Delving into the Metabolism of Sézary Cells: A Brief Review
Abstract
:1. Introduction: Primary Cutaneous Lymphomas
1.1. Sézary Syndrome
1.2. Therapy for Sézary Syndrome
| Comments | |
|---|---|
| Systemic Therapies | |
| Retinoids derive from vitamin A whose function is to interact with nuclear receptors (retinoic acid (RAR) and retinoic X receptor (RXR)). Bexarotene activates RXRs and induces apoptosis and prevents malignant T cell homing to the skin by downregulating CCR4 and E-selectin [3,28]. |
| IFN-α activates CD8+ T cells and NK cells and suppresses cytokine production from malignant lymphoma cells [29]. |
| Patients’ white blood cells are exposed ex vivo to a photosensitizing agent (8-methoxypsolaren) and then to UVA light. Cells are then reinfused in the patient. The purpose is to induce an immune response against malignant T cells [28]. |
| Mogamulizumab selectively binds to CCR4—which is highly expressed in malignant T cells—and induces antibody-dependent cellular toxicity, thus destroying tumor cells [20,30]. Lacutamab (IPH4102) binds to CD158k, a cell surface marker aberrantly expressed in patients with SS [31]. IPH4102 is designed to deplete CD158k-expressing cells via antibody-dependent cell cytotoxicity and phagocytosis [22,32]. |
| Brentuximab vedotin (BV) is an anti-CD30 antibody attached to monomethyl auristatin E (MMAE), an antitubulin agent. The binding of BV to CD30 and its internalization will allow MMAE to exert its action and inhibit the assemblage of the microtubules, induce cell cycle arrest and cause cell death due to apoptosis of tumor cells [21,33]. |
| Histone deacetylases (HDACs) are epigenetic regulators of gene expression. Their inhibitors (HDACis) are reported to induce upregulation of proapoptotic genes, DNA damage and alterations in the assembly of kinetochores [34,35]. |
| Single or combined agents can be administered and act via various mechanisms including topoisomerase inhibition, blocking DNA synthesis and interference with essential cellular processes [36]. |
2. Energy Metabolism in Cancer Cells
2.1. The Warburg Effect
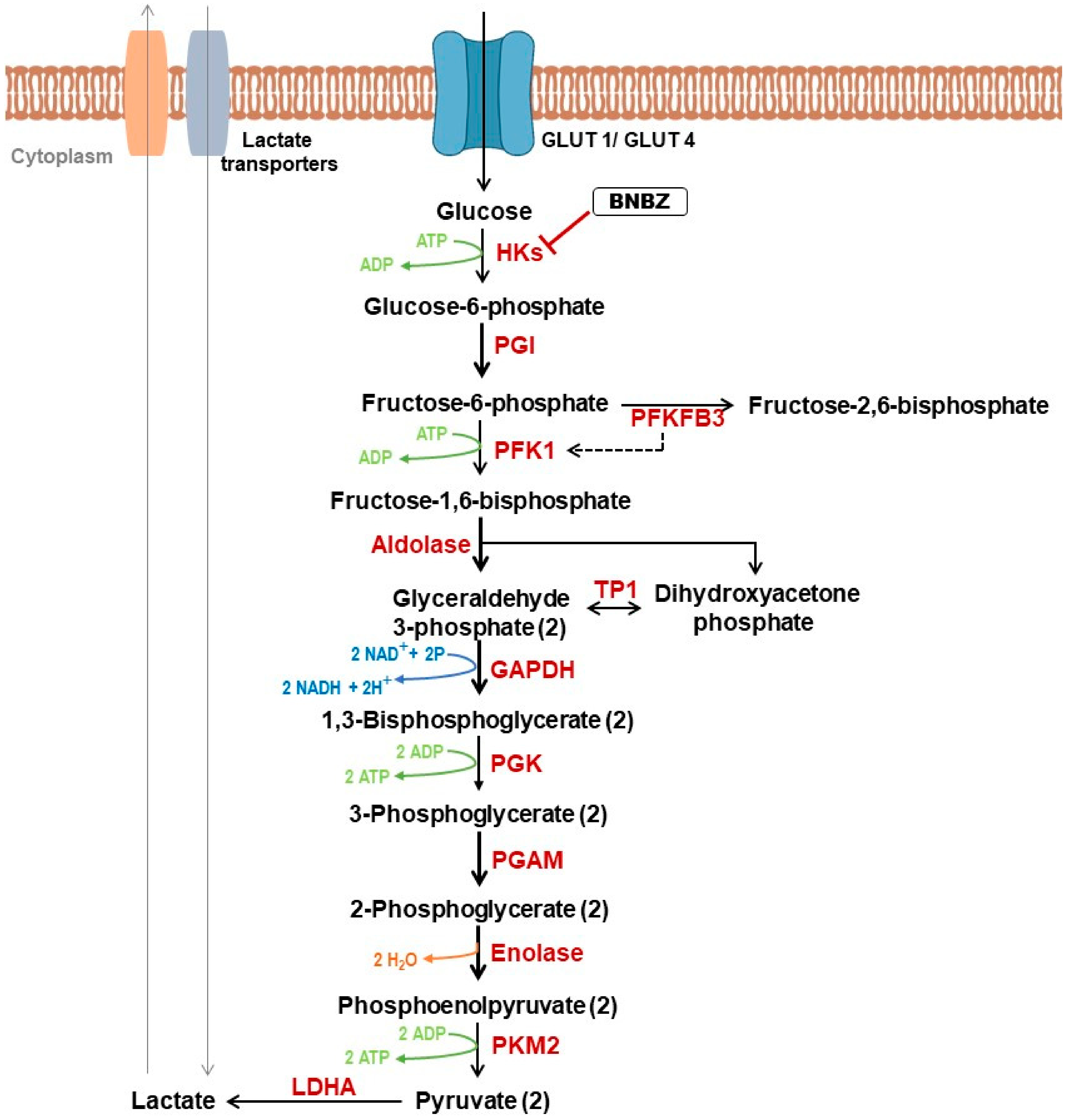
2.2. Role of Glycolysis in Cancer
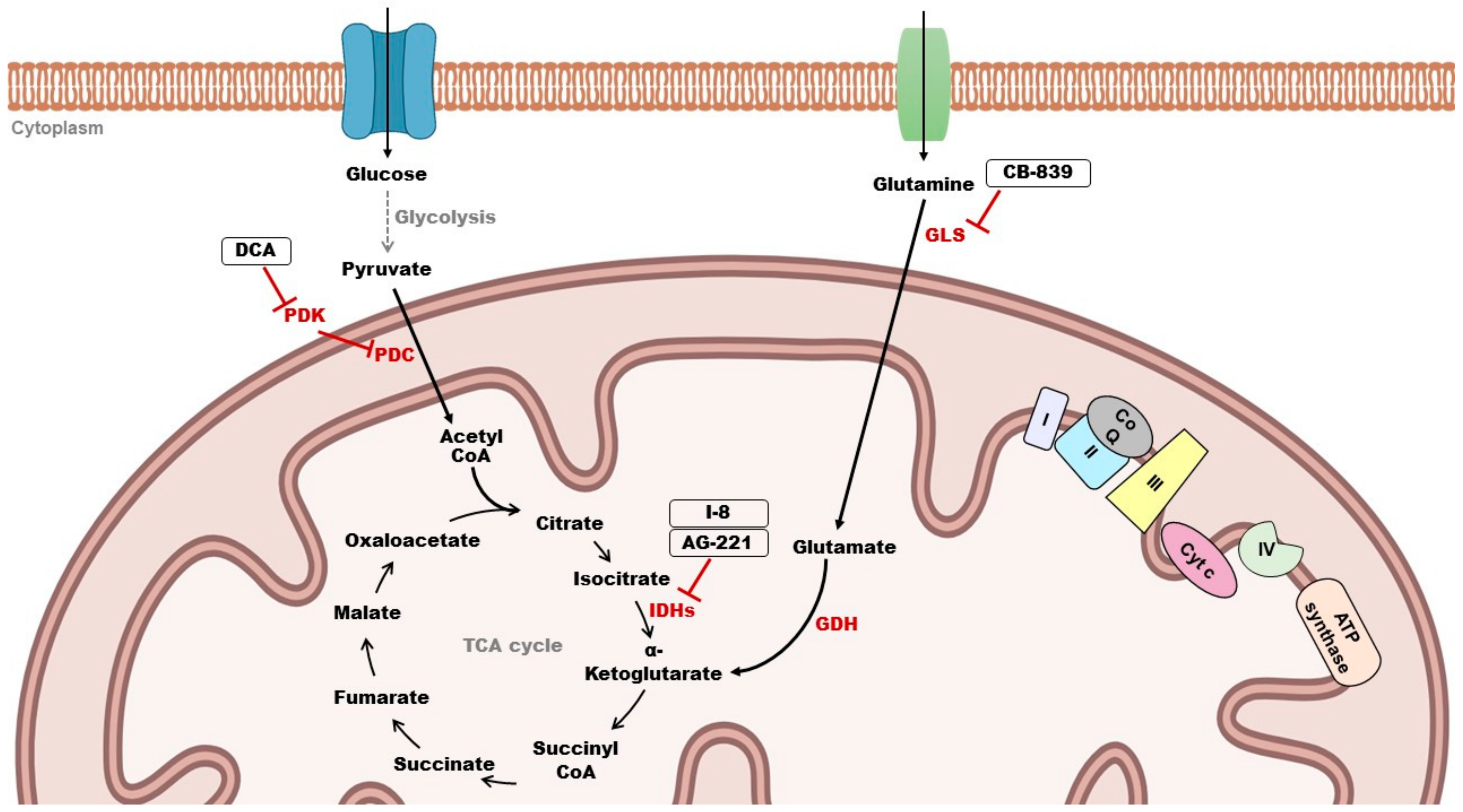
2.3. Role of Mitochondrial Metabolism in Cancer
2.4. Targeting Cancer Metabolism
2.4.1. Targeting Glycolysis
2.4.2. Targeting Mitochondrial Metabolism
3. Metabolism of Sézary Cells: State of the Art
4. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willemze, R.; Cerroni, L.; Kempf, W.; Berti, E.; Facchetti, F.; Swerdlow, S.H.; Jaffe, E.S. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood 2019, 133, 1703–1714. [Google Scholar] [CrossRef] [PubMed]
- Hristov, A.C.; Tejasvi, T.; Wilcox, R.A. Cutaneous T-cell lymphomas: 2021 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2021, 96, 1313–1328. [Google Scholar] [CrossRef] [PubMed]
- García-Díaz, N.; Piris, M.Á.; Ortiz-Romero, P.L.; Vaqué, J.P. Mycosis Fungoides and Sézary Syndrome: An Integrative Review of the Pathophysiology, Molecular Drivers, and Targeted Therapy. Cancers 2021, 13, 1931. [Google Scholar] [CrossRef] [PubMed]
- Shakeri, A. Albert Sézary-The Man, the Cell, and the Syndrome. JAMA Dermatol. 2018, 154, 496–497. [Google Scholar] [CrossRef] [PubMed]
