Transcriptomic Analysis of Alternative Splicing Events during Different Fruit Ripening Stages of Coffea arabica L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA-seq
2.3. Reads Mapping and Transcript Assembly
2.4. Identification of Differential Alternative Splicing Events and Differentially Expressed Genes
2.5. Functional Enrichment Analysis
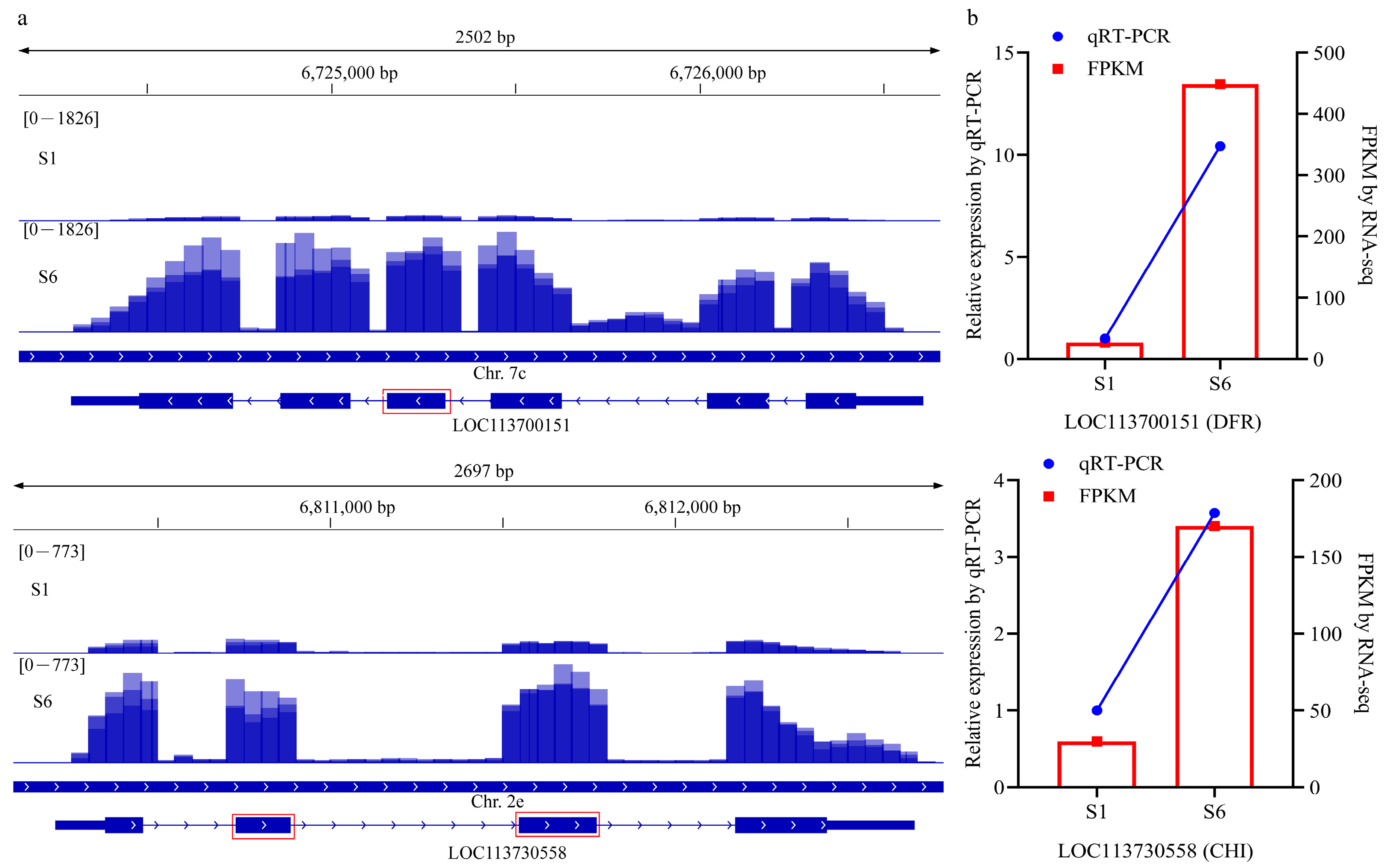
2.6. qRT-PCR Validation of AS Events
2.7. Data Presentation
3. Results
3.1. Basic Information on Transcripts
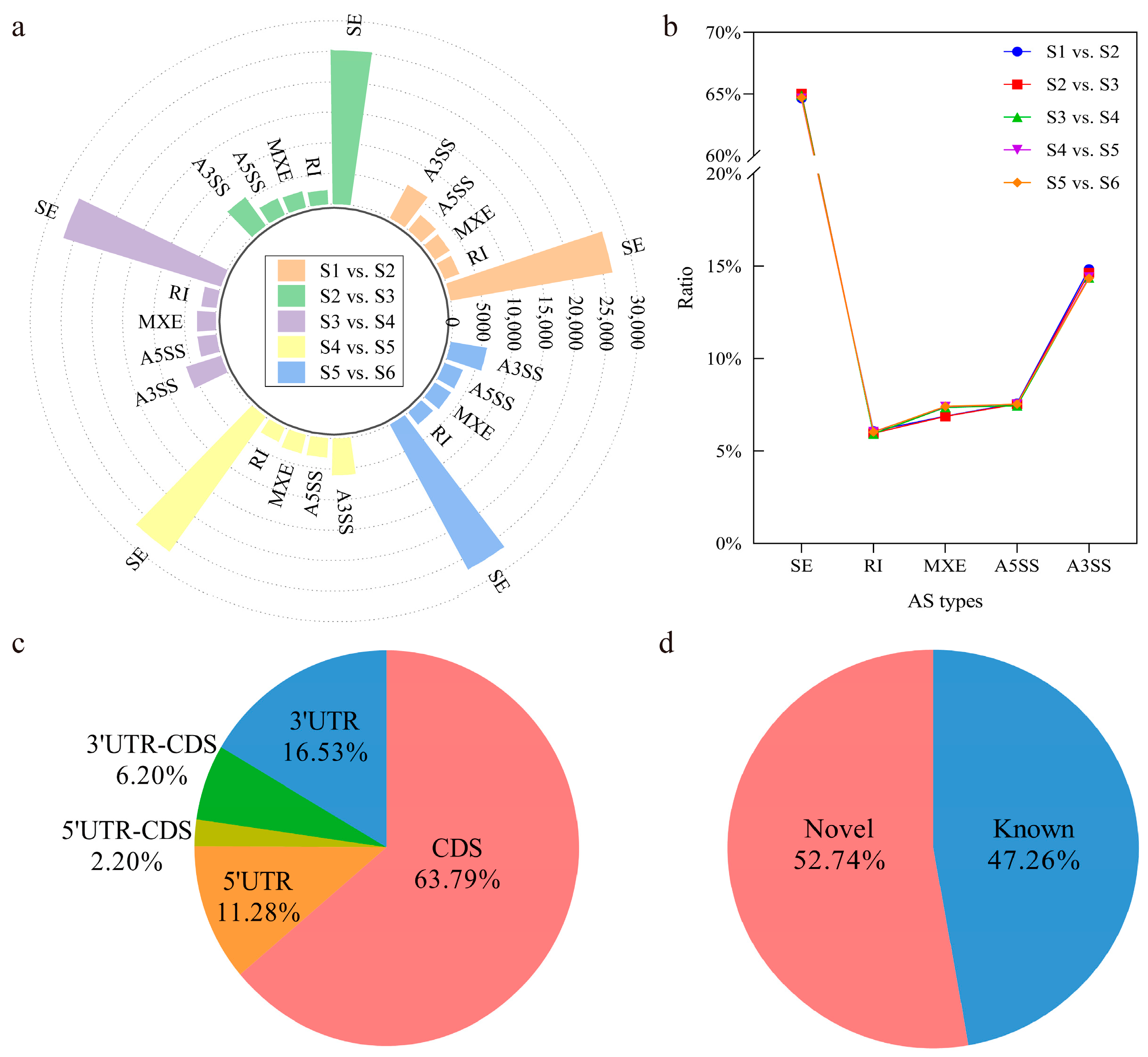
3.2. Identification and Classification of Alternative Splicing Events
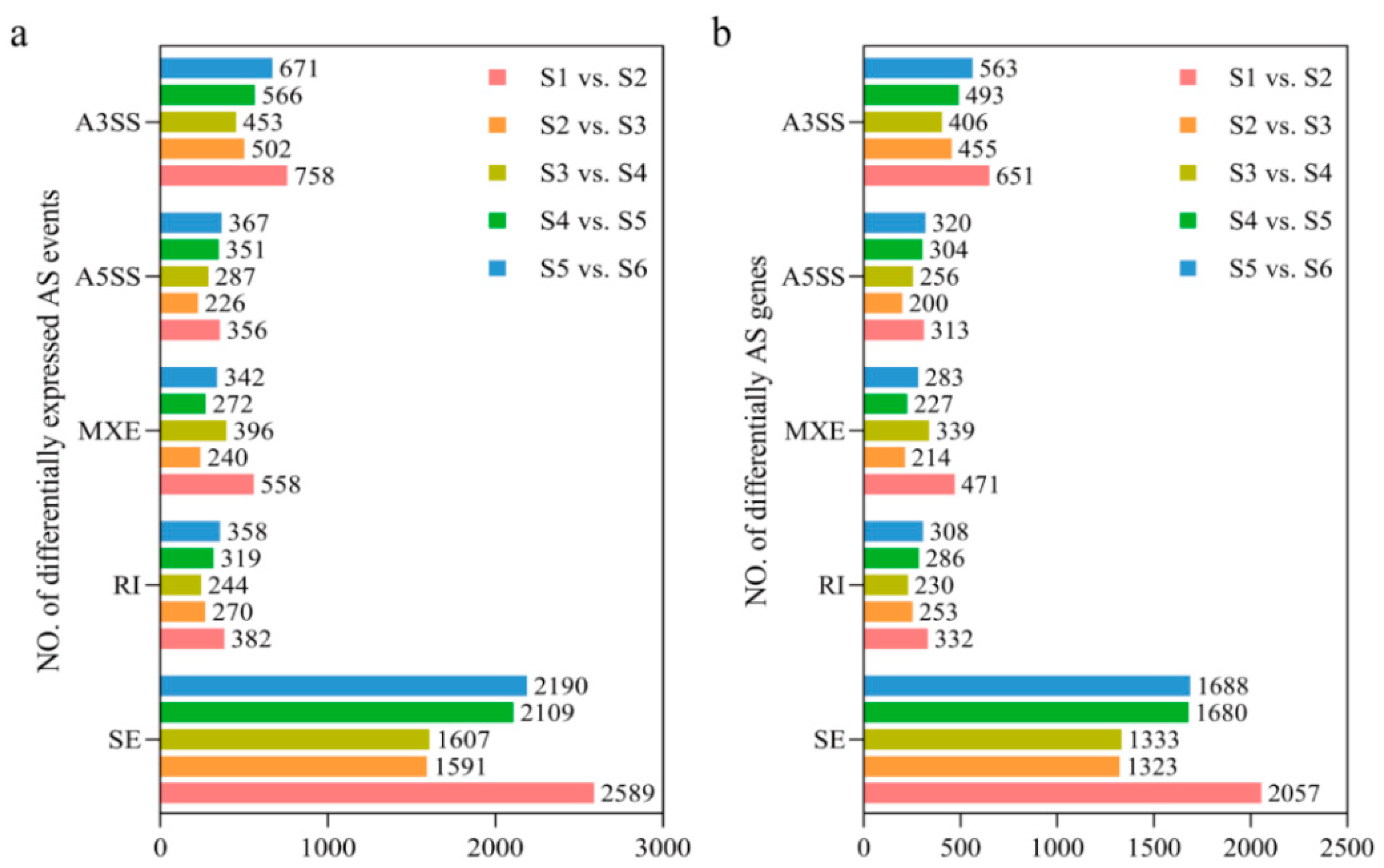
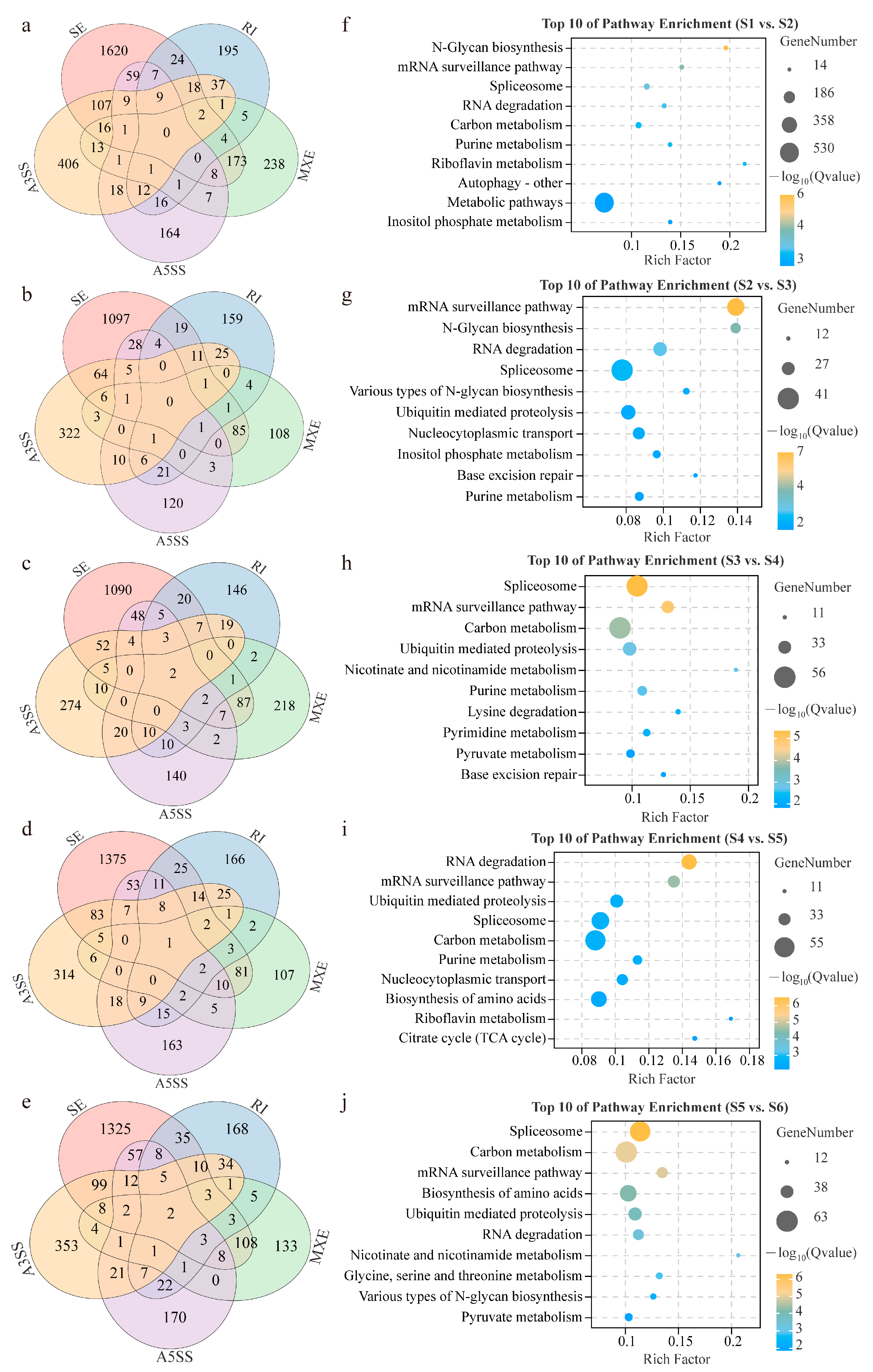
3.3. Dynamic Characterization and Function Analysis of Alternative Splicing Events
3.4. Comparative Analysis of DSGs and DEGs
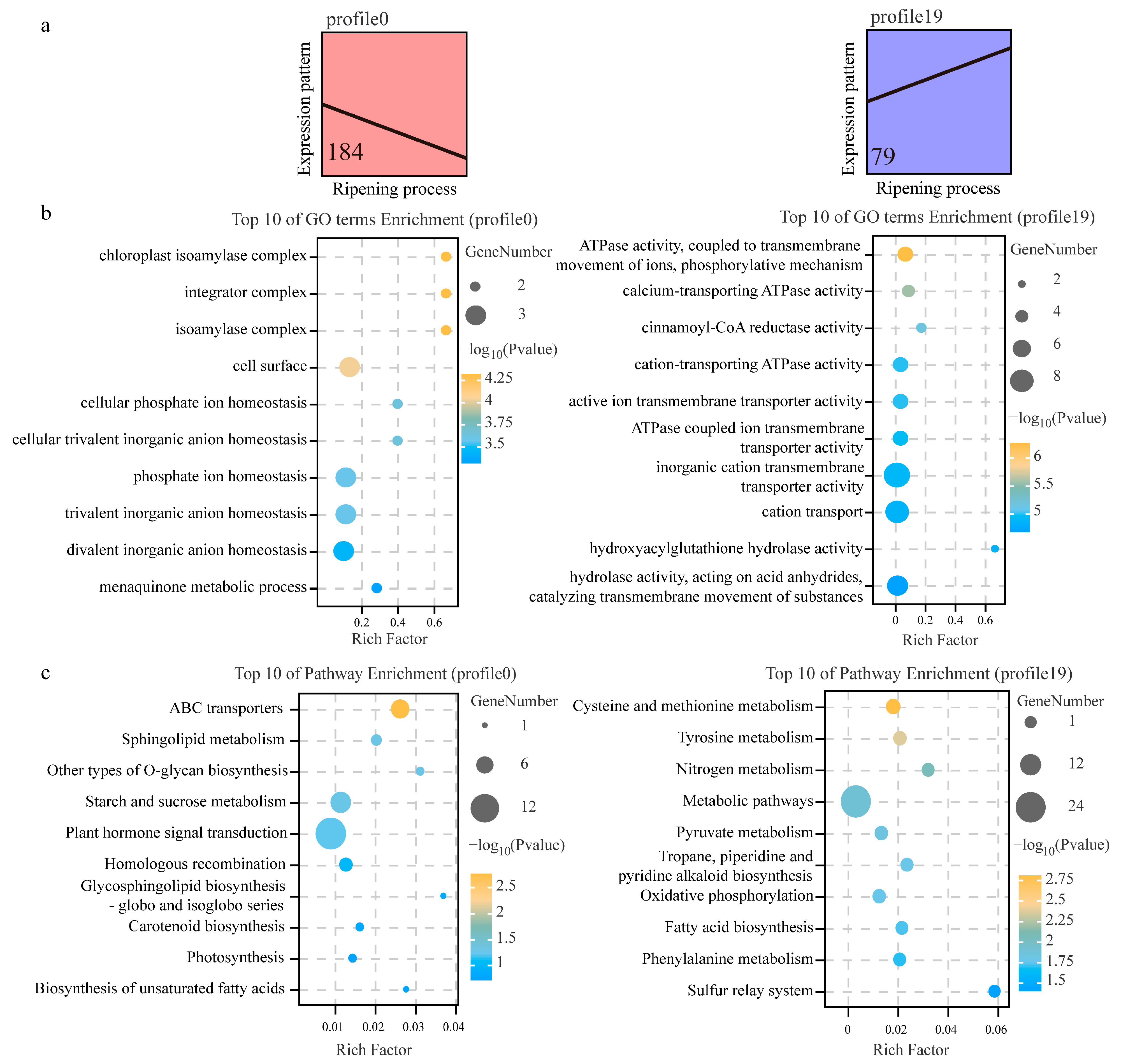
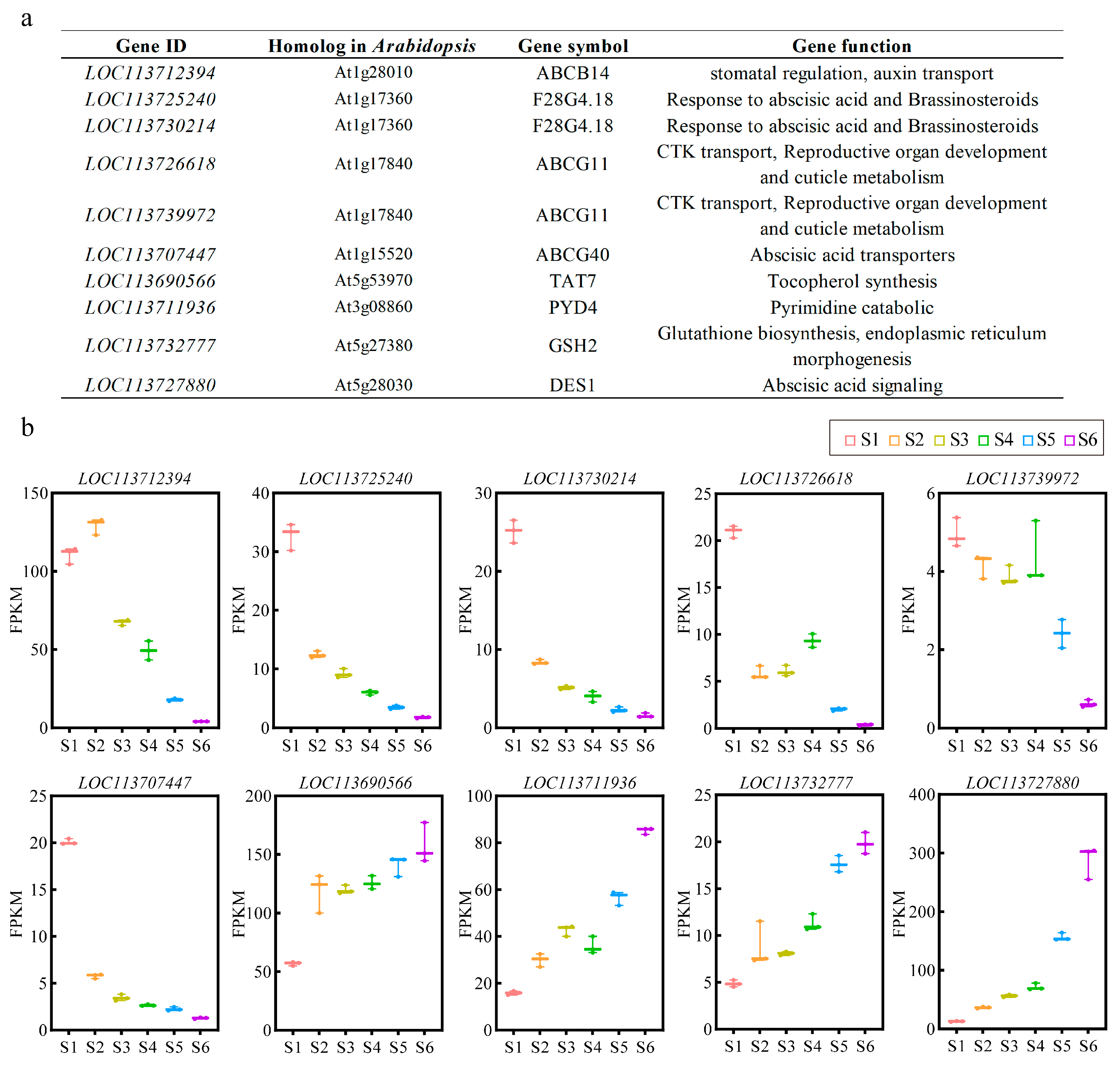
3.5. Biological Function Analysis between DSGs and DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davis, A.P.; Tosh, J.; Ruch, N.; Fay, M.F. Growing coffee: Psilanthus (Rubiaceae) subsumed on the basis of molecular and morphological data; implications for the size, morphology, distribution and evolutionary history of Coffea. Bot. J. Linnean Soc. 2011, 167, 357–377. [Google Scholar] [CrossRef]
- Geleta, M.; Herrera, I.; Monzón, A.; Bryngelsson, T. Genetic Diversity of Arabica Coffee (Coffea arabica L.) in Nicaragua as Estimated by Simple Sequence Repeat Markers. Sci. World J. 2012, 11, 939820. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Pruvot-Woehl, S.; Davis, A.P.; Schilling, T.; Moat, J.; Solano, W.; Al Hakimi, A.; Montagnon, C. Validating South Sudan as a Center of Origin for Coffea arabica: Implications for Conservation and Coffee Crop Improvement. Front. Sustain. Food Syst. 2021, 5, 11. [Google Scholar] [CrossRef]
- Rodrigues, W.P.; Machado, J.A.; da Silva, J.R.; Figueiredo, F.; Ferraz, T.M.; Ferreira, L.S.; Bezerra, L.B.D.; de Abreu, D.P.; Bernado, W.D.; Passos, L.C.; et al. Whole-canopy gas exchanges in Coffea sp. is affected by supra-optimal temperature and light distribution within the canopy: The insights from an improved multi-chamber system. Sci. Hortic. 2016, 211, 194–202. [Google Scholar] [CrossRef]
