.; Jalanka-Tuovinen, J.; Palva, A.; Orešič, M.; De Vos, W.M. Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. PeerJ 2013, 1, e32. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, L.; Mu, C.; Sun, Y.; Gu, Y.; Chen, D.; Liu, T.; Cao, H. Gut microbial metabolome in inflammatory bowel disease: From association to therapeutic perspectives. Comput. Struct. Biotechnol. J. 2022, 20, 2402–2414. [Google Scholar] [CrossRef] [PubMed]
- Hatami, S.; Yavarmanesh, M.; Sankian, M. Comparison of the effects of probiotic strains (Lactobacillus gasseri, Lactiplantibacillus plantarum, Lactobacillus acidophilus, and Limosilactobacillus fermentum) isolated from human and food products on the immune response of CT26 tumor–bearing mice. Braz. J. Microbiol. 2023, 54, 2047–2062. [Google Scholar] [CrossRef] [PubMed]
- Bove, P.; Gallone, A.; Russo, P.; Capozzi, V.; Albenzio, M.; Spano, G.; Fiocco, D. Probiotic features of Lactobacillus plantarum mutant strains. J. Appl. Microbiol. 2012, 96, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Bull, M.; Plummer, S.; Marchesi, J.; Mahenthiralingam, E. The life history of Lactobacillus acidophilus as a probiotic: A tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiol. Lett. 2013, 349, 77–87. [Google Scholar] [CrossRef]
- Baiandina, I.; Kuleshova, O.; Krivenko, O. Light perception in Beroidae ctenophores: Evidence from laboratory experiments and genomics data. Bioinformatics of Genome Regulation and Structure/Systems Biology (BGRS/SB-2022). 2022, pp. 29–30. Available online: https://elibrary.ru/item.asp?id=49238119 (accessed on 26 March 2024).
- Richard, C.; Chevrier, S.; Chalus, L.; Forget, J.; Pinteur, B.; Bravetti, P.; Boidot, R. Sensitizing potential of a novel stimulated and haptenized vaccine in immunotherapy resistant metastatic colorectal cancer. Cancer Res. 2022, 82, 3566. [Google Scholar] [CrossRef]
- Ye, X.; Huang, X.; Fu, X.; Zhang, X.; Lin, R.; Zhang, W.; Zhang, J.; Lu, Y. Myeloid-like tumor hybrid cells in bone marrow promote progression of prostate cancer bone metastasis. J. Hematol. Oncol. 2023, 16, 46. [Google Scholar] [CrossRef]
- Xia, X.; Yu, H.; Li, Y.; Liang, Y.; Li, G.; Huang, F. Transcriptome Analysis Identifies Biomarkers for the Diagnosis and Management of Psoriasis Complicated with Depression. Clin. Cosmet. Investig. Dermatol. 2023, 16, 1287–1301. [Google Scholar] [CrossRef]
- Shehata, H.R.; Newmaster, S.G. A validated real-time PCR method for the specific identification of probiotic strain Lactobacillus rhamnosus GG (ATCC 53103). J. AOAC Int. 2020, 103, 1604–1609. [Google Scholar] [CrossRef]
- Ni, Z.; Sun, S.; Bi, Y.; Ding, J.; Cheng, W.; Yu, J.; Zhou, L.; Li, M.; Yu, C. Correlation of fecal metabolomics and gut microbiota in mice with endometriosis. Am. J. Reprod. Immunol. 2020, 84, e13307. [Google Scholar] [CrossRef]
- Yu, M.; Jia, H.; Zhou, C.; Yang, Y.; Zhao, Y.; Yang, M.; Zou, Z. Variations in gut microbiota and fecal metabolic phenotype associated with depression by 16S rRNA gene sequencing and LC/MS-based metabolomics. J. Pharm. Biomed. Anal. 2017, 138, 231–239. [Google Scholar] [CrossRef]