- Najidh, S.; Tensen, C.P.; van der Sluijs-Gelling, A.J.; Teodosio, C.; Cats, D.; Mei, H.; Kuipers, T.B.; Out-Luijting, J.J.; Zoutman, W.H.; van Hall, T.; et al. Improved Sézary cell detection and novel insights into immunophenotypic and molecular heterogeneity in Sézary syndrome. Blood 2021, 138, 2539–2554. [Google Scholar] [CrossRef] [PubMed]
- Cristofoletti, C.; Narducci, M.G.; Russo, G. Sézary Syndrome, recent biomarkers and new drugs. Chin. Clin. Oncol. 2019, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Scarisbrick, J.J.; Hodak, E.; Bagot, M.; Stranzenbach, R.; Stadler, R.; Ortiz-Romero, P.L.; Papadavid, E.; Evison, F.; Knobler, R.; Quaglino, P.; et al. Blood classification and blood response criteria in mycosis fungoides and Sézary syndrome using flow cytometry: Recommendations from the EORTC cutaneous lymphoma task force. Eur. J. Cancer 2018, 93, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Hristov, A.C.; Tejasvi, T.; Wilcox, R.A. Cutaneous T-cell lymphomas: 2023 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2023, 98, 193–209. [Google Scholar] [CrossRef] [PubMed]
- Dobos, G.; De Cevins, C.; Ly Ka So, S.; Jean-Louis, F.; Mathieu, S.; Ram-Wolff, C.; Resche-Rigon, M.; Bensussan, A.; Bagot, M.; Michel, L. The value of five blood markers in differentiating mycosis fungoides and Sézary syndrome: A validation cohort. Br. J. Dermatol. 2021, 185, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Michel, L.; Jean-Louis, F.; Begue, E.; Bensussan, A.; Bagot, M. Use of PLS3, Twist, CD158k/KIR3DL2, and NKp46 gene expression combination for reliable Sézary syndrome diagnosis. Blood 2013, 121, 1477–1478. [Google Scholar] [CrossRef] [PubMed]
- Moins-Teisserenc, H.; Daubord, M.; Clave, E.; Douay, C.; Félix, J.; Marie-Cardine, A.; Ram-Wolff, C.; Maki, G.; Beldjord, K.; Homyrda, L.; et al. CD158k is a reliable marker for diagnosis of Sézary syndrome and reveals an unprecedented heterogeneity of circulating malignant cells. J. Investig. Dermatol. 2015, 135, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.J.; Clark, R.A.; Watanabe, R.; Kupper, T.S. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: A biologic rationale for their distinct clinical behaviors. Blood 2010, 116, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Roelens, M.; Delord, M.; Ram-Wolff, C.; Marie-Cardine, A.; Alberdi, A.; Maki, G.; Homyrda, L.; Bensussan, A.; Bagot, M.; Toubert, A.; et al. Circulating and skin-derived Sézary cells: Clonal but with phenotypic plasticity. Blood 2017, 130, 1468–1471. [Google Scholar] [CrossRef] [PubMed]
- Chevret, E.; Merlio, J.-P. Sézary Syndrome: Translating Genetic Diversity into Personalized Medicine. J. Investig. Dermatol. 2016, 136, 1319–1324. [Google Scholar] [CrossRef] [PubMed]
- Chevret, E.; Andrique, L.; Prochazkova-Carlotti, M.; Ferrer, J.; Cappellen, D.; Laharanne, E.; Idrissi, Y.; Boettiger, A.; Sahraoui, W.; Ruiz, F.; et al. Telomerase functions beyond telomere maintenance in primary cutaneous T-cell lymphoma. Blood 2014, 123, 1850–1859. [Google Scholar] [CrossRef] [PubMed]
