Genomic and Transcriptome Analysis Reveals the Biosynthesis Network of Cordycepin in Cordyceps militaris
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Cultivation
2.2. Determination of Cordycepin
2.3. Genomic DNA Extraction and Purification
2.4. De Novo Genome Sequencing, Assembly, and Annotation
2.5. Whole-Genome Resequencing and Data Analysis
2.6. Orthology Comparisons and Population Phylogenetic Analysis
2.7. Linkage Disequilibrium Analysis and Selection Elimination Analysis
2.8. Transcriptome Sequencing and Differential Expression Analysis
2.9. qRT-PCR Validation
3. Results
3.1. Analysis of Cordycepin Production Regularity
3.2. Genome Component of C. militaris
3.3. Genome Annotation
3.4. Prediction of the Secondary Metabolite Synthesis Genes
3.5. Genome Mutation
3.6. Analysis of Phylogenetic Relationships and Population Structure
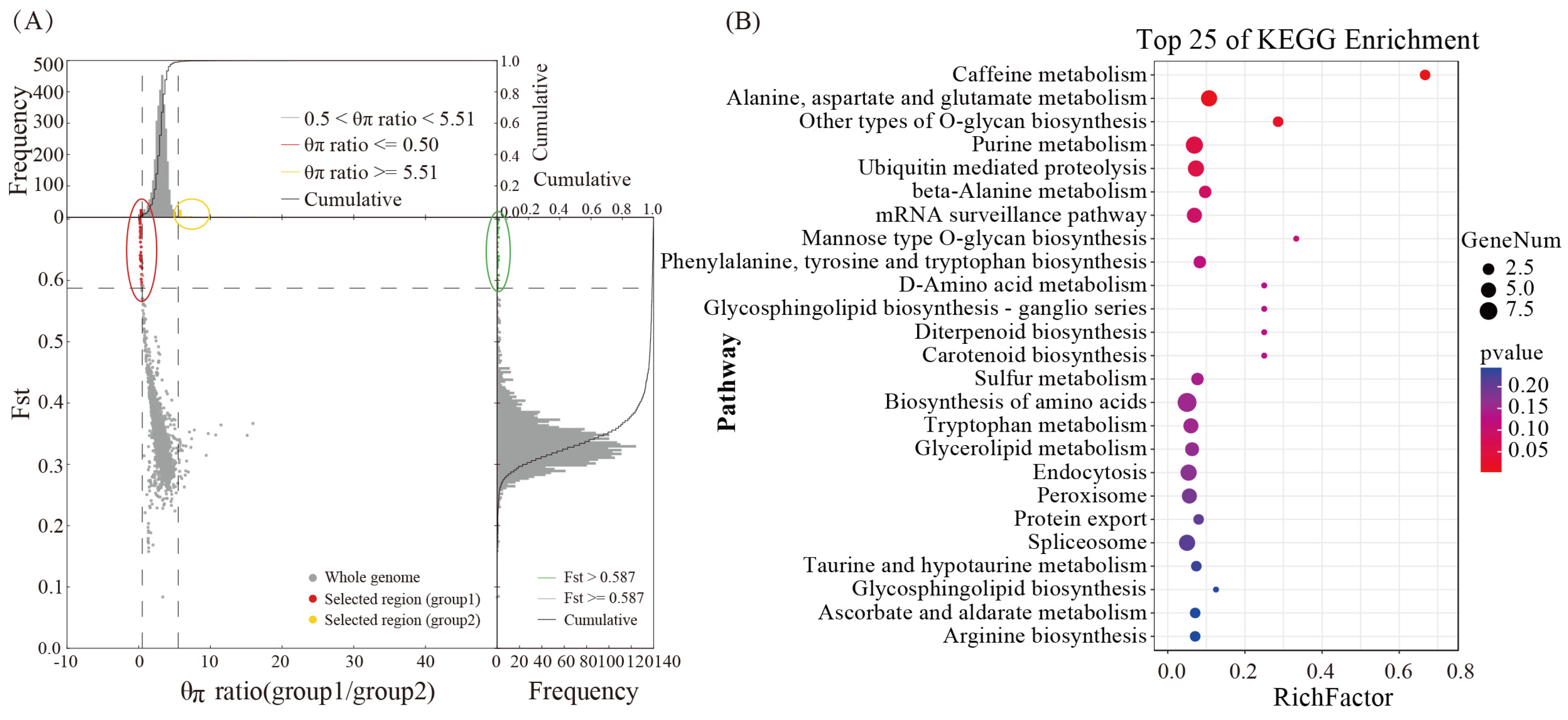
3.7. Analysis of the Selective Elimination of Functional Genes Related to Cordycepin Synthesis
3.8. Transcriptome Studies of Cordycepin Anabolism
3.9. Prediction of Hypothetical Metabolic Pathways of Cordycepin
3.10. Differential Gene Expression Measured Based on qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qu, S.L.; Li, S.S.; Li, D.; Zhao, P.J. Metabolites and Their Bioactivities from the Genus Cordyceps. Microorganisms 2022, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Yue, K.; Ye, M.; Zhou, Z.; Sun, W.; Lin, X. The genus Cordyceps: A chemical and pharmacological review. J. Pharm. Pharmacol. 2013, 65, 474–493. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, T.; Huang, A.; Shen, Y.Y.; Zhang, X.Y.; Song, W.J. New Insights into the Biosynthesis of Typical Bioactive Components in the Traditional Chinese Medicinal Fungus Cordyceps militaris. Front. Bioeng. Biotechnol. 2021, 9, 801721. [Google Scholar] [CrossRef] [PubMed]
- Basith, M.; Madelin, M.F. Studies on the production of perithecial stromata by Cordyceps militaris in artificial culture. Can. J. Bot. 1968, 46, 473–480. [Google Scholar] [CrossRef]
- Wongsa, B.; Raethong, N.; Chumnanpuen, P.; Wong, E.J.; Laoteng, K.; Vongsangnak, W. Alternative metabolic routes in channeling xylose to cordycepin production of Cordyceps militaris identified by comparative transcriptome analysis. Genomics 2020, 112, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wei, T.; Xue, L.; Zheng, Q.W.; Ye, Z.W.; Zou, Y.; Yang, Y.; Yun, F.; Guo, L.Q.; Lin, J.F. Transcriptome analysis reveals the flexibility of cordycepin network in Cordyceps militaris activated by L-Alanine addition. Front. Microbiol. 2020, 11, 577. [Google Scholar] [CrossRef]
- Xia, Y.; Luo, F.; Shang, Y.; Chen, P.L.; Lu, Y.Z.; Wang, C.S. Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.S.; Liang, Z.C.; Wang, Y.C.; Liang, C.H. Effect of light-emitting diodes on the production of cordycepin, mannitol and adenosine in solid-state fermented rice by Cordyceps militaris. J. Food Compos. Anal. 2017, 60, 51–56. [Google Scholar] [CrossRef]
