MALAT1: A Long Non-Coding RNA with Multiple Functions and Its Role in Processes Associated with Fat Deposition
Abstract
1. Long Non-Coding RNAs
2. Molecular Structure and Expression of MALAT1, a Highly Interesting lncRNA
3. Upstream Regulation of MALAT1 Gene Expression
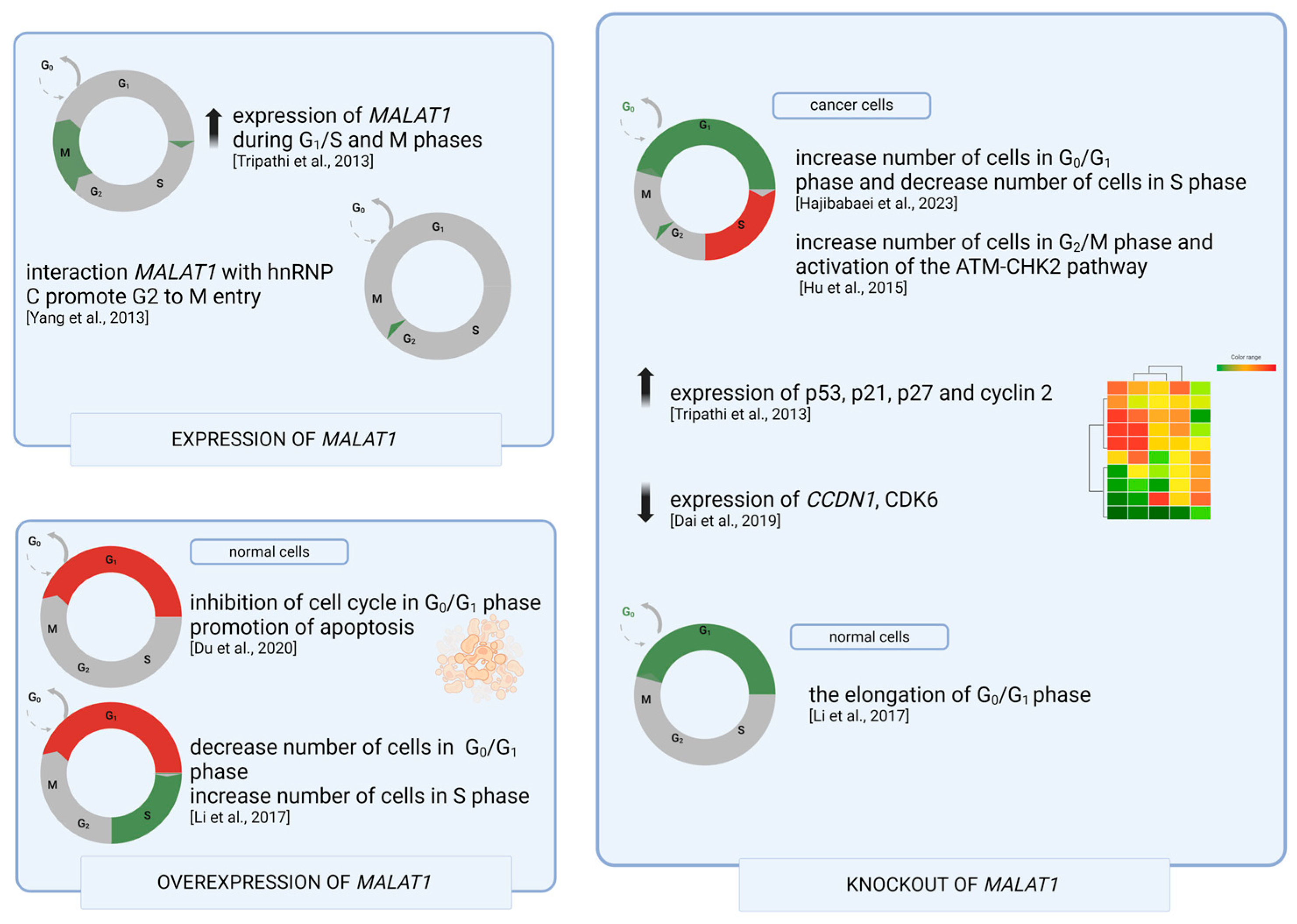
4. MALAT1 in Cell Cycle Regulation
5. Diseases Related to the MALAT1 Gene
6. MALAT1 Is Involved in Molecular Process Related to Carcinogenesis and Cancer Progression
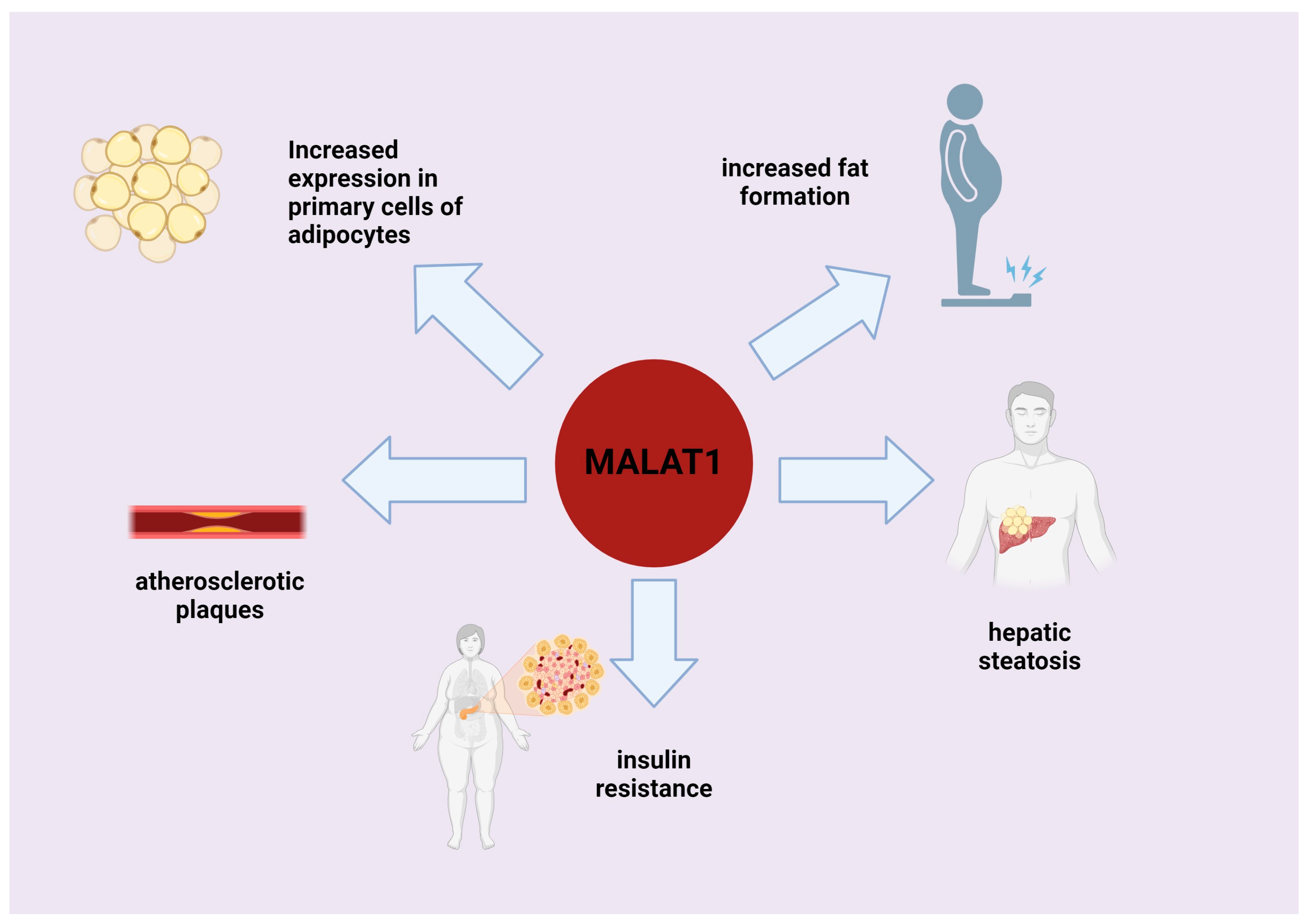
7. Role of MALAT1 in Processes Associated with Fat Deposition and Adipogenesis
8. Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Patraquim, P.; Magny, E.G.; Pueyo, J.I.; Platero, A.I.; Couso, J.P. Translation and natural selection of micropeptides from long non-canonical RNAs. Nat. Commun. 2022, 13, 6515. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, Z. The contribution of databases towards understanding the universe of long non-coding RNAs. Nat. Rev. Mol. Cell Biol. 2023, 24, 601–602. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Long, W.; Yang, L.; Zhao, Y.; Wu, X.; Li, M.; Du, F.; Chen, Y.; Yang, Z.; Wen, Q.; et al. Functional Peptides Encoded by Long Non-Coding RNAs in Gastrointestinal Cancer. Front. Oncol. 2021, 11, 777374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Hu, C.; Yi, H. Shiny transcriptional junk: lncRNA-derived peptides in cancers and immune responses. Life Sci. 2023, 316, 121434. [Google Scholar] [CrossRef] [PubMed]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grützner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.L.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Haerty, W.; Ponting, C.P. Mutations within lncRNAs are effectively selected against in fruitfly but not in human. Genome Biol. 2013, 14, R49. [Google Scholar] [CrossRef]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556. [Google Scholar] [CrossRef]
- Kapusta, A.; Feschotte, C. Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef]
- Liu, L.; Li, Z.; Liu, C.; Zou, D.; Li, Q.; Feng, C.; Jing, W.; Luo, S.; Zhang, Z.; Ma, L. LncRNAWiki 2.0: A knowledgebase of human long non-coding RNAs with enhanced curation model and database system. Nucleic Acids Res. 2022, 50, D190–D195. [Google Scholar] [CrossRef] [PubMed]
- Böhmdorfer, G.; Wierzbicki, A.T. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef]
- Smith, J.E.; Alvarez-Dominguez, J.R.; Kline, N.; Huynh, N.J.; Geisler, S.; Hu, W.; Coller, J.; Baker, K.E. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 2014, 7, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hamblin, M.H.; Yin, K.J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017, 14, 1705. [Google Scholar] [CrossRef]
- Masoumi, F.; Ghorbani, S.; Talebi, F.; Branton, W.G.; Rajaei, S.; Power, C.; Noorbakhsh, F. Malat1 long noncoding RNA regulates inflammation and leukocyte differentiation in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2019, 328, 50–59. [Google Scholar] [CrossRef]
- Gao, Q.; Wang, Y. Long noncoding RNA MALAT1 regulates apoptosis in ischemic stroke by sponging miR-205-3p and modulating PTEN expression. Am. J. Transl. Res. 2020, 12, 2738. [Google Scholar] [PubMed]
