Karyotype Description and Comparative Chromosomal Mapping of 5S rDNA in 42 Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chromosome and Oligonucleotide Probe Preparation
2.2. FISH
2.3. Karyotype Analysis
3. Results
3.1. Karyotype Analysis Revealed Differences among 42 Species
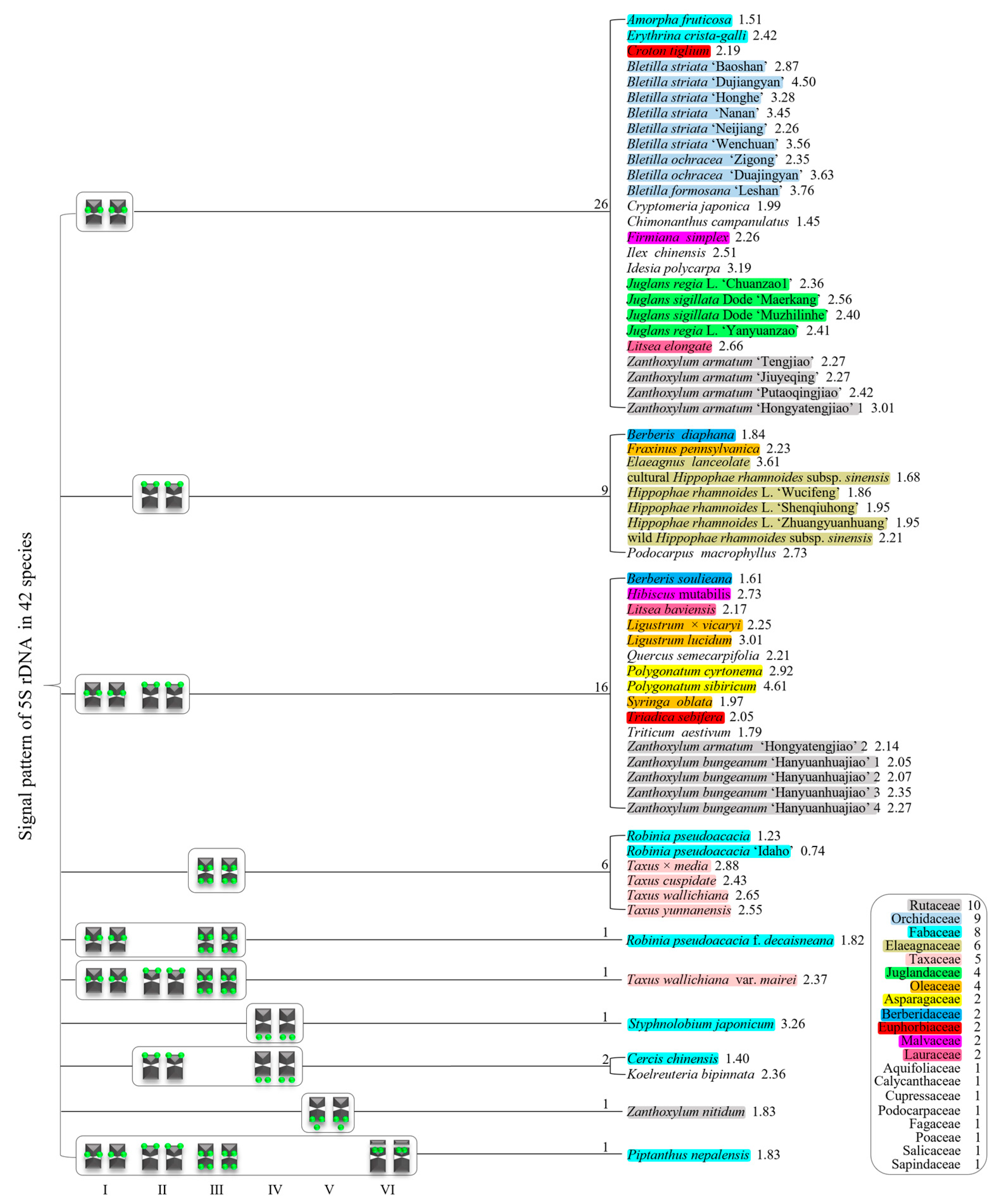
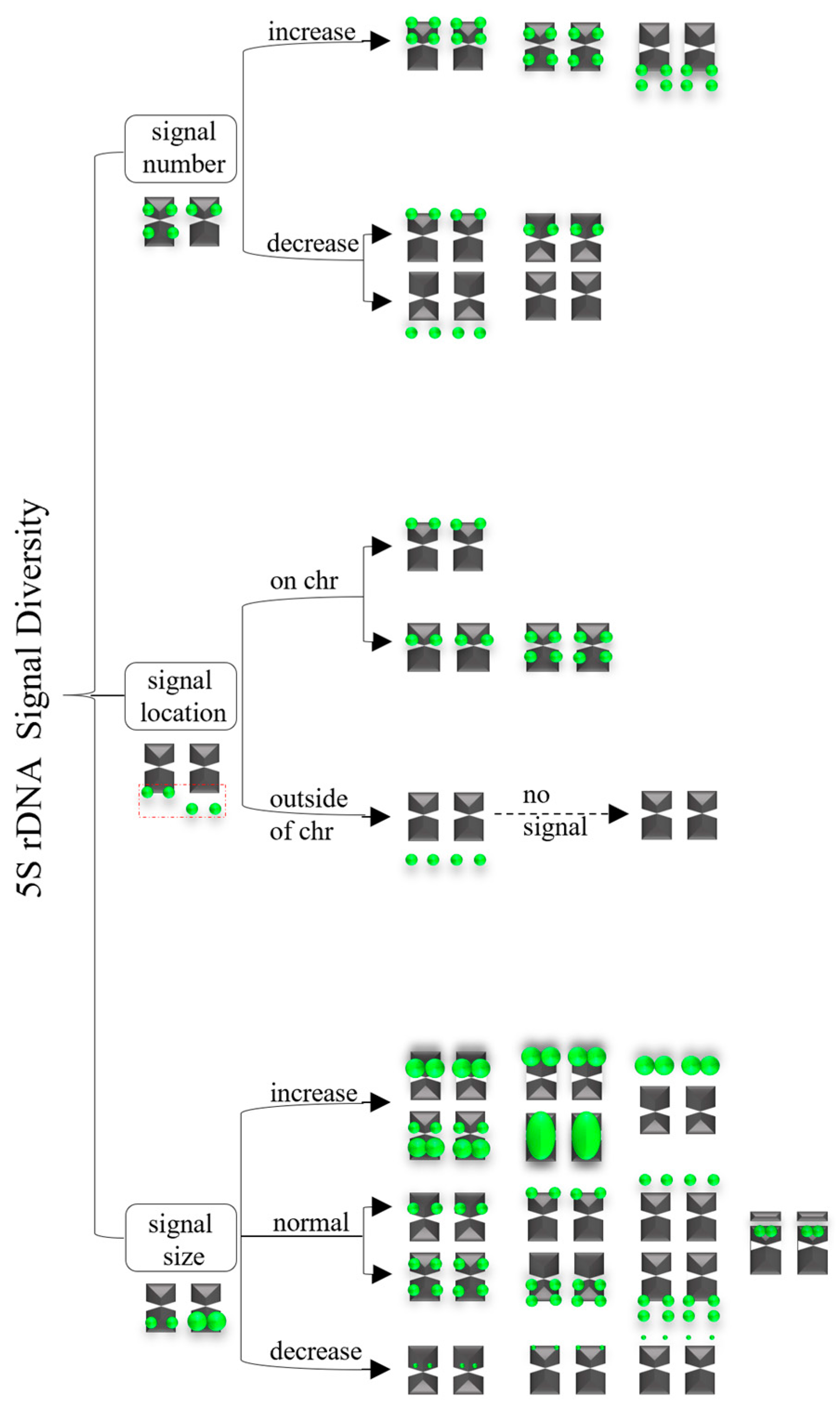
3.2. Diverse Signal Patterns of 5S rDNA Reveal a Complex Genome Architecture
4. Discussion
4.1. Karyotype Analysis of the 42 Species
4.2. Occurrence and Diversity of 5S rDNA in Plants
4.3. Potential Origin of 5S rDNA Diversity in Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slidler, C. Chapter 29—Genome stability and Aging: Causes and Consequences. In Genome Stability: From Virus to Human Application; Kovalchuk, I., Kovalchuk, O., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 511–525. ISBN 9780128033098. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S. Comparative genome organization in plants: From sequence and markers to chromatin and chromosomes. Plant Cell 2000, 12, 617–636. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-González, R.; Gutiérrez, M.L.; Fuentes, I.; Gálvez-Prada, F.; Sochorová, J.; Kovařík, A.; Garcia, S. Release 4.0 of the plant rDNA database: A database on plant ribosomal DNA loci number, their position, and organization: An information source for comparative cytogenetics. Methods Mol. Biol. 2023, 2703, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Kamisugi, Y.; Nakayama, S.; Nakajima, R.; Ohtsubo, H.; Ohtsubo, E.; Fukui, K. Physical mapping of the 5S ribosomal RNA genes on rice chromosome 11. Mol. Gen. Genet. 1994, 245, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Chen, J. Distinguishing Sichuan walnut cultivars and examining their relationships with Juglans regia and J. sigillata by FISH, early-fruiting gene analysis, and SSR analysis. Front. Plant Sci. 2020, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, J.; Wang, J.; Gong, W.; Chen, L.; Wan, W. FISH analysis of Zanthoxylum armatum based on oligonucleotides for 5S rDNA and (GAA)6. Genome 2018, 61, 699–702. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Lei, Y.; Gong, W.; Ye, M.; Luo, X. Karyotype and phylogenetic relationship analysis of five varieties and cultivars of Zanthoxylum armatum based on oligo-FISH. Genes 2023, 14, 1459. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xu, Z.; Fan, R.; Wang, G.; Wang, F.; Qin, X.; Yan, L.; Ji, X.; Meng, M.; Sim, S.; et al. The complex genome and adaptive evolution of polyploid Chinese pepper (Zanthoxylum armatum and Zanthoxylum bungeanum). Plant Biotechnol. J. 2023, 21, 78–96. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Chen, J. Physical map of FISH 5S rDNA and (AG3T3)3 signals displays Chimonanthus campanulatus R.H. Chang & C.S. Ding chromosomes, reproduces its metaphase dynamics and distinguishes its chromosomes. Genes 2019, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.J.; Brown, T.A. Abridged 5S rDNA units in sea beet (Beta vulgaris subsp. maritima). Genome 2005, 48, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Song, M.; Xia, Y.; Zeng, X. A tandemly arranged pattern of two 5S rDNA arrays in Amolops mantzorum (Anura, Ranidae). Cytogenet. Genome Res. 2017, 151, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, J.; Zhao, A.; Chen, X.; Wan, W.; Chen, L. Karyotype analysis of Piptanthus concolor based on FISH with an oligonucleotide for rDNA 5S. Sci. Hortic. 2017, 226, 361–365. [Google Scholar] [CrossRef]
- Glugoski, L.; Deon, G.; Schott, S.; Vicari, M.R.; Nogaroto, V.; Moreira-Filho, O. Comparative cytogenetic analyses in Ancistrus species (Siluriformes: Loricariidae). Neotrop. Ichthyol. 2020, 18, e200013. [Google Scholar] [CrossRef]
- Zhang, Z.T.; Yang, S.Q.; Li, Z.A.; Zhang, Y.X.; Wang, Y.Z.; Cheng, C.Y.; Li, J.; Chen, J.F.; Lou, Q.F. Comparative chromosomal localization of 45S and 5S rDNAs and implications for genome evolution in Cucumis. Genome 2016, 59, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Khensuwan, S.; Sassi, F.M.C.; Moraes, R.L.R.; Jantarat, S.; Seetapan, K.; Phintong, K.; Thongnetr, W.; Kaewsri, S.; Jumrusthanasan, S.; Supiwong, W.; et al. Chromosomes of Asian cyprinid fishes: Genomic differences in conserved karyotypes of ‘Poropuntiinae’ (Teleostei, Cyprinidae). Animals 2023, 13, 1415. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, O.S.; Romanov, D.V.; Divashuk, M.G.; Razumova, O.V.; Ulyanov, D.S.; Karlov, G.I. Study and physical mapping of the species-specific tandem repeat CS-237 linked with 45S ribosomal DNA intergenic spacer in Cannabis sativa L. Plants 2022, 11, 1396. [Google Scholar] [CrossRef] [PubMed]