- Chebly, A.; Ropio, J.; Peloponese, J.-M.; Poglio, S.; Prochazkova-Carlotti, M.; Cherrier, F.; Ferrer, J.; Idrissi, Y.; Segal-Bendirdjian, E.; Chouery, E.; et al. Exploring hTERT promoter methylation in cutaneous T-cell lymphomas. Mol. Oncol. 2022, 16, 1931–1946. [Google Scholar] [CrossRef] [PubMed]
- Herrera, A.; Cheng, A.; Mimitou, E.P.; Seffens, A.; George, D.; Bar-Natan, M.; Heguy, A.; Ruggles, K.V.; Scher, J.U.; Hymes, K.; et al. Multimodal single-cell analysis of cutaneous T-cell lymphoma reveals distinct subclonal tissue-dependent signatures. Blood 2021, 138, 1456–1464. [Google Scholar] [CrossRef]
- Poglio, S.; Prochazkova-Carlotti, M.; Cherrier, F.; Gros, A.; Laharanne, E.; Pham-Ledard, A.; Beylot-Barry, M.; Merlio, J.-P. Xenograft and cell culture models of Sézary syndrome reveal cell of origin diversity and subclonal heterogeneity. Leukemia 2021, 35, 1696–1709. [Google Scholar] [CrossRef]
- Buus, T.B.; Willerslev-Olsen, A.; Fredholm, S.; Blümel, E.; Nastasi, C.; Gluud, M.; Hu, T.; Lindahl, L.M.; Iversen, L.; Fogh, H.; et al. Single-cell heterogeneity in Sézary syndrome. Blood Adv. 2018, 2, 2115–2126. [Google Scholar] [CrossRef] [PubMed]
- Beylot-Barry, M.; Quereux, G.; Nardin, C.; Duval-Modeste, A.-B.; Dereure, O.; Dalac-Rat, S.; Dobos, G.; Pham-Ledard, A.; Ram-Wolff, C.; D’Incan, M.; et al. Effectiveness of mogamulizumab in patients with Mycosis Fungoides or Sézary syndrome: A multicentre, retrospective, real-world French study. J. Eur. Acad. Dermatol. Venereol. JEADV 2023, 37, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.J.; Haun, P.L.; Samimi, S.S.; Vittorio, C.C.; Villasenor-Park, J.; Barta, S.K.; Landsburg, D.J.; Svoboda, J.; Nasta, S.D.; Schuster, S.J.; et al. Brentuximab Vedotin for Relapsed or Refractory Sézary Syndrome. JAMA Dermatol. 2021, 157, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Bagot, M.; Porcu, P.; Marie-Cardine, A.; Battistella, M.; William, B.M.; Vermeer, M.; Whittaker, S.; Rotolo, F.; Ram-Wolff, C.; Khodadoust, M.S.; et al. IPH4102, a first-in-class anti-KIR3DL2 monoclonal antibody, in patients with relapsed or refractory cutaneous T-cell lymphoma: An international, first-in-human, open-label, phase 1 trial. Lancet Oncol. 2019, 20, 1160–1170. [Google Scholar] [CrossRef] [PubMed]
- de Masson, A.; Beylot-Barry, M.; Ram-Wolff, C.; Mear, J.-B.; Dalle, S.; d’Incan, M.; Ingen-Housz-Oro, S.; Orvain, C.; Abraham, J.; Dereure, O.; et al. Allogeneic transplantation in advanced cutaneous T-cell lymphomas (CUTALLO): A propensity score matched controlled prospective study. Lancet 2023, 401, 1941–1950. [Google Scholar] [CrossRef] [PubMed]
- Latzka, J.; Assaf, C.; Bagot, M.; Cozzio, A.; Dummer, R.; Guenova, E.; Gniadecki, R.; Hodak, E.; Jonak, C.; Klemke, C.-D.; et al. EORTC consensus recommendations for the treatment of mycosis fungoides/Sézary syndrome—Update 2023. Eur. J. Cancer 2023, 195, 113343. [Google Scholar] [CrossRef] [PubMed]