- Privat, I.; Foucrier, S.; Prins, A.; Epalle, T.; Eychenne, M.; Kandalaft, L.; Caillet, V.; Lin, C.W.; Tanksley, S.; Foyer, C.; et al. Differential regulation of grain sucrose accumulation and metabolism in Coffea arabica (Arabica) and Coffea canephora (Robusta) revealed through gene expression and enzyme activity analysis. New Phytol. 2008, 178, 781–797. [Google Scholar] [CrossRef] [PubMed]
- Freitas, V.V.; Borges, L.L.R.; Castro, G.A.D.; dos Santos, M.H.; Vidigal, M.; Fernandes, S.A.; Stringheta, P.C. Impact of different roasting conditions on the chemical composition, antioxidant activities, and color of Coffea canephora and Coffea arabica L. samples. Heliyon 2023, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Toniutti, L.; Breitler, J.C.; Etienne, H.; Campa, C.; Doulbeau, S.; Urban, L.; Lambot, C.; Pinilla, J.C.H.; Bertrand, B. Influence of Environmental Conditions and Genetic Background of Arabica Coffee (C. arabica L.) on Leaf Rust (Hemileia vastatrix) Pathogenesis. Front. Plant Sci. 2017, 8, 12. [Google Scholar] [CrossRef]
- Torres, L.F.; Reichel, T.; Déchamp, E.; de Aquino, S.O.; Duarte, K.E.; Alves, G.S.C.; Silva, A.T.; Cotta, M.G.; Costa, T.S.; Diniz, L.E.C.; et al. Expression of DREB-Like Genes in Coffea canephora and C. arabica Subjected to Various Types of Abiotic Stress. Trop. Plant Biol. 2019, 12, 98–116. [Google Scholar] [CrossRef]
- Noirot, M.; Poncet, V.; Barre, P.; Hamon, P.; Hamon, S.; De Kochko, A. Genome size variations in diploid African Coffea species. Ann. Bot. 2003, 92, 709–714. [Google Scholar] [CrossRef]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar]
- Ságio, S.A.; Lima, A.A.; Barreto, H.G.; de Carvalho, C.H.S.; Paiva, L.V.; Chalfun, A. Physiological and molecular analyses of early and late Coffea arabica cultivars at different stages of fruit ripening. Acta Physiol. Plant. 2013, 3, 3091–3098. [Google Scholar] [CrossRef]