- Wang, L.; Li, S.; Stone, S.S.; Liu, N.; Gong, K.; Ren, C.; Sun, K.; Zhang, C.; Shao, G. The Role of the lncRNA MALAT1 in Neuroprotection against Hypoxic/Ischemic Injury. Biomolecules 2022, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- Piórkowska, K.; Żukowski, K.; Ropka-Molik, K.; Tyra, M. New long-non coding RNAs related to fat deposition based on pig model. Ann. Anim. Sci. 2022, 22, 1211–1224. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, R.; Zhu, S.; Li, X.; Li, H.; Yu, H.; Li, K. Systematic Identification and Molecular Characteristics of Long Noncoding RNAs in Pig Tissues. Biomed. Res. Int. 2017, 2017, 6152582. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Freier, S.M.; Spector, D.L. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 2008, 135, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Bulkley, D.; Wang, J.; Valenstein, M.L.; Yario, T.A.; Steitz, T.A.; Steitz, J.A. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014, 21, 633–640. [Google Scholar] [CrossRef]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885. [Google Scholar] [CrossRef]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 Long Non-Coding RNA: Functional Implications. Non-Coding RNA 2020, 6, 22. [Google Scholar] [CrossRef]
- Lin, S.; Lin, Y.; Nery, J.R.; Urich, M.A.; Breschi, A.; Davis, C.A.; Dobin, A.; Zaleski, C.; Beer, M.A.; Chapman, W.C.; et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc. Natl. Acad. Sci. USA 2014, 111, 17224–17229. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, L.; Shen, S.; Li, J.; Lu, H.; Mo, W.; Dang, Y.; Luo, D.; Chen, G.; Feng, Z. Sp1 cooperates with Sp3 to upregulate MALAT1 expression in human hepatocellular carcinoma. Oncol. Rep. 2015, 34, 2403–2412. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, Y.; Trovik, J.; Sun, K.; Zhou, L.; Jiang, P.; Lau, T.S.; Hoivik, E.A.; Salvesen, H.B.; Sun, H.; et al. A novel wnt regulatory axis in endometrioid endometrial cancer. Cancer Res. 2014, 74, 5103–5117. [Google Scholar] [CrossRef] [PubMed]
- Wenger, R.; Lelli, A.; Nolan, K.; Santambrogio, S.; Marti, H.; Hoogewijs, D.; Frew, I.; Goncalves, A.F.; Schonenberger, M.; Guinot, A. Induction of long noncoding RNA MALAT1 in hypoxic mice. Hypoxia 2015, 3, 45. [Google Scholar] [CrossRef] [PubMed]
- Sallé-Lefort, S.; Miard, S.; Nolin, M.A.; Boivin, L.; Paré, M.È.; Debigaré, R.; Picard, F. Hypoxia upregulates Malat1 expression through a CaMKK/AMPK/HIF-1α axis. Int. J. Oncol. 2016, 49, 1731–1736. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Sun, B.; Li, H.; Xu, Y.; Liu, Y.; Liu, X.; Lu, L.; Li, J.; Wang, Q.; Wei, S.; et al. A MALAT1/HIF-2α feedback loop contributes to arsenite carcinogenesis. Oncotarget 2016, 7, 5769–5787. [Google Scholar] [CrossRef]
- Fuschi, P.; Carrara, M.; Voellenkle, C.; Garcia-Manteiga, J.M.; Righini, P.; Maimone, B.; Sangalli, E.; Villa, F.; Specchia, C.; Picozza, M.; et al. Central role of the p53 pathway in the noncoding-RNA response to oxidative stress. Aging 2017, 9, 2559–2586. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.Y.; Wang, J.H.; Wang, J.L.; Ma, C.X.; Wang, X.C.; Liu, F.S. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genom. 2015, 16, 676. [Google Scholar] [CrossRef]
- Sun, H.; Lin, D.C.; Cao, Q.; Pang, B.; Gae, D.D.; Lee, V.K.M.; Lim, H.J.; Doan, N.; Said, J.W.; Gery, S.; et al. Identification of a Novel SYK/c-MYC/MALAT1 Signaling Pathway and Its Potential Therapeutic Value in Ewing Sarcoma. Clin. Cancer Res. 2017, 23, 4376–4387. [Google Scholar] [CrossRef]