- Huang, F.; Fu, M.; Li, J.; Chen, L.; Feng, K.; Huang, T.; Cai, Y.-D. Analysis and prediction of protein stability based on interaction network, gene ontology, and kegg pathway enrichment scores. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2023, 1871, 140889. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.-P.; Michel, M.-L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Pan, S.; Zhang, K.; Michiels, J.; Zeng, Q.; Ding, X.; Wang, J.; Peng, H.; Bai, J.; Xuan, Y. Impact of dietary manganese on intestinal barrier and inflammatory response in broilers challenged with Salmonella typhimurium. Microorganisms 2020, 8, 757. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.J.; Winter, S.E. Salmonella finds a way: Metabolic versatility of Salmonella enterica serovar typhimurium in diverse host environments. PLoS Pathog. 2020, 16, e1008540. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Fischer, J.; Ganesan, R.; Hos, N.J.; Cildir, G.; Wolke, M.; Pessia, A.; Frommolt, P.; Desiderio, V.; Velagapudi, V. Salmonella typhimurium impairs glycolysis-mediated acidification of phagosomes to evade macrophage defense. PLoS Pathog. 2021, 17, e1009943. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yang, M.; Cai, H.; Liu, Y.; Gorris, L.; Aslam, M.Z.; Jia, K.; Sun, T.; Wang, X.; Dong, Q. Antibiotic Resistance of Salmonella typhimurium Monophasic Variant 1,4,[5],12:i:-in China: A Systematic Review and Meta-Analysis. Antibiotics 2022, 11, 532. [Google Scholar] [CrossRef]
- Latif, A.; Shehzad, A.; Niazi, S.; Zahid, A.; Ashraf, W.; Iqbal, M.W.; Rehman, A.; Riaz, T.; Aadil, R.M.; Khan, I.M. Probiotics: Mechanism of action, health benefits and their application in food industries. Front. Microbiol. 2023, 14, 1216674. [Google Scholar] [CrossRef]
- Mazziotta, C.; Tognon, M.; Martini, F.; Torreggiani, E.; Rotondo, J.C. Probiotics mechanism of action on immune cells and beneficial effects on human health. Cells 2023, 12, 184. [Google Scholar] [CrossRef]
- Wolfe, W.; Xiang, Z.; Yu, X.; Li, P.; Chen, H.; Yao, M.; Fei, Y.; Huang, Y.; Yin, Y.; Xiao, H.; et al. The challenge of applications of probiotics in gastrointestinal diseases. Adv. Gut Microbiome Res. 2023, 2023, 1984200. [Google Scholar] [CrossRef]
- Sherman, P.; Mayer, E. An update on the use and investigation of probiotics in health and disease. Gut 2013, 62, 787796Satin. [Google Scholar]
- Wu, Y.; Jha, R.; Li, A.; Liu, H.; Zhang, Z.; Zhang, C.; Zhai, Q.; Zhang, J. Probiotics (Lactobacillus plantarum HNU082) supplementation relieves ulcerative colitis by affecting intestinal barrier functions, immunity-related gene expression, gut microbiota, and metabolic pathways in mice. Microbiol. Spectr. 2022, 10, e01651-22. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Jiang, S.; Lv, L.; Wu, W.; Wang, Q.; Xu, Q.; Ye, J.; Fang, D.; Li, Y.; Wu, J.; et al. Modulation of the immune response and metabolism in germ-free rats colonized by the probiotic Lactobacillus salivarius LI01. Appl. Microbiol. Biotechnol. 2021, 105, 1629–1645. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Vlasova, A.N.; Liu, Z.; Chattha, K.S.; Kandasamy, S.; Esseili, M.; Zhang, X.; Rajashekara, G.; Saif, L. In vivo gut transcriptome responses to Lactobacillus rhamnosus GG and Lactobacillus acidophilus in neonatal gnotobiotic piglets. Gut Microbes 2014, 5, 152–164. [Google Scholar] [CrossRef]
- Dunislawska, A.; Slawinska, A.; Bednarczyk, M.; Siwek, M. Transcriptome modulation by in ovo delivered Lactobacillus synbiotics in a range of chicken tissues. Gene 2019, 698, 27–33. [Google Scholar] [CrossRef]
- Memon, F.U.; Yang, Y.; Leghari, I.H.; Lv, F.; Soliman, A.M.; Zhang, W.; Si, H. Transcriptome analysis revealed ameliorative effects of Bacillus based probiotic on immunity, gut barrier system, and metabolism of chicken under an experimentally induced Eimeria tenella infection. Genes 2021, 12, 536. [Google Scholar] [CrossRef]