- Beylot-Barry, M.; Dereure, O.; Vergier, B.; Barete, S.; Laroche, L.; Machet, L.; Delfau-Larue, M.-H.; D’Incan, M.; Grange, F.; Ortonne, N.; et al. Management of cutaneous T-cell lymphomas: Recommendations of the French Cutaneous Lymphoma Group. Ann. Dermatol. Venereol. 2010, 137, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Schug, Z.T.; Salvino, J.M.; Dang, C.V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022, 21, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Sethi, T.K.; Montanari, F.; Foss, F.; Reddy, N. How we treat advanced stage cutaneous T-cell lymphoma—Mycosis fungoides and Sézary syndrome. Br. J. Haematol. 2021, 195, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Yoo, E.K.; Cassin, M.; Lessin, S.R.; Rook, A.H. Complete molecular remission during biologic response modifier therapy for Sézary syndrome is associated with enhanced helper T type 1 cytokine production and natural killer cell activity. J. Am. Acad. Dermatol. 2001, 45, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Duvic, M.; Evans, M.; Wang, C. Mogamulizumab for the treatment of cutaneous T-cell lymphoma: Recent advances and clinical potential. Ther. Adv. Hematol. 2016, 7, 171–174. [Google Scholar] [CrossRef]
- Battistella, M.; Leboeuf, C.; Ram-Wolff, C.; Hurabielle, C.; Bonnafous, C.; Sicard, H.; Bensussan, A.; Bagot, M.; Janin, A. KIR3DL2 expression in cutaneous T-cell lymphomas: Expanding the spectrum for KIR3DL2 targeting. Blood 2017, 130, 2900–2902. [Google Scholar] [CrossRef]
- Marie-Cardine, A.; Viaud, N.; Thonnart, N.; Joly, R.; Chanteux, S.; Gauthier, L.; Bonnafous, C.; Rossi, B.; Bléry, M.; Paturel, C.; et al. IPH4102, a humanized KIR3DL2 antibody with potent activity against cutaneous T-cell lymphoma. Cancer Res. 2014, 74, 6060–6070. [Google Scholar] [CrossRef] [PubMed]
- Sanches, J.A.; Cury-Martins, J.; Abreu, R.M.; Miyashiro, D.; Pereira, J. Mycosis fungoides and Sézary syndrome: Focus on the current treatment scenario. An. Bras. Dermatol. 2021, 96, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.T.; Bates, S.; Geskin, L. Current Status of HDAC Inhibitors in Cutaneous T-cell Lymphoma. Am. J. Clin. Dermatol. 2018, 19, 805–819. [Google Scholar] [CrossRef]
- Whittaker, S.J.; Demierre, M.-F.; Kim, E.J.; Rook, A.H.; Lerner, A.; Duvic, M.; Scarisbrick, J.; Reddy, S.; Robak, T.; Becker, J.C.; et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 4485–4491. [Google Scholar] [CrossRef] [PubMed]
- Alpdogan, O.; Kartan, S.; Johnson, W.; Sokol, K.; Porcu, P. Systemic therapy of cutaneous T-cell lymphoma (CTCL). Chin. Clin. Oncol. 2019, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Cancer metabolism: A therapeutic perspective. Nat. Rev. Clin. Oncol. 2017, 14, 11–31. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Ashton, T.M.; McKenna, W.G.; Kunz-Schughart, L.A.; Higgins, G.S. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 2482–2490. [Google Scholar] [CrossRef]
- Greene, J.; Segaran, A.; Lord, S. Targeting OXPHOS and the electron transport chain in cancer; Molecular and therapeutic implications. Semin. Cancer Biol. 2022, 86, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Smolková, K.; Plecitá-Hlavatá, L.; Bellance, N.; Benard, G.; Rossignol, R.; Ježek, P. Waves of gene regulation suppress and then restore oxidative phosphorylation in cancer cells. Int. J. Biochem. Cell Biol. 2011, 43, 950–968. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Kepp, O.; Vander Heiden, M.G.; Kroemer, G. Metabolic targets for cancer therapy. Nat. Rev. Drug Discov. 2013, 12, 829–846. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, C. Oncometabolites in Cancer: Current Understanding and Challenges. Cancer Res. 2021, 81, 2820–2823. [Google Scholar] [CrossRef]