- Sun, D.; Li, X.; He, Y.; Li, W.; Wang, Y.; Wang, H.; Jiang, S.; Xin, Y. YAP1 enhances cell proliferation, migration, and invasion of gastric cancer in vitro and in vivo. Oncotarget 2016, 7, 81062–81076. [Google Scholar] [CrossRef]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef]
- Radhakrishnan, S.K.; Lee, C.S.; Young, P.; Beskow, A.; Chan, J.Y.; Deshaies, R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 2010, 38, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Manasanch, E.E.; Orlowski, R.Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2017, 14, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.H.; Hsu, C.M.; Hsiao, S.Y.; Hsiao, H.H. Pathogenic Mechanisms of Myeloma Bone Disease and Possible Roles for NRF2. Int. J. Mol. Sci. 2020, 21, 6723. [Google Scholar] [CrossRef] [PubMed]
- Kuo, I.Y.; Wu, C.C.; Chang, J.M.; Huang, Y.L.; Lin, C.H.; Yan, J.J.; Sheu, B.S.; Lu, P.J.; Chang, W.L.; Lai, W.W.; et al. Low SOX17 expression is a prognostic factor and drives transcriptional dysregulation and esophageal cancer progression. Int. J. Cancer 2014, 135, 563–573. [Google Scholar] [CrossRef]
- Wang, X.; Li, M.; Wang, Z.; Han, S.; Tang, X.; Ge, Y.; Zhou, L.; Zhou, C.; Yuan, Q.; Yang, M. Silencing of Long Noncoding RNA MALAT1 by miR-101 and miR-217 Inhibits Proliferation, Migration, and Invasion of Esophageal Squamous Cell Carcinoma Cells. J. Biol. Chem. 2014, 290, 3925–3935. [Google Scholar] [CrossRef]
- Koshimizu, T.; Fujiwara, Y.; Sakai, N.; Shibata, K.; Tsuchiya, H. Oxytocin stimulates expression of a noncoding RNA tumor marker in a human neuroblastoma cell line. Life Sci. 2010, 86, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014, 20, 1531–1541. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chang, L.; Chen, Z.; Liu, Z.; Wang, Y.; Ye, Q. The role of lncRNA MALAT1 in the regulation of hepatocyte proliferation during liver regeneration. Int. J. Mol. Med. 2017, 39, 347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xue, J.; Peng, F. The regulatory activities of MALAT1 in the development of bone and cartilage diseases. Front. Endocrinol. 2022, 13, 1054827. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, D.-Y.; Sha, W.-G.; Shen, L.; Lu, G.-Y. Long non-coding RNA MALAT1 interacts with transcription factor Foxo1 to regulate SIRT1 transcription in high glucose-induced HK-2 cells injury. Biochem. Biophys. Res. Commun. 2018, 503, 849–855. [Google Scholar] [CrossRef]
- Leucci, E.; Patella, F.; Waage, J.; Holmstrøm, K.; Lindow, M.; Porse, B.; Kauppinen, S.; Lund, A.H. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 2013, 3, 2535. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, K.; Berneman, Z.N.; Van Bockstaele, D.R. Cell cycle and apoptosis. Cell Prolif. 2003, 36, 165–175. [Google Scholar] [CrossRef]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013, 9, e1003368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, J.; Guo, L.; Peng, M. Long non-coding RNA MALAT1 regulates cell proliferation, invasion and apoptosis by modulating the Wnt signaling pathway in squamous cell carcinoma. Am. J. Transl. Res. 2021, 13, 9233. [Google Scholar]
- Dai, X.; Liu, L.; Liang, Z.; Guo, K.; Xu, S.; Wang, H. Silencing of lncRNA MALAT1 inhibits cell cycle progression via androgen receptor signaling in prostate cancer cells. Pathol. Res. Pract. 2019, 215, 712–721. [Google Scholar] [CrossRef]