- Moraes, J.N.; Viana, P.F.; Favarato, R.M.; Pinheiro-Figliuolo, V.S.; Feldberg, E. Karyotype variability in six Amazonian species of the family Curimatidae (Characiformes) revealed by repetitive sequence mapping. Genet. Mol. Biol. 2022, 45, e20210125. [Google Scholar] [CrossRef] [PubMed]
- Yurkevich, O.Y.; Samatadze, T.E.; Selyutina, I.Y.; Suprun, N.A.; Suslina, S.N.; Zoshchuk, S.A.; Amosova, A.V.; Muravenko, O.V. Integration of genomic and cytogenetic data on tandem DNAs for analyzing the genome diversity within the genus Hedysarum L. (Fabaceae). Front. Plant Sci. 2022, 13, 865958. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.B.; Lysak, M.A.; Schubert, I. Chromosomal localization of rDNA in the Brassicaceae. Genome 2005, 48, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Albert, V.A. Dynamic distribution patterns of ribosomal DNA and chromosomal evolution in Paphiopedilum, a lady’s slipper orchid. BMC Plant Biol. 2011, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Y.; Wang, Y.; Zhang, J.; Chen, Q.; Liu, Z.; Liu, C.; He, W.; Wang, H.; Yang, S.; et al. accurate chromosome identification in the Prunus subgenus Cerasus (Prunus pseudocerasus) and its relatives by oligo-FISH. Int. J. Mol. Sci. 2022, 23, 13213. [Google Scholar] [CrossRef] [PubMed]
- de Barros, D.; Montenegro, C.; Gomes, M.; Ferraz, M.E.; Miotto, S.T.S.; Pedrosa-Harand, A. Cytogenetic characterization and karyotype evolution in six Macroptilium species (Leguminosae). Genome 2023, 66, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Amosova, A.V.; Yurkevich, O.Y.; Bolsheva, N.L.; Samatadze, T.E.; Zoshchuk, S.A.; Muravenko, O.V. Repeatome analyses and satellite DNA chromosome patterns in Deschampsia sukatschewii, D. cespitosa, and D. antarctica (Poaceae). Genes 2022, 13, 762. [Google Scholar] [CrossRef] [PubMed]
- Samatadze, T.E.; Yurkevich, O.Y.; Khazieva, F.M.; Basalaeva, I.V.; Konyaeva, E.A.; Burova, A.E.; Zoshchuk, S.A.; Morozov, A.I.; Amosova, A.V.; Muravenko, O.V. Agro-morphological and cytogenetic characterization of colchicine-induced tetraploid plants of Polemonium caeruleum L. (Polemoniaceae). Plants 2022, 11, 2585. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zhang, D.; Liu, Z.L.; Wang, X.R. Chromosomal localization of 5S and 18S rDNA in five species of subgenus Strobus and their implications for genome evolution of Pinus. Ann. Bot. 2006, 97, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, V.; Carlson, J.E. Physical mapping and characterization of 5S rRNA genes in Douglas-fir. J. Hered. 1998, 89, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Campomayor, N.B.; Waminal, N.E.; Kang, B.Y.; Nguyen, T.H.; Lee, S.S.; Huh, J.H.; Kim, H.H. Subgenome discrimination in Brassica and raphanus allopolyploids using microsatellites. Cells 2021, 10, 2358. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, W.; Luo, X.; Huan, J. Five Fabaceae karyotype and phylogenetic relationship analysis based on oligo-FISH for 5S rDNA and (AG3T3)3. Genes 2022, 13, 768. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, J. Fluorescence In Situ Hybridization (FISH) analysis of the locations of the oligonucleotides 5S rDNA, (AGGGTTT)3, and (TTG)6 in three genera of Oleaceae and their phylogenetic framework. Genes 2019, 10, 375. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Luo, X. First report of bicolour FISH of Berberis diaphana and B. soulieana reveals interspecific differences and co-localization of (AGGGTTT)3 and rDNA 5S in B. diaphana. Hereditas 2019, 156, 13. [Google Scholar] [CrossRef] [PubMed]
- Islam-Faridi, N.; Sakhanokho, H.F.; Dana, N.C. New chromosome number and cyto-molecular characterization of the African Baobab (Adansonia digitata L.)—“The Tree of Life”. Sci. Rep. 2020, 10, 13174. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; He, Z. Distribution of FISH oligo-5S rDNA and oligo-(AGGGTTT)3 in Hibiscus mutabilis L. Genome 2021, 64, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, J.; He, Z. Oligo-FISH can identify chromosomes and distinguish Hippophaë rhamnoides L. taxa. Genes 2022, 13, 195. [Google Scholar] [CrossRef] [PubMed]
- Beliveau, B.J.; Joyce, E.F.; Apostolopoulos, N.; Yilmaz, F.; Fonseka, C.Y.; McCole, R.B.; Chang, Y.; Li, J.B.; Senaratne, T.N.; Williams, B.R.; et al. Versatile design and synthesis platform for visualizing genomes with oligopaint FISH probes. Proc. Natl. Acad. Sci. USA 2012, 109, 21301–21306. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhao, H.; He, J.; Yang, Z.; Guan, B.; Chen, K.; Hong, Q.; Wang, J.; Liu, J.; Jiang, J. Extraordinarily conserved chromosomal synteny of Citrus species revealed by chromosome-specific painting. Plant J. 2020, 103, 2225–2235. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Zhang, T.; Wu, Y.; Zhang, W.; Zhang, P.; Xi, M.; Jiang, J. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J. 2020, 101, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; He, Z.; Liu, J.; Wu, H.; Gong, X. FISH mapping of telomeric and non-telomeric (AG3T3)3 reveal the chromosome numbers and chromosome rearrangements of 41 Woody plants. Genes 2022, 13, 1239. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Luo, X.; Lei, Y.; Zhang, W. Five species of Taxus karyotype based on oligo-FISH for 5S rDNA and (AG3T3)3. Genes 2022, 13, 2209. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yang, L.; Zhang, Y.; Liang, Q. Ploidy level, karyotype, and genome size of Bletilla species (Orchidaceae) from China. HortScience 2022, 57, 48–55. [Google Scholar] [CrossRef]
- Huan, J.; He, Z.; Lei, Y.; Li, W.; Jiang, L.; Luo, X. The genetic diversity of Bletilla spp. based on SLAF-seq and oligo-FISH. Genes 2022, 13, 1118. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. Available online: https://www.ncbi.nlm.nih.gov/books/NBK21054/ (accessed on 2 February 2024).
- Sergeeva, E.M.; Shcherban, A.B.; Adonina, I.G.; Nesterov, M.A.; Beletsky, A.V.; Rakitin, A.L.; Mardanov, A.V.; Ravin, N.V.; Salina, E.A. Fine organization of genomic regions tagged to the 5S rDNA locus of the bread wheat 5B chromosome. BMC Plant Biol. 2017, 17 (Suppl. S1), 183. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.G. Preliminary study of the kayotype of Polygonatum anhuiense and P. langaense. J. Anhui Nor. Univ. (Nat. Sci.) 1996, 2, 144–150. [Google Scholar] [CrossRef]
- Chen, C.W.; Chen, S.B. Karyotype analysis of three species of Polygonatum Mill from Tiantangzhai, West Anhui. J. Anhui Nor. Univ. (Nat. Sci.) 2005, 28, 324–327. [Google Scholar]
- Wang, Z.; Du, F.; Zhang, L.; Wang, S.; Wei, S. Advances in classification and identification of the plants of Polygonati rhizoma and its adulterants. North Hortic. 2019, 24, 130–136. Available online: http://qikan.cqvip.com/Qikan/Article/Detail?id=7100712585 (accessed on 2 February 2024).