- Buitrago-Osorio, J.; Tinoco, H.A.; Perdomo-Hurtado, L.; Rincon-Jimenez, A.; Ocampo, O.; Berrio, L.V.; Pineda, M.F.; Lopez-Guzman, J. Physical-mechanical characterization of coffee fruits Coffea arabica L. var. Castillo classified by a colorimetry approach. Materialia 2022, 21, 11. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, B.; Zheng, T.; Zhao, C.; Shen, X.; Wang, X.; Qiu, M.; Fan, J. Integrating Metabolomics and Proteomics Technologies Provides Insights into the Flavor Precursor Changes at Different Maturity Stages of Arabica Coffee Cherries. Foods 2023, 12, 1432. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Yu, H.; Hu, F.; Fu, X.; Li, Y.; Li, Y.; Yang, Y.; Liu, D.; Li, G.; Shi, R.; et al. A Systematic Analysis of the Correlation between Flavor Active Differential Metabolites and Multiple Bean Ripening Stages of Coffea arabica L. Molecules 2024, 29, 180. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.F.; Chen, J.Y.; Liu, X.C.; Han, Y.C.; Xiao, Y.Y.; Shan, W.; Tang, Y.; Wu, K.Q.; He, J.X.; Lu, W.J. The transcriptional regulatory network mediated by banana (Musa acuminata) dehydration-responsive element binding (MaDREB) transcription factors in fruit ripening. New Phytol. 2017, 214, 762–781. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, K.S.; Grierson, D. A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 2019, 221, 1724–1741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Lang, Z.B.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Wang, Y.Y.; Chen, T.; Qin, G.Z.; Tian, S.P. Current insights into posttranscriptional regulation of fleshy fruit ripening. Plant Physiol. 2023, 192, 1785–1798. [Google Scholar] [CrossRef]
- Zhou, L.L.; Tian, S.P.; Qin, G.Z. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 23. [Google Scholar] [CrossRef]
- Zhou, L.L.; Tang, R.K.; Li, X.J.; Tian, S.P.; Li, B.B.; Qin, G.Z. N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner. Genome Biol. 2021, 22, 32. [Google Scholar] [CrossRef]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Barbazuk, W.B.; Fu, Y.; McGinnis, K.M. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008, 18, 1381–1392. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.F.; Xiao, B.Z. Comparative alternative splicing analysis of two contrasting rice cultivars under drought stress and association of differential splicing genes with drought response QTLs. Euphytica 2018, 214, 16. [Google Scholar] [CrossRef]