- Sadasivam, S.; Duan, S.; DeCaprio, J.A. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012, 26, 474–489. [Google Scholar] [CrossRef]
- Yang, F.; Yi, F.; Han, X.; Du, Q.; Liang, Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013, 587, 3175–3181. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, G.; Wang, X.; Wang, Y.; Wang, K. Functions and mechanisms of lncRNA MALAT1 in cancer chemotherapy resistance. Biomark. Res. 2023, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Wang, J.; Zang, S.; Mao, X.; Du, Y. Long non-coding RNA MALAT1 suppresses the proliferation and migration of endothelial progenitor cells in deep vein thrombosis by regulating the Wnt/β-catenin pathway. Exp. Ther. Med. 2020, 20, 3138–3146. [Google Scholar] [CrossRef]
- Hajibabaei, S.; Nafissi, N.; Azimi, Y.; Mahdian, R.; Rahimi-Jamnani, F.; Valizadeh, V.; Rafiee, M.H.; Azizi, M. Targeting long non-coding RNA MALAT1 reverses cancerous phenotypes of breast cancer cells through microRNA-561-3p/TOP2A axis. Sci. Rep. 2023, 13, 8652. [Google Scholar] [CrossRef]
- Hu, L.; Wu, Y.; Tan, D.; Meng, H.; Wang, K.; Bai, Y.; Yang, K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 7. [Google Scholar] [CrossRef] [PubMed]
- Guffanti, A.; Iacono, M.; Pelucchi, P.; Kim, N.; Soldà, G.; Croft, L.J.; Taft, R.J.; Rizzi, E.; Askarian-Amiri, M.; Bonnal, R.J.; et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genom. 2009, 10, 163. [Google Scholar] [CrossRef]
- Chen, Q.; Su, Y.; He, X.; Zhao, W.; Wu, C.; Zhang, W.; Si, X.; Dong, B.; Zhao, L.; Gao, Y.; et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol. Lett. 2016, 12, 1361–1366. [Google Scholar] [CrossRef]
- Shi, D.; Zhang, Y.; Lu, R.; Zhang, Y. The long non-coding RNA MALAT1 interacted with miR-218 modulates choriocarcinoma growth by targeting Fbxw8. Biomed. Pharmacother. 2018, 97, 543–550. [Google Scholar] [CrossRef]
- Wang, J.; Su, L.; Chen, X.; Li, P.; Cai, Q.; Yu, B.; Liu, B.; Wu, W.; Zhu, Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed. Pharmacother. 2014, 68, 557–564. [Google Scholar] [CrossRef]
- Qi, Y.; Ooi, H.S.; Wu, J.; Chen, J.; Zhang, X.; Tan, S.; Yu, Q.; Li, Y.Y.; Kang, Y.; Li, H.; et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget 2016, 7, 12693–12703. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Hu, Z.Y.; Xu, C.; Xie, L.Y.; Wang, X.Y.; Chen, S.Y.; Li, Z.G. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim. Biophys. Acta 2015, 1852, 166–174. [Google Scholar] [CrossRef]
- Zhang, T.H.; Liang, L.Z.; Liu, X.L.; Wu, J.N.; Su, K.; Chen, J.Y.; Zheng, Q.Y.; Huang, H.Z.; Liao, G.Q. Long non-coding RNA MALAT1 interacts with miR-124 and modulates tongue cancer growth by targeting JAG1. Oncol. Rep. 2017, 37, 2087–2094. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Huang, Z.; Li, X. Long Non-Coding RNA MALAT1 Interacts With miR-204 to Modulate Human Hilar Cholangiocarcinoma Proliferation, Migration, and Invasion by Targeting CXCR4. J. Cell. Biochem. 2017, 118, 3643–3653. [Google Scholar] [CrossRef]
- Han, Y.; Liu, Y.; Nie, L.; Gui, Y.; Cai, Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology 2013, 81, 209.e1–209.e7. [Google Scholar] [CrossRef]
- Liu, X.; Lv, R.; Zhang, L.; Xu, G.; Bi, J.; Gao, F.; Zhang, J.; Xue, F.; Wang, F.; Wu, Y.; et al. Long noncoding RNA expression profile of infantile hemangioma identified by microarray analysis. Tumour Biol. 2016, 37, 15977–15987. [Google Scholar] [CrossRef]