- Thananusak, R.; Laoteng, K.; Raethong, N.; Zhang, Y.; Vongsangnak, W. Metabolic responses of carotenoid and cordycepin biosynthetic pathways in Cordyceps militaris under light-programming exposure through genome-wide transcriptional analysis. Biology 2020, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.J.; Zhu, Y.; Li, Y.M.; Li, J.; Sun, Y.F.; Shen, H.T. Effect of strain separated parts, solid-state substrate and light condition on yield and bioactive compounds of Cordyceps militaris fruiting bodies. J. Food 2018, 16, 916–922. [Google Scholar] [CrossRef]
- Lou, H.W.; Ye, Z.W.; Yu, Y.H.; Lin, J.F.; Guo, L.Q.; Chen, B.X.; Tang, H.B.; Chen, L.R.; Yun, F. The efficient genetic transformation of Cordyceps militaris by using mononuclear protoplasts. Sci. Hortic. 2019, 243, 307–313. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.D.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.B.; Fang, X.D.; Shi, Z.B.; Li, Y.G.; Li, S.T.; Shan, G.; Kristiansen, K.; et al. De novo assembly of human genomes with massiverly parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMark S: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifis in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Belton, J.M.; Mccord, R.; Gibcus, J.; Naumova, N.; Zhan, Y.; Dekker, J. Hi-C: A comprehensive technique to capture the conformation of genomes. Methods 2012, 58, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Keller, O.; Gunduz, L.; Hayse, A.; Waack, S. AUGUSTUS: Ab initio prediction of alternative transcripts. Nuleic Acids Res. 2006, 34, 435–439. [Google Scholar] [CrossRef]
- Sun, L.; Fu, Y.; Yang, Y.; Wang, X.X.; Cui, W.J.; Li, D.; Yuan, X.H.; Zhang, Z.W.; Fu, Y.P.; Li, Y. Genomic Analyses Reveal Evidence of Independent Evolution, Demographic History, and Extreme Environment Adaptation of Tibetan Plateau Agaricus bisporus. Front. Microbiol. 2019, 10, 1786. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 4, 4.10.1–4.10.14. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Sam, G.J.; Eddy, S.R.; et al. Rfam: Updates to the RNA familiyes database. Nucleic Acids Res. 2009, 37, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.; Chen, Y.; He, F.C. Integrated nr database in protein annotation system and its localization. Comput. Ind. Eng. 2006, 32, 71–74. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Li, R.; Li, R.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Bathke, J.; Luhken, G. OVarFlow. A resource optimized GATK 4 based open source Variant calling workFlow. BMC Bioinform. 2021, 22, 402. [Google Scholar] [CrossRef]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigen analysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef]