- Marcobal, A.; Yusufaly, T.; Higginbottom, S.; Snyder, M.; Sonnenburg, J.L.; Mias, G.I. Metabolome progression during early gut microbial colonization of gnotobiotic mice. Sci. Rep. 2015, 5, 11589. [Google Scholar] [CrossRef]
- Gensollen, T.; Blumberg, R.S. Correlation between early-life regulation of the immune system by microbiota and allergy development. J. Allergy Clin. Immunol. 2017, 139, 1084–1091. [Google Scholar] [CrossRef]
- Wang, J.; Han, C.; Lu, Z.; Ge, P.; Cui, Y.; Zhao, D.; Yang, X.; Wu, B.; Qiang, L.; Zhang, Y. Simulated microgravity suppresses MAPK pathway-mediated innate immune response to bacterial infection and induces gut microbiota dysbiosis. FASEB J. 2020, 34, 14631–14644. [Google Scholar] [CrossRef] [PubMed]
- Symons, A.; Beinke, S.; Ley, S.C. MAP kinase kinase kinases and innate immunity. Trends Immunol. 2006, 27, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-G.; Ohta, T.; Takahashi, T.; Kushiro, A.; Nomoto, K.; Yokokura, T.; Okada, N.; Danbara, H. Probiotic Lactobacillus casei activates innate immunity via NF-κB and p38 MAP kinase signaling pathways. Microbes 2006, 8, 994–1005. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.-C.; Huang, S.-C. The different effects of probiotics treatment on Salmonella-induced interleukin-8 response in intestinal epithelia cells via PI3K/Akt and NOD2 expression. Benef. Microbes 2016, 7, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Hu, A.; Shu, X.; Huang, W.; Zhang, R.; Xu, Y.; Yang, C. Lactobacillus plantarum postbiotics trigger AMPK-dependent autophagy to suppress Salmonella intracellular infection and NLRP3 inflammasome activation. J. Cell. Physiol. 2023, 238, 1336–1353. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.-S.; He, Z.-M.; Xia, J.-B. FoxO3 Regulates the Progress and Development of Aging and Aging-Related Diseases. Curr. Mol. Med. 2023, 23, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Karamanos, N.K.; Theocharis, A.D.; Piperigkou, Z.; Manou, D.; Passi, A.; Skandalis, S.S.; Vynios, D.H.; Orian-Rousseau, V.; Ricard-Blum, S.; Schmelzer, C.E. A guide to the composition and functions of the extracellular matrix. FEBS J. 2021, 288, 6850–6912. [Google Scholar] [CrossRef] [PubMed]
- Mishra, Y.G.; Manavathi, B. Focal adhesion dynamics in cellular function and disease. Cell. Signal. 2021, 85, 110046. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, M.; Cossart, P. Impenetrable barriers or entry portals? The role of cell–cell adhesion during infection. J. Cell Biol. 2011, 195, 349–358. [Google Scholar] [CrossRef]
- Wu, S.; Liu, M.; Chen, H.; Song, Q.; Wu, Z.; Dai, Z. Tryptophan regulates bile and nitrogen metabolism in two pig gut lactobacilli species in vitro based on metabolomics study. Amino Acids 2022, 54, 1421–1435. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, Y.; Li, Y.; Ma, D.; Zhang, H.; Kwok, L.-Y. Lacticaseibacillus rhamnosus Probio-M9-driven mouse mammary tumor-inhibitory effect is accompanied by modulation of host gut microbiota, immunity, and serum metabolome. Nutrients 2022, 15, 5. [Google Scholar] [CrossRef]
- Lin, C.-C.; Huang, W.-C.; Su, C.-H.; Lin, W.-D.; Wu, W.-T.; Yu, B.; Hsu, Y.-M. Effects of multi-strain probiotics on immune responses and metabolic balance in helicobacter pylori-infected mice. Nutrients 2020, 12, 2476. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.H.; Zabed, H.M.; Yun, J.; Zhang, G.; Qi, X. An insight into the roles of dietary tryptophan and its metabolites in intestinal inflammation and inflammatory bowel disease. Mol. Nutr. 2021, 65, 2000461. [Google Scholar] [CrossRef]