- Villar, V.H.; Merhi, F.; Djavaheri-Mergny, M.; Durán, R.V. Glutaminolysis and autophagy in cancer. Autophagy 2015, 11, 1198–1208. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Verdone, J.E.; Huang, J.; Kahlert, U.D.; Hernandez, J.R.; Torga, G.; Zarif, J.C.; Epstein, T.; Gatenby, R.; McCartney, A.; et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget 2015, 6, 130–143. [Google Scholar] [CrossRef]
- Romero-Garcia, S.; Moreno-Altamirano, M.M.B.; Prado-Garcia, H.; Sánchez-García, F.J. Lactate Contribution to the Tumor Microenvironment: Mechanisms, Effects on Immune Cells and Therapeutic Relevance. Front. Immunol. 2016, 7, 52. [Google Scholar] [CrossRef]
- Hunt, T.K.; Aslam, R.; Hussain, Z.; Beckert, S. Lactate, with oxygen, incites angiogenesis. Adv. Exp. Med. Biol. 2008, 614, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef]
- Baumann, F.; Leukel, P.; Doerfelt, A.; Beier, C.P.; Dettmer, K.; Oefner, P.J.; Kastenberger, M.; Kreutz, M.; Nickl-Jockschat, T.; Bogdahn, U.; et al. Lactate promotes glioma migration by TGF-beta2-dependent regulation of matrix metalloproteinase-2. Neuro-oncology 2009, 11, 368–380. [Google Scholar] [CrossRef]
- Fogal, V.; Richardson, A.D.; Karmali, P.P.; Scheffler, I.E.; Smith, J.W.; Ruoslahti, E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol. Cell. Biol. 2010, 30, 1303–1318. [Google Scholar] [CrossRef]
- LeBleu, V.S.; O’Connell, J.T.; Gonzalez Herrera, K.N.; Wikman, H.; Pantel, K.; Haigis, M.C.; de Carvalho, F.M.; Damascena, A.; Domingos Chinen, L.T.; Rocha, R.M.; et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014, 16, 992–1003, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ippolito, L.; Marini, A.; Cavallini, L.; Morandi, A.; Pietrovito, L.; Pintus, G.; Giannoni, E.; Schrader, T.; Puhr, M.; Chiarugi, P.; et al. Metabolic shift toward oxidative phosphorylation in docetaxel resistant prostate cancer cells. Oncotarget 2016, 7, 61890–61904. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Banerjee, S.; Chellappan, S.; Simon, G.R. Glut-1 antibodies induce growth arrest and apoptosis in human cancer cell lines. Cancer Lett. 2007, 257, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Wang, J.; Yan, W.; Cui, Y.; Chen, Z.; Gao, X.; Wen, X.; Chen, J. GLUT1 regulates cell glycolysis and proliferation in prostate cancer. Prostate 2018, 78, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Yang, F.; Chen, C.; Liu, P.; Ren, Y.; Sun, P.; Wang, Z.; You, Y.; Zeng, Y.-X.; et al. DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis. Nat. Commun. 2021, 12, 5872. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, S.; Li, Y.; Tang, Z.; Kong, W. Hexokinase 2 overexpression promotes the proliferation and survival of laryngeal squamous cell carcinoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 3743–3753. [Google Scholar] [CrossRef] [PubMed]
- Patra, K.C.; Wang, Q.; Bhaskar, P.T.; Miller, L.; Wang, Z.; Wheaton, W.; Chandel, N.; Laakso, M.; Muller, W.J.; Allen, E.L.; et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 2013, 24, 213–228. [Google Scholar] [CrossRef]