- Yang, Y.; Huang, M.; Wang, D.; Ruan, B.; Yang, Q.; Yang, Y.; Luo, Y.; Tan, Y. Analysis of genome size and characteristics of Bletilla striata based on flow cytometry and genomic survey. Chin. Tradit. Pat. Med. 2023, 45, 3677–3682. Available online: https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=cjfdtemp&filename=zcya202311028 (accessed on 2 February 2024).
- Geukens, E.; Haegeman, A.; van Meulder, J.; van Laere, K.; Smolders, E.; Ruttink, T.; Leus, L. Exploring genetic diversity in an Ilex crenata breeding germplasm. Horticulturae 2023, 9, 485. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hartley, G.T. Zanthoxylum Linnaeus. In Flora of China; Flora of China Editorial Committee, Ed.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2008; pp. 53–66. [Google Scholar]
- Chen, R.Y.; Chen, C.B.; Song, W.Q.; Liang, G.L.; Li, X.L.; Chen, L. Chromosome atlas of major economic plants genome in China (Tomus V). In Chromosome Atlas of Medicinal Plants in China; Li, S.W., Wang, J., Eds.; Science Press: Beijing, China, 2009; p. 636. ISBN 978-7-03-022915-1. Available online: https://www.hceis.com/home/book_view.aspx?id=6683 (accessed on 2 February 2024).
- Yu, L.Y.; Tan, X.M.; Zhou, Y.Q. Karyotype analysis of Zanthoxylum nitidum. Lishizhen Med. Mater. Med. Res. 2010, 21, 3284–3285. Available online: http://qikan.cqvip.com/Qikan/Article/Detail?id=2000017695 (accessed on 2 February 2024).
- Miduno, T. Chromosome studies an Orchidaceae V. The study of cytology between Bletilla striata (n = 16) and Bl. formosana (n = 18). Cytologia 1954, 19, 239–248. [Google Scholar] [CrossRef]
- Woodworth, R.H. Meiosis of micro-sporogenesis in the Juglandaceae. Am. J. Bot. 1930, 17, 863–869. [Google Scholar] [CrossRef]
- Hans, A.S. Chromosomal numbers in Juglandeceae. J. Arnold. Abroretum. 1970, 51, 534–539. Available online: https://www.jstor.org/stable/43781708 (accessed on 2 February 2024). [CrossRef]
- Tulecke, W.; McGranahan, G.; Ahmadi, H. Regeneration by somatic embryogenesis of triploid plants from endosperm of walnut, Juglans regia L. cv manregian. Plant Cell Rep. 1988, 7, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.L.; Xi, R.T. Microsporogenesis studying and karyotype analysis of Juglans regia L. and J. hopeiensis Hu. J. Agric. Univ. Hebei 1988, 11, 48–55. [Google Scholar]
- Mu, Y.L.; Xi, R.T.; Lv, Z.S. Microsporogenesis observation and karyotype analysis of some species in genus Juglans L. J. Wuhan Bot. Res. 1990, 8, 301–310. Available online: https://plantscience.cn/cn/article/id/1255 (accessed on 2 February 2024).
- García, M.A.; Castroviejo, S. Estudios citotaxonómicos en las especies Ibéricas del género Cuscuta (Convolvulaceae). An. Jard. Bot. Madr. 2003, 60, 33–44. [Google Scholar]
- Ibiapino, A.; García, M.A.; Amorim, B.; Baez, M.; Costea, M.; Stefanović, S.; Pedrosa-Harand, A. The evolution of cytogenetic traits in Cuscuta (Convolvulaceae), the genus with the most diverse chromosomes in angiosperms. Front. Plant Sci. 2022, 13, 842260. [Google Scholar] [CrossRef] [PubMed]
- Damas, J.; Corbo, M.; Kim, J.; Turner-Maier, J.; Farré, M.; Larkin, D.M.; Ryder, O.A.; Steiner, C.; Houck, M.L.; Hall, S.; et al. Evolution of the ancestral mammalian karyotype and syntenic regions. Proc. Natl. Acad. Sci. USA 2022, 119, e2209139119. [Google Scholar] [CrossRef] [PubMed]
- Damas, J.; Corbo, M.; Lewin, H.A. Vertebrate chromosome evolution. Annu. Rev. Anim. Biosci. 2021, 9, 1–27. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhao, Y.; Zhang, S.; He, Y.; Jiang, J.; Chen, S.; Fang, W.; Guan, Z.; Liao, Y.; Wang, Z.; et al. Uneven levels of 5S and 45S rDNA site number and loci variations across wild Chrysanthemum accessions. Genes 2022, 13, 894. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.S.; Xue, C.J.; Li, R.P. Karyotype analysis of sea buckthorn. J. Shanxi Univ. (Nat. Sci. Ed.) 1989, 12, 323–330. Available online: https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=cjfd1989&filename=sxdr198903014&dbcode=cjfq (accessed on 2 February 2024).