- Jia, Z.C.; Yang, X.; Hou, X.X.; Nie, Y.X.; Wu, J. The Importance of a Genome-Wide Association Analysis in the Study of Alternative Splicing Mutations in Plants with a Special Focus on Maize. Int. J. Mol. Sci. 2022, 23, 4201. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xiao, H. Identification of alternative splicing events by RNA sequencing in early growth tomato fruits. BMC Genom. 2015, 16, 13. [Google Scholar] [CrossRef]
- Gupta, V.; Estrada, A.D.; Blakley, I.; Reid, R.; Patel, K.; Meyer, M.D.; Andersen, S.U.; Brown, A.F.; Lila, M.A.; Loraine, A.E. RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience 2015, 4, 22. [Google Scholar] [CrossRef]
- Chen, X.D.; Huang, S.Q.; Jiang, M.Q.; Chen, Y.K.; Xu, X.H.; Zhang, Z.H.; Lin, Y.L.; Lai, Z.X. Genome-wide identification and expression analysis of the SR gene family in longan (Dimocarpus longan Lour.). PloS ONE 2020, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.X.; Zhang, D.D.; Li, Z.W.; Liang, H.Z.; Deng, R.F.; Su, X.G.; Jiang, Y.M.; Duan, X.W. Alternative splicing of MaMYB16L regulates starch degradation in banana fruit during ripening. J. Integr. Plant Biol. 2021, 63, 1341–1352. [Google Scholar] [CrossRef]
- Maillot, P.; Velt, A.; Rustenholz, C.; Butterlin, G.; Merdinoglu, D.; Duchêne, E. Alternative splicing regulation appears to play a crucial role in grape berry development and is also potentially involved in adaptation responses to the environment. BMC Plant Biol. 2021, 21, 20. [Google Scholar] [CrossRef]
- Rawoof, A.; Ahmad, I.; Islam, K.; Momo, J.; Kumar, A.; Jaiswal, V.; Ramchiary, N. Integrated omics analysis identified genes and their splice variants involved in fruit development and metabolites production in Capsicum species. Funct. Integr. Genom. 2022, 22, 1189–1209. [Google Scholar] [CrossRef]
- Deng, H.H.; Xia, H.; Guo, Y.Q.; Liu, X.L.; Lin, L.J.; Wang, J.; Xu, K.F.; Lv, X.L.; Hu, R.P.; Liang, D. Dynamic Changes in Ascorbic Acid Content during Fruit Development and Ripening of Actinidia latifolia (an Ascorbate-Rich Fruit Crop) and the Associated Molecular Mechanisms. Int. J. Mol. Sci. 2022, 23, 5808. [Google Scholar] [CrossRef]