- Fan, K.; Fan, Z.; Cheng, H.; Huang, Q.; Yang, C.; Jin, K.; Luo, G.; Yu, X.; Liu, C. Hexokinase 2 dimerization and interaction with voltage-dependent anion channel promoted resistance to cell apoptosis induced by gemcitabine in pancreatic cancer. Cancer Med. 2019, 8, 5903–5915. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Wu, C.; Yang, K.; Yang, Y.; Liu, Y.; Gao, S.; Wang, Q.; Li, C.; Chen, L.; Li, H. Novel selective hexokinase 2 inhibitor Benitrobenrazide blocks cancer cells growth by targeting glycolysis. Pharmacol. Res. 2021, 164, 105367. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, P.; Yarani, R.; Dokaneheifard, S.; Mansouri, K. The emerging role of targeting cancer metabolism for cancer therapy. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2020, 42, 1010428320965284. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, T.; Goldhardt, T.; Edelmann, M.; Rogge, T.; Rauch, K.; Kyuchukov, N.D.; Menck, K.; Bleckmann, A.; Kalucka, J.; Khan, S.; et al. Effects of the Novel PFKFB3 Inhibitor KAN0438757 on Colorectal Cancer Cells and Its Systemic Toxicity Evaluation In Vivo. Cancers 2021, 13, 1011. [Google Scholar] [CrossRef]
- Deng, J.; Cheng, Y.; Li, H.; He, X.; Yu, S.; Ma, J.; Li, X.; Chen, J.; Xiao, H.; Guan, H.; et al. PFKFB3 facilitates cell proliferation and migration in anaplastic thyroid carcinoma via the WNT/β-catenin signaling pathway. Endocrine 2024. [Google Scholar] [CrossRef] [PubMed]
- Zahra, K.; Dey, T.; Ashish; Mishra, S.P.; Pandey, U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front. Oncol. 2020, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.S.; Sharp, P.A. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J. Exp. Med. 2012, 209, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Qiao, T.; Zhuang, X.; Chen, W.; Xing, N.; Zhang, Q. Knockdown of the M2 Isoform of Pyruvate Kinase (PKM2) with shRNA Enhances the Effect of Docetaxel in Human NSCLC Cell Lines In Vitro. Yonsei Med. J. 2016, 57, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, S.-L.; Hu, X.; Tam, K.Y. Targeting Tumor Metabolism for Cancer Treatment: Is Pyruvate Dehydrogenase Kinases (PDKs) a Viable Anticancer Target? Int. J. Biol. Sci. 2015, 11, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Michelakis, E.D.; Sutendra, G.; Dromparis, P.; Webster, L.; Haromy, A.; Niven, E.; Maguire, C.; Gammer, T.-L.; Mackey, J.R.; Fulton, D.; et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci. Transl. Med. 2010, 2, 31ra34. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef] [PubMed]
- Sainero-Alcolado, L.; Liaño-Pons, J.; Ruiz-Pérez, M.V.; Arsenian-Henriksson, M. Targeting mitochondrial metabolism for precision medicine in cancer. Cell Death Differ. 2022, 29, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S. Enasidenib: First Global Approval. Drugs 2017, 77, 1705–1711. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Wu, Y.; Du, H.; Yang, L.; Zhang, Z.; Ma, T.; Li, S.; Yuan, S.; Lu, L.; Zha, X. I-8, a novel inhibitor of mutant IDH1, inhibits cancer progression in vitro and in vivo. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2019, 140, 105072. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Vidal, C.; Dey, S.; Zhang, L. Mitochondria Targeting as an Effective Strategy for Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 3363. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Rohlena, J.; Dong, L.-F.; Ralph, S.J.; Neuzil, J. Anticancer drugs targeting the mitochondrial electron transport chain. Antioxid. Redox Signal. 2011, 15, 2951–2974. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.R.; Sun, Y.; Protopopova, M.; Gera, S.; Bandi, M.; Bristow, C.; McAfoos, T.; Morlacchi, P.; Ackroyd, J.; Agip, A.-N.A.; et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 2018, 24, 1036–1046. [Google Scholar] [CrossRef] [PubMed]
- Bergers, G.; Fendt, S.-M. The metabolism of cancer cells during metastasis. Nat. Rev. Cancer 2021, 21, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Corazao-Rozas, P.; Guerreschi, P.; Jendoubi, M.; André, F.; Jonneaux, A.; Scalbert, C.; Garçon, G.; Malet-Martino, M.; Balayssac, S.; Rocchi, S.; et al. Mitochondrial oxidative stress is the Achille’s heel of melanoma cells resistant to Braf-mutant inhibitor. Oncotarget 2013, 4, 1986–1998. [Google Scholar] [CrossRef]
- Haq, R.; Shoag, J.; Andreu-Perez, P.; Yokoyama, S.; Edelman, H.; Rowe, G.C.; Frederick, D.T.; Hurley, A.D.; Nellore, A.; Kung, A.L.; et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 2013, 23, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A.; Vultur, A.; Bogeski, I.; Wang, H.; Zimmermann, K.M.; Speicher, D.; Körbel, C.; Laschke, M.W.; Gimotty, P.A.; Philipp, S.E.; et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 2013, 23, 811–825. [Google Scholar] [CrossRef]
- Yuan, P.; Ito, K.; Perez-Lorenzo, R.; Del Guzzo, C.; Lee, J.H.; Shen, C.-H.; Bosenberg, M.W.; McMahon, M.; Cantley, L.C.; Zheng, B. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc. Natl. Acad. Sci. USA 2013, 110, 18226–18231. [Google Scholar] [CrossRef] [PubMed]
- Jaune, E.; Rocchi, S. Metformin: Focus on Melanoma. Front. Endocrinol. 2018, 9, 472. [Google Scholar] [CrossRef] [PubMed]
- Baenke, F.; Chaneton, B.; Smith, M.; Van Den Broek, N.; Hogan, K.; Tang, H.; Viros, A.; Martin, M.; Galbraith, L.; Girotti, M.R.; et al. Resistance to BRAF inhibitors induces glutamine dependency in melanoma cells. Mol. Oncol. 2016, 10, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Bishop, E.L.; Gudgeon, N.; Dimeloe, S. Control of T Cell Metabolism by Cytokines and Hormones. Front. Immunol. 2021, 12, 653605. [Google Scholar] [CrossRef]
- Gaydosik, A.M.; Stonesifer, C.J.; Khaleel, A.E.; Geskin, L.J.; Fuschiotti, P. Single-Cell RNA Sequencing Unveils the Clonal and Transcriptional Landscape of Cutaneous T-Cell Lymphomas. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2022, 28, 2610–2622. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Cui, H. Sirtuins and cellular metabolism in cancers. In Sirtuin Biology in Cancer and Metabolic Disease; Elsevier: Amsterdam, The Netherlands, 2021; pp. 195–217. ISBN 978-0-12-822467-0. [Google Scholar]
- Mei, Z.; Zhang, X.; Yi, J.; Huang, J.; He, J.; Tao, Y. Sirtuins in metabolism, DNA repair and cancer. J. Exp. Clin. Cancer Res. CR 2016, 35, 182. [Google Scholar] [CrossRef] [PubMed]