- Huang, J.K.; Ma, L.; Song, W.H.; Lu, B.Y.; Huang, Y.B.; Dong, H.M.; Ma, X.K.; Zhu, Z.Z.; Zhou, R. LncRNA-MALAT1 Promotes Angiogenesis of Thyroid Cancer by Modulating Tumor-Associated Macrophage FGF2 Protein Secretion. J. Cell. Biochem. 2017, 118, 4821–4830. [Google Scholar] [CrossRef]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell. Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Tao, Z.F.; Li, X.M.; Zhang, H.; Yao, J.; Jiang, Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 941–951. [Google Scholar] [CrossRef]
- Zhou, R.M.; Wang, X.Q.; Yao, J.; Shen, Y.; Chen, S.N.; Yang, H.; Jiang, Q.; Yan, B. Identification and characterization of proliferative retinopathy-related long noncoding RNAs. Biochem. Biophys. Res. Commun. 2015, 465, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef]
- Zhuo, Y.; Zeng, Q.; Zhang, P.; Li, G.; Xie, Q.; Cheng, Y. Functional polymorphism of lncRNA MALAT1 contributes to pulmonary arterial hypertension susceptibility in Chinese people. Clin. Chem. Lab. Med. 2017, 55, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ding, L.; Wang, L.; Zhao, Y.; Sun, Z.; Karnes, R.J.; Zhang, J.; Huang, H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 2015, 6, 41045. [Google Scholar] [CrossRef]
- Zheng, X.; Ren, J.; Peng, B.; Ye, J.; Wu, X.; Zhao, W.; Li, Y.; Chen, R.; Gong, X.; Bai, C.; et al. MALAT1 overexpression promotes the growth of colon cancer by repressing β-catenin degradation. Cell. Signal. 2020, 73, 109676. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.H.; Yang, W.I.; Kim, S.J.; Yoon, S.O. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget 2017, 8, 31305. [Google Scholar] [CrossRef]
- Arratia, F.; Fierro, C.; Blanco, A.; Fuentes, S.; Nahuelquen, D.; Montecino, M.; Rojas, A.; Aguilar, R. Selective Concurrence of the Long Non-Coding RNA MALAT1 and the Polycomb Repressive Complex 2 to Promoter Regions of Active Genes in MCF7 Breast Cancer Cells. Curr. Issues Mol. Biol. 2023, 45, 4735–4748. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liang, L.; Ouyang, K.; Li, Z.; Yi, X. MALAT1 induces tongue cancer cells’ EMT and inhibits apoptosis through Wnt/β-catenin signaling pathway. J. Oral Pathol. Med. 2017, 46, 98–105. [Google Scholar] [CrossRef]
- Li, Z.; Wu, Y.; Ma, C.; Nie, S.; Mao, X.; Shi, Y. Down-regulation of c-Myc expression inhibits the invasion of bile duct carcinoma cells. Cell Biol. Int. 2011, 35, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gill, A.J.M.; Issacs, J.D.; Atmore, B.; Johns, A.; Delbridge, L.W.; Lai, R.; McMullen, T.P.W. The Wnt/β-catenin pathway drives increased cyclin D1 levels in lymph node metastasis in papillary thyroid cancer. Hum. Pathol. 2012, 43, 1044–1050. [Google Scholar] [CrossRef]
- Yang, X.; Du, X.; Sun, L.; Zhao, X.; Zhu, J.; Li, G.; Tian, J.; Li, X.; Wang, Z. SULT2B1b promotes epithelial-mesenchymal transition through activation of the β-catenin/MMP7 pathway in hepatocytes. Biochem. Biophys. Res. Commun. 2019, 510, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Zeilstra, J.; Joosten, S.P.J.; Dokter, M.; Verwiel, E.; Spaargaren, M.; Pals, S.T. Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 2008, 68, 3655–3661. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Feng, S.J.; Qiu, S.; Shao, N.; Zheng, J.H. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3176–3184. [Google Scholar]