- Zhou, H.; Ren, F.; Wang, X.; Qiu, K.; Sheng, Y.; Xie, Q.; Shi, P.; Zhang, J.; Pan, H. Genome-wide identification and characterization of long noncoding RNAs during peach (Prunus persica) fruit development and ripening. Sci. Rep. 2022, 12, 11044. [Google Scholar] [CrossRef]
- Xanthopoulou, A.; Moysiadis, T.; Bazakos, C.; Karagiannis, E.; Karamichali, I.; Stamatakis, G.; Samiotaki, M.; Manioudaki, M.; Michailidis, M.; Madesis, P. The perennial fruit tree proteogenomics atlas: A spatial map of the sweet cherry proteome and transcriptome. Plant J. 2022, 109, 1319–1336. [Google Scholar] [CrossRef]
- Yu, Y.T.; Liufu, Y.X.; Ren, Y.; Zhang, J.; Li, M.Y.; Tian, S.W.; Wang, J.F.; Liao, S.J.; Gong, G.Y.; Zhang, H.Y.; et al. Comprehensive Profiling of Alternative Splicing and Alternative Polyadenylation during Fruit Ripening in Watermelon (Citrullus lanatus). Int. J. Mol. Sci. 2023, 24, 15333. [Google Scholar] [CrossRef]
- Yan, X.M.; Bai, D.; Song, H.T.; Lin, K.; Pang, E.L. Alternative splicing during fruit development among fleshy fruits. BMC Genom. 2021, 22, 14. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.G.; Bi, X.F.; Liu, H.M.; Fu, X.F.; Li, Y.A.; Yang, Y.; Zhang, X.F.; Wu, R.R.; Li, G.P.; Lv, Y.L.; et al. Transcriptome and carotenoid profiling of different varieties of Coffea arabica provides insights into fruit color formation. Plant Divers. 2022, 44, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.G.; Shi, R.; Fu, X.F.; Li, Y.; Li, G.P.; Yang, Y.; Liu, D.X.; Luo, X.P.; Bi, X.F.; Dong, W. Transcriptome and metabolome profiling provides insight into the regulatory network of fruit coloration in Coffea arabica L. Sci. Hortic. 2024, 326, 14. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Zuo, D.D.; He, G.Q.; Sun, H.T.; Guo, D.L. Methylation-related alternative splicing events in H2O2-treated Kyoho grape berries during development. Sci. Hortic. 2023, 321, 11. [Google Scholar] [CrossRef]
- Montazerinezhad, S.; Solouki, M.; Emamjomeh, A.; Kavousi, K.; Taheri, A.; Shiri, Y. Transcriptomic analysis of alternative splicing events for different stages of growth and development in Sistan Yaghooti grape clusters. Gene 2024, 896, 12. [Google Scholar] [CrossRef] [PubMed]