- Kaiser, J.C.; Heinrichs, D.E. Branching out: Alterations in bacterial physiology and virulence due to branched-chain amino acid deprivation. mBio 2018, 9, e01188-18. [Google Scholar] [CrossRef]
- Shao, Y.; Zhen, W.; Guo, F.; Hu, Z.; Zhang, K.; Kong, L.; Guo, Y.; Wang, Z. Pretreatment with probiotics Enterococcus faecium NCIMB 11181 attenuated Salmonella typhimurium-induced gut injury through modulating intestinal microbiome and immune responses with barrier function in broiler chickens. J. Anim. Sci. Biotechnol. 2022, 13, 130. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Huang, S.; Cai, S.; Yu, H.; Wang, Y.; Zeng, X.; Qiao, S. Lactobacillus reuteri ameliorates intestinal inflammation and modulates gut microbiota and metabolic disorders in dextran sulfate sodium-induced colitis in mice. Nutrients 2020, 12, 2298. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, W.; Ci, W.; Zheng, Y.; Han, X.; Huang, J.; Zhu, J. Effects of dietary supplementation with Lactobacillus acidophilus and Bacillus subtilis on mucosal immunity and intestinal barrier are associated with its modulation of gut metabolites and microbiota in late-phase laying hens. Probiotics Antimicrob. Proteins 2023, 15, 912–924. [Google Scholar] [CrossRef]
- Martí i Líndez, A.-A.; Reith, W. Arginine-dependent immune responses. Cell. Mol. Life Sci. 2021, 78, 5303–5324. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wu, G. Important roles of amino acids in immune responses. Br. J. Nutr. 2022, 127, 398–402. [Google Scholar] [CrossRef]
- Jahan-Mihan, A.; Luhovyy, B.L.; Khoury, D.E.; Anderson, G.H. Dietary proteins as determinants of metabolic and physiologic functions of the gastrointestinal tract. Nutrients 2011, 3, 574–603. [Google Scholar] [CrossRef]
- Zhao, D.-D.; Gai, Y.-D.; Li, C.; Fu, Z.-Z.; Yin, D.-Q.; Xie, M.; Dai, J.-Y.; Wang, X.-X.; Li, Y.-X.; Wu, G.-F. Dietary taurine effect on intestinal barrier function, colonic microbiota and metabolites in weanling piglets induced by LPS. Front. Microbiol. 2023, 14, 1259133. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Junaid, M.; Lu, H.; Li, Y.; Liu, Y.; Din, A.U.; Qi, Z.; Xiong, Y.; Yan, J. Novel Synergistic Probiotic Intervention: Transcriptomic and Metabolomic Analysis Reveals Ameliorative Effects on Immunity, Gut Barrier, and Metabolism of Mice during Salmonella typhimurium Infection. Genes 2024, 15, 435. https://doi.org/10.3390/genes15040435
Junaid M, Lu H, Li Y, Liu Y, Din AU, Qi Z, Xiong Y, Yan J. Novel Synergistic Probiotic Intervention: Transcriptomic and Metabolomic Analysis Reveals Ameliorative Effects on Immunity, Gut Barrier, and Metabolism of Mice during Salmonella typhimurium Infection. Genes. 2024; 15(4):435. https://doi.org/10.3390/genes15040435
Chicago/Turabian StyleJunaid, Muhammad, Hongyu Lu, Yixiang Li, Yu Liu, Ahmad Ud Din, Zhongquan Qi, Yi Xiong, and Jianhua Yan. 2024. "Novel Synergistic Probiotic Intervention: Transcriptomic and Metabolomic Analysis Reveals Ameliorative Effects on Immunity, Gut Barrier, and Metabolism of Mice during Salmonella typhimurium Infection" Genes 15, no. 4: 435. https://doi.org/10.3390/genes15040435
APA StyleJunaid, M., Lu, H., Li, Y., Liu, Y., Din, A. U., Qi, Z., Xiong, Y., & Yan, J. (2024). Novel Synergistic Probiotic Intervention: Transcriptomic and Metabolomic Analysis Reveals Ameliorative Effects on Immunity, Gut Barrier, and Metabolism of Mice during Salmonella typhimurium Infection. Genes, 15(4), 435. https://doi.org/10.3390/genes15040435