- Peng, N.; He, J.; Li, J.; Huang, H.; Huang, W.; Liao, Y.; Zhu, S. Long noncoding RNA MALAT1 inhibits the apoptosis and autophagy of hepatocellular carcinoma cell by targeting the microRNA-146a/PI3K/Akt/mTOR axis. Cancer Cell Int. 2020, 20, 165. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.L.; Liu, W.; Tian, L.H.; Chai, T.T.; Liu, Y.; Feng, Z.; Fu, H.Y.; Zhou, H.R.; Shen, J.Z. Upregulation of long non-coding RNA MALAT-1 confers poor prognosis and influences cell proliferation and apoptosis in acute monocytic leukemia. Oncol. Rep. 2017, 38, 1353–1362. [Google Scholar] [CrossRef]
- Guo, F.; Li, Y.; Liu, Y.; Wang, J.; Li, Y.; Li, G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim. Biophys. Sin. 2010, 42, 224–229. [Google Scholar] [CrossRef]
- Ji, D.G.; Guan, L.Y.; Luo, X.; Ma, F.; Yang, B.; Liu, H.Y. Inhibition of MALAT1 sensitizes liver cancer cells to 5-flurouracil by regulating apoptosis through IKKα/NF-κB pathway. Biochem. Biophys. Res. Commun. 2018, 501, 33–40. [Google Scholar] [CrossRef]
- Si, Y.; Yang, Z.; Ge, Q.; Yu, L.; Yao, M.; Sun, X.; Ren, Z.; Ding, C. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell. Mol. Biol. Lett. 2019, 24, 50. [Google Scholar] [CrossRef]
- Xie, H.; Liao, X.; Chen, Z.; Fang, Y.; He, A.; Zhong, Y.; Gao, Q.; Xiao, H.; Li, J.; Huang, W.; et al. LncRNA MALAT1 Inhibits Apoptosis and Promotes Invasion by Antagonizing miR-125b in Bladder Cancer Cells. J. Cancer 2017, 8, 3803. [Google Scholar] [CrossRef]
- Sun, Y.; Qin, B. Long noncoding RNA MALAT1 regulates HDAC4-mediated proliferation and apoptosis via decoying of miR-140-5p in osteosarcoma cells. Cancer Med. 2018, 7, 4584–4597. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lu, X.; Yu, R. Lncrna malat1 promotes emt process and cisplatin resistance of oral squamous cell carcinoma via pi3k/akt/m-tor signal pathway. Onco. Targets. Ther. 2020, 13, 4049–4061. [Google Scholar] [CrossRef]
- Zhang, Z.C.; Tang, C.; Dong, Y.; Zhang, J.; Yuan, T.; Tao, S.C.; Li, X.L. Targeting the long noncoding RNA MALAT1 blocks the pro-angiogenic effects of osteosarcoma and suppresses tumour growth. Int. J. Biol. Sci. 2017, 13, 1398. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; Lamond, A.I. Nuclear Speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P.; Shilo, A.; Mogilevsky, A.; Stein, I.; Pikarsky, E.; Nevo, Y.; Benyamini, H.; Elgavish, S.; Zong, X.; Prasanth, K.V.; et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017, 77, 1155–1167. [Google Scholar] [CrossRef]
- Miao, H.; Wu, F.; Li, Y.; Qin, C.; Zhao, Y.; Xie, M.; Dai, H.; Yao, H.; Cai, H.; Wang, Q.; et al. MALAT1 modulates alternative splicing by cooperating with the splicing factors PTBP1 and PSF. Sci. Adv. 2022, 8, eabq7289. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Hu, K.; Qiu, J.; Hu, Y.; Zhou, M.; Zhang, S. Elevated long noncoding RNA MALAT-1 expression is predictive of poor prognosis in patients with breast cancer: A meta-analysis. Biosci. Rep. 2020, 40, 20200215. [Google Scholar] [CrossRef]