- Finley, L.W.S.; Carracedo, A.; Lee, J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.I.; Cardoso, S.M.; Clish, C.B.; et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 2011, 19, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Torrens-Mas, M.; Hernández-López, R.; Pons, D.-G.; Roca, P.; Oliver, J.; Sastre-Serra, J. Sirtuin 3 silencing impairs mitochondrial biogenesis and metabolism in colon cancer cells. Am. J. Physiol. Cell Physiol. 2019, 317, C398–C404. [Google Scholar] [CrossRef]
- Yakymiv, Y.; Marchisio, S.; Ortolan, E.; Bracci, C.; Senetta, R.; Rumore, M.R.; Tampieri, C.; Fia, M.; Ribero, S.; Funaro, A.; et al. CD39/CD73 dysregulation and adenosine metabolism contribute to T-cell immunosuppression in patients with Sézary syndrome. Blood 2023, 141, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Longhi, M.S.; Robson, S.C.; Bernstein, S.H.; Serra, S.; Deaglio, S. Biological functions of ecto-enzymes in regulating extracellular adenosine levels in neoplastic and inflammatory disease states. J. Mol. Med. 2013, 91, 165–172. [Google Scholar] [CrossRef]
- Leone, R.D.; Emens, L.A. Targeting adenosine for cancer immunotherapy. J. Immunother. Cancer 2018, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Wartewig, T.; Daniels, J.; Schulz, M.; Hameister, E.; Joshi, A.; Park, J.; Morrish, E.; Venkatasubramani, A.V.; Cernilogar, F.M.; Van Heijster, F.H.A.; et al. PD-1 instructs a tumor-suppressive metabolic program that restricts glycolysis and restrains AP-1 activity in T cell lymphoma. Nat. Cancer 2023, 4, 1508–1525. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-Y.; Zhu, Y.; Shen, Y.-Y.; Xu, Q.-Y.; Tang, H.-Y.; Cui, N.-X.; Jiang, L.; Dai, X.-M.; Chen, W.-Q.; Lin, Q.; et al. The role of PD-1 signaling in health and immune-related diseases. Front. Immunol. 2023, 14, 1163633. [Google Scholar] [CrossRef]
- Cristofoletti, C.; Bresin, A.; Picozza, M.; Picchio, M.C.; Monzo, F.; Helmer Citterich, M.; Passarelli, F.; Frezzolini, A.; Scala, E.; Monopoli, A.; et al. Blood and skin-derived Sezary cells: Differences in proliferation-index, activation of PI3K/AKT/mTORC1 pathway and its prognostic relevance. Leukemia 2019, 33, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Carracedo, A.; Pandolfi, P.P. Tenets of PTEN tumor suppression. Cell 2008, 133, 403–414. [Google Scholar] [CrossRef]
- Lizcano, J.M.; Göransson, O.; Toth, R.; Deak, M.; Morrice, N.A.; Boudeau, J.; Hawley, S.A.; Udd, L.; Mäkelä, T.P.; Hardie, D.G.; et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004, 23, 833–843. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cherfan, C.; Chebly, A.; Rezvani, H.R.; Beylot-Barry, M.; Chevret, E. Delving into the Metabolism of Sézary Cells: A Brief Review. Genes 2024, 15, 635. https://doi.org/10.3390/genes15050635
Cherfan C, Chebly A, Rezvani HR, Beylot-Barry M, Chevret E. Delving into the Metabolism of Sézary Cells: A Brief Review. Genes. 2024; 15(5):635. https://doi.org/10.3390/genes15050635
Chicago/Turabian StyleCherfan, Carel, Alain Chebly, Hamid Reza Rezvani, Marie Beylot-Barry, and Edith Chevret. 2024. "Delving into the Metabolism of Sézary Cells: A Brief Review" Genes 15, no. 5: 635. https://doi.org/10.3390/genes15050635
APA StyleCherfan, C., Chebly, A., Rezvani, H. R., Beylot-Barry, M., & Chevret, E. (2024). Delving into the Metabolism of Sézary Cells: A Brief Review. Genes, 15(5), 635. https://doi.org/10.3390/genes15050635