- Lee, T.H.; Guo, H.; Wang, X.Y.; Kim, C.; Paterson, A.H. SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genom. 2014, 15, 162. [Google Scholar] [CrossRef]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martinez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Sun, L.; Yin, X.; Gao, M.; Zhao, Y.T.; Jia, P.S.; Yuan, X.H.; Fu, Y.P.; Li, Y. Pleurotus eryngii Genomes Reveal Evolution and Adaptation to the Gobi Desert Environment. Front. Microbiol. 2009, 10, 2024. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Kofler, R.; Pandey, R.V.; Schlotterer, C. PoPoolation2: Identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 2011, 27, 3435–3436. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Kenneth, J.L.; Thomas, D.S. Analysis of relative gene expression data using real-time quantitative PCR and 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.-M.; Luangsa-Ard, J.; Shresth, B.; Spatafora, J.W. Phylogenetic Classification of Cordyceps and the Clavicipitaceous Fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef]
- Gu, C.M.; Zhang, D.B.; Zhai, W.J.; Zhang, H.P. Research progress on Cordyceps militaris polysaccharides. Food Biosci. 2022, 45, 101503. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, Z.J.; Zhou, X.W. Chinese Cordyceps: Bioactive Components, Antitumor Effects and Underlying Mechanism-A Review. Molecules 2022, 27, 6576. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, P.; Xu, L.; Xu, D.; Hu, W.D.; Cheng, Y.; Yang, S.L. Construction of Cordycepin High-Production Strain and Optimization of Culture Conditions. Curr. Microbiol. 2023, 80, 12. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.Y.; Yang, H.; Wang, C.; Liu, H.H.; Lu, X.Y.; Tian, Y. Microbial synthesis of cordycepin, current systems and future perspectives. Trends Food Sci. Technol. 2023, 132, 162–170. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, Y.; Liu, L.; Feng, J.H.; Zhang, T.C.; Qin, S.; Zhao, X.Y.; Wang, C.X.; Li, D.M.; Han, W.; et al. Study of the whole genome, methylome and transcriptome of Cordyceps militaris. Sci. Rep. 2019, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.H.; Hu, X.; Zhang, S.W.; Zheng, H.J.; Huang, Y.; Zhou, Y.; Wang, S.Y.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef] [PubMed]
- Kramer, G.J.; Nodwell, J.R. Chromosome level assembly and secondary metabolite potential of the parasitic fungus Cordyceps militaris. BMC Genom. 2017, 18, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Bushly, K.E.; Raja, R.; Jaiswal, P.; Cumbie, J.S.; Nonogaki, M.; Boyd, A.E.; Owensby, C.A.; Knaus, B.J.; Elser, J.; Miller, D.; et al. The genome of tolypocladium inflatum: Evolution, organization and expression of the cyclosporin biosynthetic gene cluster. PLoS Genet. 2013, 9, e1000349. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, Y.; Khatri, I.; Subramanian, S.; Shenoy, B.D. Genome sequence, comparative analysis and evolutionary insights into Chitinases of Entomopathogenic fungus Hirsutella thompsonii. Genome Biol. Evol. 2015, 25, 916–946. [Google Scholar] [CrossRef]
- Larriba, E.; Jaime, M.; Carbonell, J.; Conesa, A.; Dopazo, J.; Nislow, C.; Nieto, J.M.; Lopez-Llorca, L.V. Sequencing and functional analysis of the genome of a nematode egg-parastic fungus, Pochonia chlamydosporia. Fungal Genet Biol. 2014, 65, 69–80. [Google Scholar] [CrossRef]