- Liu, H.Z.; Sheng, L.X. Studies on karyotype of three sea buckthorn. J. Jilin Agr. Univ. 2011, 33, 628–631, 636. Available online: http://qikan.cqvip.com/Qikan/Article/Detail?id=39921155 (accessed on 2 February 2024).
- Said, M.; Hřibová, E.; Danilova, T.V.; Karafiátová, M.; Čížková, J.; Friebe, B.; Doležel, J.; Gill, B.S.; Vrána, J. The Agropyron cristatum karyotype, chromosome structure and cross-genome homoeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes. Theor. Appl. Genet. 2018, 131, 2213–2227. [Google Scholar] [CrossRef] [PubMed]
- de Moraes, R.L.R.; Sassi, F.M.C.; Marinho, M.M.F.; Ráb, P.; Porto, J.I.R.; Feldberg, E.; Cioffi, M.B. Small body, large chromosomes: Centric fusions shaped the karyotype of the Amazonian miniature fish Nannostomus anduzei (Characiformes, Lebiasinidae). Genes 2023, 14, 192. [Google Scholar] [CrossRef] [PubMed]
- Kroupin, P.Y.; Ulyanov, D.S.; Karlov, G.I.; Divashuk, M.G. The launch of satellite: DNA repeats as a cytogenetic tool in discovering the chromosomal universe of wild Triticeae. Chromosoma 2023, 132, 65–88. [Google Scholar] [CrossRef] [PubMed]
- Röser, M.; Winterfeld, G.; Grebenstein, B.; Hemleben, V. Molecular diversity and physical mapping of 5S rDNA in wild and cultivated oat grasses (Poaceae: Aveneae). Mol. Phylogenet. Evol. 2001, 21, n198–n217. [Google Scholar] [CrossRef] [PubMed]
- Kulak, S.; Hasterok, R.; Maluszynska, J. Karyotyping of Brassica amphidiploids using 5S and 25S rDNA as chromosome markers. Hereditas 2002, 136, 144–150, Erratum in Hereditas 2002, 137, 79–80. [Google Scholar] [CrossRef] [PubMed]
- Kovács, Z.; Mlinarec, J.; Höhn, M. Living on the edge: Morphological, karyological and genetic diversity studies of the Hungarian Plantago maxima populations and established ex situ collection. Bot. Stud. 2023, 64, 2. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Matyasek, R.; Kovarik, A.; Leitch, A. Parental origin and genome evolution in the allopolyploid Iris versicolor. Ann. Bot. 2007, 100, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, A.; Chen, G.; Hoang, P.T.N.; Fuchs, J.; Schubert, I.; Borisjuk, N. The ribosomal DNA loci of the ancient monocot Pistia stratiotes L. (Araceae) contain different variants of the 35S and 5S ribosomal RNA gene units. Front. Plant Sci. 2022, 13, 819750. [Google Scholar] [CrossRef] [PubMed]
- Mishima, M.; Ohmido, N.; Fukui, K.; Yahara, T. Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 2002, 110, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Rosa, K.O.; Ziemniczak, K.; de Barros, A.V.; Nogaroto, V.; Almeida, M.C.; Cestari, M.M.; Artoni, R.F.; Vicari, M.R. Numeric and structural chromosome polymorphism in Rineloricaria lima (Siluriformes: Loricariidae): Fusion points carrying 5S rDNA or telomere sequence vestiges. Rev. Fish Biol. Fish. 2012, 22, 739–749. [Google Scholar] [CrossRef]