- Marquez, Y.; Brown, J.W.S.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Zheng, Y.; Dong, J.; Yu, J.; Yue, J.; Liu, F.; Guo, X.; Huang, S.; Wisniewski, M.; Sun, J.; et al. Comprehensive Transcriptome Profiling Reveals Long Noncoding RNA Expression and Alternative Splicing Regulation during Fruit Development and Ripening in Kiwifruit (Actinidia chinensis). Front. Plant Sci. 2016, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Pfaff, C.; Ehrnsberger, H.F.; Flores-Tornero, M.; Sørensen, B.B.; Schubert, T.; Längst, G.; Griesenbeck, J.; Sprunck, S.; Grasser, M.; Grasser, K.D. ALY RNA-Binding Proteins Are Required for Nucleocytosolic mRNA Transport and Modulate Plant Growth and Development. Plant Physiol. 2018, 177, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive Expression Profile Analysis of the Arabidopsis Hsp70 Gene Family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.F.; Szakonyi, D.; Simpson, C.G.; Barbosa, I.C.R.; Brown, J.W.S.; Baena-González, E.; Duque, P. The Arabidopsis SR45 Splicing Factor, a Negative Regulator of Sugar Signaling, Modulates SNF1-Related Protein Kinase 1 Stability. Plant Cell 2016, 28, 1910–1925. [Google Scholar] [CrossRef] [PubMed]
- Laloum, T.; Carvalho, S.D.; Martin, G.; Richardson, D.N.; Cruz, T.M.D.; Carvalho, R.F.; Stecca, K.L.; Kinney, A.J.; Zeidler, M.; Barbosa, I.C.R.; et al. The SCL30a SR protein regulates ABA-dependent seed traits and germination under stress. Plant Cell Environ. 2023, 46, 2112–2127. [Google Scholar] [CrossRef] [PubMed]
- Durán-Soria, S.; Pott, D.M.; Osorio, S.; Vallarino, J.G. Sugar Signaling During Fruit Ripening. Front. Plant Sci. 2020, 11, 564917. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Chiu, C.H.; Do, Y.Y.; Huang, P.L. Expression Fluctuations of Genes Involved in Carbohydrate Metabolism Affected by Alterations of Ethylene Biosynthesis Associated with Ripening in Banana Fruit. Plants 2020, 9, 1120. [Google Scholar] [CrossRef]
- Liu, C.J.; Yang, N.; Yang, Q.; Ayed, C.; Linforth, R.; Fisk, I.D. Enhancing Robusta coffee aroma by modifying flavour precursors in the green coffee bean. Food Chem. 2019, 281, 8–17. [Google Scholar] [CrossRef]