- Sun, L.; Li, S.D.; Li, Y.; Wang, L.; Pu, X.M.; Ge, Y.P.; Na, Q.; Li, W.H.; Cheng, X.H. Population genetics provides insights into the important agronomic traits of Ganoderma cultivation varieties in China. Gene 2024, 893, 147938. [Google Scholar] [CrossRef]
- Zhang, J.C.; Shen, N.; Li, C.; Xiang, X.J.; Liu, G.L.; Gui, Y.; Patev, S.; Hibbett, D.S.; Barry, K.; Anderopoulos, W.; et al. Population genomics provides insights into the genetic basis of adaptive evolution in the mushroom-forming fungus Lentinula edodes. J. Adv. Res. 2022, 3, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Song, X.; Ren, Y.; Wang, M.; Guo, C.J.; Guo, D.; Gu, Y.C.; Li, Y.Z.; Cao, Z.X.; Deng, Y.; et al. Anti-inflammatory effects of cordycepin: A review. Phytother. Res. 2021, 35, 1284–1297. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yan, H.; Zeng, B.; Hu, Z. Research Progress on Cordycepin Synthesis and Methods for Enhancement of Cordycepin Production in Cordyceps militaris. Bioengineering 2022, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.H.; Xia, Y.L.; Zheng, P.; Shi, S.H.; Wang, C.S. Developmental stage-specific gene expression profiling for a medicinal fungus Cordyceps militaris. Mycology 2010, 1, 25–66. [Google Scholar] [CrossRef]
- Kang, N.; Lee, H.H.; Park, I.; Seo, Y.S. Development of High cordycepin-producing Cordyceps militaris Strains. Mycobiology 2017, 45, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Yoon, D.H.; Shrestha, B.; Choi, H.K.; Sung, G.H. Metabolomic profiling reveals enrichment of cordycepin in senescence process of Cordyceps militaris fruit bodier. J. Microbiol. 2019, 57, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Galina, Z.; Oleinik, E.; Shibaev, M.; Ignatev, K. Adenosine-Metabolizing Enzymes, Adenosine Kinase and Adenosine Deaminase, in Cancer. Biomolecules 2022, 12, 418. [Google Scholar] [CrossRef] [PubMed]
- Cicero, A.F.G.; Fogacci, F.; Di, M.V.; Angeloni, C.; Giovannini, M.; Borghi, C. Purine Metabolism Dysfunctions: Experimental Methods of Detection and Diagnostic Potential. Int. J. Mol. Sci. 2023, 24, 7027. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Li, X.; Sun, L.; Feng, Y.T.; Sa, F.P.; Ge, Y.P.; Yang, S.D.; Liu, Y.; Li, W.H.; et al. Transcriptome and metabolome profiling unveils the mechanisms of naphthalene acetic acid in promoting cordycepin synthesis in Cordyceps militaris. Front. Nutr. 2023, 10, 1104446. [Google Scholar] [CrossRef]
- Yang, L.Y.; Li, G.L.; Chai, Z.; Gong, Q.; Guo, J.Q. Synthesis of cordycepin: Current scenario and future perspectives. Fungal Genet. Biol. 2020, 143, 103431. [Google Scholar] [CrossRef]