- Schmidt, L.H.; Spieker, T.; Koschmieder, S.; Humberg, J.; Jungen, D.; Bulk, E.; Hascher, A.; Wittmer, D.; Marra, A.; Hillejan, L.; et al. The Long Noncoding MALAT-1 RNA Indicates a Poor Prognosis in Non-small Cell Lung Cancer and Induces Migration and Tumor Growth. J. Thorac. Oncol. 2011, 6, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.X.; Wang, H.J.; Li, X.R.; Li, T.; Su, G.; Yang, P.; Wu, J.W. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumor Biol. 2015, 36, 3355–3359. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Jin, E.H.; Chang, I.A.; Kang, H.; Lee, S.I.; Sung, J.K. Association of long noncoding RNA MALAT1 polymorphisms with gastric cancer risk in Korean individuals. Mol. Genet. Genom. Med. 2020, 8, e1541. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Chen, L.; Tian, H.; Li, J.; Zhang, M.; Cao, Q.; Zhang, W.; Chen, S.; Shi, L. Effect of MALAT1 Polymorphisms on Papillary Thyroid Cancer in a Chinese Population. J. Cancer 2019, 10, 5714. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.T.; Chang, J.H.; Lee, H.L.; Yang, Y.C.; Su, S.C.; Lin, C.L.; Yang, S.F.; Chien, M.H. Genetic Variants of lncRNA MALAT1 Exert Diverse Impacts on the Risk and Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma. J. Clin. Med. 2019, 8, 1406. [Google Scholar] [CrossRef]
- Zheng, L.; Rong, L.; Cheng, Z. Association between LncRNA MALAT1 Polymorphisms and Cancer Risk: A Meta-Analysis Based on7007 Cases and 8791 Controls. 2020. Available online: https://www.researchsquare.com/article/rs-42022/v1 (accessed on 15 November 2023).
- Wu, S.; Sun, H.; Wang, Y.; Yang, X.; Meng, Q.; Yang, H.; Zhu, H.; Tang, W.; Li, X.; Aschner, M.; et al. MALAT1 rs664589 Polymorphism Inhibits Binding to miR-194-5p, Contributing to Colorectal Cancer Risk, Growth, and Metastasis. Cancer Res. 2019, 79, 5432–5441. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, S.; He, Y.; Li, X.; Zhu, Y.; Lin, X.; Chen, L.; Zhao, Y.; Niu, L.; Zhang, S.; et al. LncRNA-Mediated Adipogenesis in Different Adipocytes. Int. J. Mol. Sci. 2022, 23, 7488. [Google Scholar] [CrossRef]
- Powell-Wiley, T.M.; Poirier, P.; Burke, L.E.; Després, J.P.; Gordon-Larsen, P.; Lavie, C.J.; Lear, S.A.; Ndumele, C.E.; Neeland, I.J.; Sanders, P.; et al. Obesity and Cardiovascular Disease: A Scientific Statement From the American Heart Association. Circulation 2021, 143, E984–E1010. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, P.; Weiskirchen, R. The Role of Obesity in Type 2 Diabetes Mellitus—An Overview. Int. J. Mol. Sci. 2024, 25, 1882. [Google Scholar] [CrossRef]
- Shariq, O.A.; Mckenzie, T.J. Obesity-related hypertension: A review of pathophysiology, management, and the role of metabolic surgery. Gland Surg. 2020, 9, 80. [Google Scholar] [CrossRef]
- Avgerinos, K.I.; Spyrou, N.; Mantzoros, C.S.; Dalamaga, M. Obesity and cancer risk: Emerging biological mechanisms and perspectives. Metabolism 2019, 92, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, S.N.; Nicholson, T.; Tsintzas, K.; Jones, S.W. Involvements of long noncoding RNAs in obesity-associated inflammatory diseases. Obes. Rev. 2021, 22, e13156. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, X.; Qin, J.; Liu, S.; Zhao, R.; Yu, T.; Chu, G.; Yang, G.; Pang, W. Comparative Analysis of Long Noncoding RNAs Expressed during Intramuscular Adipocytes Adipogenesis in Fat-Type and Lean-Type Pigs. J. Agric. Food Chem. 2018, 66, 12122–12130. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Tai, L.; Zhang, L.; Chu, Y.; Li, Y.; Zhou, L. Comparative analyses of long non-coding RNA in lean and obese pig. Oncotarget 2017, 8, 41440–41450. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cai, R.; Wang, Y.; Zhao, R.; Qin, J.; Pang, W. A newly identified LNcRNA LncIMF4 controls adipogenesis of porcine intramuscular preadipocyte through attenuating autophagy to inhibit lipolysis. Animals 2020, 10, 926. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.; Miard, S.; Boivin, L.; Sallé-Lefort, S.; Picard, F. Loss of Malat1 does not modify age- or diet-induced adipose tissue accretion and insulin resistance in mice. PLoS ONE 2018, 13, e0196603. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Patel, N.A.; Chalfant, C.E.; Cooper, D.R. Ceramide synthesis regulates biogenesis and packaging of exosomal MALAT1 from adipose derived stem cells, increases dermal fibroblast migration and mitochondrial function. Cell Commun. Signal. 2023, 21, 221. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Shen, L.; Zhan, Z.; Liu, Y.; Zhang, C.; Guo, R.; Luo, Y.; Xie, Z.; Feng, Y.; Wu, G. The long noncoding RNA MALAT1 modulates adipose loss in cancer-associated cachexia by suppressing adipogenesis through PPAR-γ. Nutr. Metab. 2021, 18, 27. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.S.; Carter, G.; El Bassit, G.; Patel, A.A.; Cooper, D.R.; Murr, M.; Patel, N.A. Adipose-derived stem cells from lean and obese humans show depot specific differences in their stem cell markers, exosome contents and senescence: Role of protein kinase C delta (PKCδ) in adipose stem cell niche. Stem Cell Investig. 2016, 3, 2. [Google Scholar] [CrossRef]
- Ebrahimi, R.; Toolabi, K.; Jannat Ali Pour, N.; Mohassel Azadi, S.; Bahiraee, A.; Zamani-Garmsiri, F.; Emamgholipour, S. Adipose tissue gene expression of long non-coding RNAs; MALAT1, TUG1 in obesity: Is it associated with metabolic profile and lipid homeostasis-related genes expression? Diabetol. Metab. Syndr. 2020, 12, 36. [Google Scholar] [CrossRef]
- Rasaei, N.; Gholami, F.; Samadi, M.; Shiraseb, F.; Khadem, A.; Yekaninejad, M.S.; Emamgholipour, S.; Mirzaei, K. The interaction between MALAT1 and TUG1 with dietary fatty acid quality indices on visceral adiposity index and body adiposity index. Sci. Rep. 2024, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tan, L.; Yao, J.; Yang, L. Long non-coding RNA MALAT1 regulates cholesterol accumulation in ox-LDL-induced macrophages via the microRNA-17-5p/ABCA1 axis. Mol. Med. Rep. 2020, 21, 1761. [Google Scholar] [CrossRef] [PubMed]
- Van Solingen, C.; Scacalossi, K.R.; Moore, K.J. Long noncoding RNAs in lipid metabolism. Curr. Opin. Lipidol. 2018, 29, 224. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piórkowska, K.; Zygmunt, K.; Hunter, W.; Wróblewska, K. MALAT1: A Long Non-Coding RNA with Multiple Functions and Its Role in Processes Associated with Fat Deposition. Genes 2024, 15, 479. https://doi.org/10.3390/genes15040479
Piórkowska K, Zygmunt K, Hunter W, Wróblewska K. MALAT1: A Long Non-Coding RNA with Multiple Functions and Its Role in Processes Associated with Fat Deposition. Genes. 2024; 15(4):479. https://doi.org/10.3390/genes15040479
Chicago/Turabian StylePiórkowska, Katarzyna, Karolina Zygmunt, Walter Hunter, and Ksenia Wróblewska. 2024. "MALAT1: A Long Non-Coding RNA with Multiple Functions and Its Role in Processes Associated with Fat Deposition" Genes 15, no. 4: 479. https://doi.org/10.3390/genes15040479
APA StylePiórkowska, K., Zygmunt, K., Hunter, W., & Wróblewska, K. (2024). MALAT1: A Long Non-Coding RNA with Multiple Functions and Its Role in Processes Associated with Fat Deposition. Genes, 15(4), 479. https://doi.org/10.3390/genes15040479