- Velasquez, S.; Pena, N.; Bohorquez, J.C.; Gutierrez, N.; Sacks, G.L. Volatile and sensory characterization of roast coffees—Effects of cherry maturity. Food Chem. 2019, 274, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Badia, M.B.; Arias, C.L.; Tronconi, M.A.; Maurino, V.G.; Andreo, C.S.; Drincovich, M.F.; Wheeler, M.C.G. Enhanced cytosolic NADP-ME2 activity in A-thaliana affects plant development, stress tolerance and specific diurnal and nocturnal cellular processes. Plant Sci. 2015, 240, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Rylott, E.L.; Gilday, A.D.; Graham, I.A. The Gluconeogenic Enzyme Phosphoenolpyruvate Carboxykinase in Arabidopsis Is Essential for Seedling Establishment. Plant Physiol. 2003, 131, 1834–1842. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.; Schuetz, M.; Lin, B.S.P.; Chanis, C.; Hamberger, B.; Western, T.L.; Ehlting, J.; Samuels, A.L. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 2011, 62, 2063–2077. [Google Scholar] [CrossRef]
- Depuydt, T.; Vandepoele, K. Multi-omics network-based functional annotation of unknown Arabidopsis genes. Plant J. 2021, 108, 1193–1212. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Park, C.H.; Hsu, C.C.; Kim, Y.W.; Ko, Y.W.; Zhang, Z.Z.; Zhu, J.Y.; Hsiao, Y.C.; Branon, T.; Kaasik, K.; et al. Mapping the signaling network of BIN2 kinase using TurboID-mediated biotin labeling and phosphoproteomics. Plant Cell 2023, 35, 975–993. [Google Scholar] [CrossRef] [PubMed]
- Panikashvili, D.; Shi, J.X.; Bocobza, S.; Franke, R.B.; Schreiber, L.; Aharoni, A. The Arabidopsis DSO/ABCG11 Transporter Affects Cutin Metabolism in Reproductive Organs and Suberin in Roots. Mol. Plant. 2010, 3, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, J.; Kojima, M.; Takebayashi, Y.; Uragami, T.; Kiba, T.; Sakakibara, H.; Lee, Y. ABCG11 modulates cytokinin responses in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hwang, J.U.; Lee, M.; Kim, Y.Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP Binding Cassette Protein from Grape Berry, Transports Anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef]