- Long, L.K.; Liu, Z.; Wang, Y.Z.; Lin, Q.Y.; Ding, S.J.; Li, C.H.; Deng, C.Y. High-level production of cordycepin by the xylose-utilizing Cordyceps militaris strain 147 in an optimized medium. Bioresour. Technol. 2023, 388, 129742. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.Z.; Liu, M.R.; Lei, C.; Zheng, X.J.; Wang, Y. Effects of selenium and light wavelengths on liquid culture of Cordyceps militaris Link. Appl. Biochem. Biotechnol. 2012, 166, 2030–2036. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, M.M.; Guo, S.P.; Guo, S.P.; Dong, C.H. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 2016, 100, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Ma, J.C.; Xue, Y.; Shen, J.W.; Wang, X.L.; Ma, B.J. The influence of vitamin B1, B6 and 2,4-D on the production of cordycepin in the liquid fermentation of Cordyceps militaris. Mycosystema 2014, 33, 477–482. [Google Scholar] [CrossRef]




| Species Information | C. militaris |
|---|---|
| Total counts of scaffold sequences: | 8 |
| Total length of scaffold sequences: | 30,963,950 bp |
| Scaffold N50 | 4,735,179 bp |
| Scaffold N90 | 1,818,051 bp |
| GC content (%) | 52.69 |
| N Length | 0 bp |
| N content (%) | 0 |
| Gene Number | 9781 |
| Gene density | 0.3 |
| Gene average length | 1527 |
| Exon number per Gene | 2.84 |
| Exon average length | 537 |
| Exon GC percent (%) | 58.92 |
| Type | Genome Strain Name | ||
|---|---|---|---|
| CMS19 | AMS333216 | CM01 | |
| T1PKS | 8 | 6 | 6 |
| NRPS | 7 | 7 | 7 |
| NRPS-like | 5 | 6 | 4 |
| Terpene | 4 | 3 | 4 |
| T1PKS, NRPS | 2 | 3 | 2 |
| NRPS, T1PKS | 4 | 2 | 4 |
| indole | 1 | 0 | 0 |
| fungal-RiPP | 1 | 0 | 0 |
| NRPS, NRPS-like | 0 | 1 | 1 |
| Total | 32 | 28 | 28 |
| Genes | Annotation | Log2FC | 2−△△ct |
|---|---|---|---|
| g5449 | adenylate kinase | 0.78 | 0.63 |
| g3376 | adenosine kinase | 1.77 | 1.22 |
| g4827 | oxidoreductase domain-containing protein | 1.38 | 1.13 |
| g4826 | Phosphoribosyl aminoimidazole carboxylase | 0.72 | 0.57 |
| g4825 | adenylate kinase | 0.70 | 1.05 |
| g4824 | ABC multidrug transporter | −0.19 | 0.55 |
| g9123 | adenylate kinase | −1.50 | 0.16 |
| g6539 | adenylosuccinate lyase | 0.74 | 0.71 |
| g1229 | adenylosuccinate synthetase | 1.94 | 4.03 |
| g2310 | adenylosuccinate synthetase | −2.55 | 0.03 |
| g2322 | hypothetical protein | 2.19 | 1.88 |
| g2847 | adenosine deaminase | 1.46 | 1.34 |
| g3195 | adenosine deaminase | 0.85 | 0.49 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, L.; Li, J.; Guo, L.; Zhang, S.; Chen, F.; Zhu, W.; Li, Y. Genomic and Transcriptome Analysis Reveals the Biosynthesis Network of Cordycepin in Cordyceps militaris. Genes 2024, 15, 626. https://doi.org/10.3390/genes15050626
Chai L, Li J, Guo L, Zhang S, Chen F, Zhu W, Li Y. Genomic and Transcriptome Analysis Reveals the Biosynthesis Network of Cordycepin in Cordyceps militaris. Genes. 2024; 15(5):626. https://doi.org/10.3390/genes15050626
Chicago/Turabian StyleChai, Linshan, Jianmei Li, Lingling Guo, Shuyu Zhang, Fei Chen, Wanqin Zhu, and Yu Li. 2024. "Genomic and Transcriptome Analysis Reveals the Biosynthesis Network of Cordycepin in Cordyceps militaris" Genes 15, no. 5: 626. https://doi.org/10.3390/genes15050626
APA StyleChai, L., Li, J., Guo, L., Zhang, S., Chen, F., Zhu, W., & Li, Y. (2024). Genomic and Transcriptome Analysis Reveals the Biosynthesis Network of Cordycepin in Cordyceps militaris. Genes, 15(5), 626. https://doi.org/10.3390/genes15050626