- Camejo, D.; Martí, M.C.; Román, P.; Ortiz, A.; Jiménez, A. Antioxidant System and Protein Pattern in Peach Fruits at Two Maturation Stages. J. Agric. Food Chem. 2010, 58, 11140–11147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kang, J.; Xie, Q.; Gong, J.; Shen, H.; Chen, Y.; Chen, G.; Hu, Z. The basic helix-loop-helix transcription factor bHLH95 affects fruit ripening and multiple metabolisms in tomato. J. Exp. Bot. 2020, 71, 6311–6327. [Google Scholar] [CrossRef] [PubMed]
- Scuffi, D.; Alvarez, C.; Laspina, N.; Gotor, C.; Lamattina, L.; García-Mata, C. Hydrogen Sulfide Generated by L-Cysteine Desulfhydrase Acts Upstream of Nitric Oxide to Modulate Abscisic Acid-Dependent Stomatal Closure. Plant Physiol. 2014, 166, 2065–2076. [Google Scholar] [CrossRef] [PubMed]
- Chaki, M.; de Morales, P.A.; Ruiz, C.; Begara-Morales, J.C.; Barroso, J.B.; Corpas, F.J.; Palma, J.M. Ripening of pepper (Capsicum annuum) fruit is characterized by an enhancement of protein tyrosine nitration. Ann. Bot. 2015, 116, 637–647. [Google Scholar] [CrossRef]
- Li, J.Y.; Khan, Z.U.; Tao, X.Y.; Mao, L.C.; Luo, Z.S.; Ying, T.J. Effects of exogenous auxin on pigments and primary metabolite profile of postharvest tomato fruit during ripening. Sci. Hortic. 2017, 219, 90–97. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Bi, X.; Li, Z.; Fu, X.; Li, Y.; Li, Y.; Yang, Y.; Liu, D.; Li, G.; Dong, W.; et al. Transcriptomic Analysis of Alternative Splicing Events during Different Fruit Ripening Stages of Coffea arabica L. Genes 2024, 15, 459. https://doi.org/10.3390/genes15040459
Yu H, Bi X, Li Z, Fu X, Li Y, Li Y, Yang Y, Liu D, Li G, Dong W, et al. Transcriptomic Analysis of Alternative Splicing Events during Different Fruit Ripening Stages of Coffea arabica L. Genes. 2024; 15(4):459. https://doi.org/10.3390/genes15040459
Chicago/Turabian StyleYu, Haohao, Xiaofei Bi, Zhongxian Li, Xingfei Fu, Yanan Li, Yaqi Li, Yang Yang, Dexin Liu, Guiping Li, Wenjiang Dong, and et al. 2024. "Transcriptomic Analysis of Alternative Splicing Events during Different Fruit Ripening Stages of Coffea arabica L." Genes 15, no. 4: 459. https://doi.org/10.3390/genes15040459
APA StyleYu, H., Bi, X., Li, Z., Fu, X., Li, Y., Li, Y., Yang, Y., Liu, D., Li, G., Dong, W., & Hu, F. (2024). Transcriptomic Analysis of Alternative Splicing Events during Different Fruit Ripening Stages of Coffea arabica L. Genes, 15(4), 459. https://doi.org/10.3390/genes15040459