- Barros, A.V.; Wolski, M.A.; Nogaroto, V.; Almeida, M.C.; Moreira-Filho, O.; Vicari, M.R. Fragile sites, dysfunctional telomere and chromosome fusions: What is 5S rDNA role? Gene 2017, 608, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Potapova, T.A.; Gerton, J.L. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res. 2019, 27, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Warmerdam, D.O.; Wolthuis, R.M.F. Keeping ribosomal DNA intact: A repeating challenge. Chromosome Res. 2019, 27, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Deon, G.A.; Glugoski, L.; Hatanaka, T.; Sassi, F.M.C.; Nogaroto, V.; Bertollo, L.A.C.; Liehr, T.; Al-Rikabi, A.; Moreira Filho, O.; Cioffi, M.B.; et al. Evolutionary breakpoint regions and chromosomal remodeling in Harttia (Siluriformes: Loricariidae) species diversification. Genet. Mol. Biol. 2022, 45, e20210170. [Google Scholar] [CrossRef] [PubMed]
- Deon, G.A.; Glugoski, L.; Vicari, M.R.; Nogaroto, V.; Sassi, F.M.C.; Cioffi, M.B.; Liehr, T.; Bertollo, L.A.C.; Moreira-Filho, O. Highly rearranged karyotypes and multiple sex chromosome systems in armored catfishes from the genus Harttia (Teleostei, Siluriformes). Genes 2020, 11, 1366. [Google Scholar] [CrossRef] [PubMed]
- Glugoski, L.; Giuliano-Caetano, L.; Moreira-Filho, O.; Vicari, M.R.; Nogaroto, V. Co-located hAT transposable element and 5S rDNA in an interstitial telomeric sequence suggest the formation of Robertsonian fusion in armored catfish. Gene 2018, 650, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Venancio, N.S.; Noleto, R.B.; Azambuja, M.; Gazolla, C.B.; Santos, B.R.; Nogaroto, V.; Vicari, M.R. Comparative cytogenetics among Boana species (Anura, Hylidae): Focus on evolutionary variability of repetitive DNA. Genet. Mol. Biol. 2023, 45, e20220203. [Google Scholar] [CrossRef] [PubMed]
- Coluccia, E.; Deidda, F.; Lobina, C.; Melis, R.; Porcu, C.; Agus, B.; Salvadori, S. Chromosome mapping of 5S ribosomal genes in indo-pacific and atlantic Muraenidae: Comparative analysis by dual colour Fluorescence In Situ Hybridisation. Genes 2020, 11, 1319. [Google Scholar] [CrossRef] [PubMed]
- de Melo, N.F.; Guerra, M. Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Ann. Bot. 2003, 92, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Volkov, R.A.; Panchuk, I.I.; Borisjuk, N.V.; Hosiawa-Baranska, M.; Maluszynska, J.; Hemleben, V. Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna. BMC Plant Biol. 2017, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.; Kovařík, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the Plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Glugoski, L.; Deon, G.A.; Nogaroto, V.; Moreira-Filho, O.; Vicari, M.R. Robertsonian fusion site in Rineloricaria pentamaculata (Siluriformes: Loricariidae): Involvement of 5S Ribosomal DNA and satellite sequences. Cytogenet. Genome Res. 2022, 162, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.P.; Leitch, I.J.; Bennett, M.D.; Chase, M.W.; Leitch, A.R. Ribosomal DNA evolution and phylogeny in Aloe (Asphodelaceae). Am. J. Bot. 2000, 87, 1578–1583. [Google Scholar] [CrossRef] [PubMed]
- Hasterok, R.; Wolny, E.; Hosiawa, M.; Kowalczyk, M.; Kulak-Ksiazczyk, S.; Ksiazczyk, T.; Heneen, W.K.; Maluszynska, J. Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann. Bot. 2006, 97, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Luo, L.; Yu, Z.; Lei, J.; Zhang, M.; Deng, Z. Repetitive sequence barcode probe for karyotype analysis in Tripidium arundinaceum. Int. J. Mol. Sci. 2022, 23, 6726. [Google Scholar] [CrossRef] [PubMed]
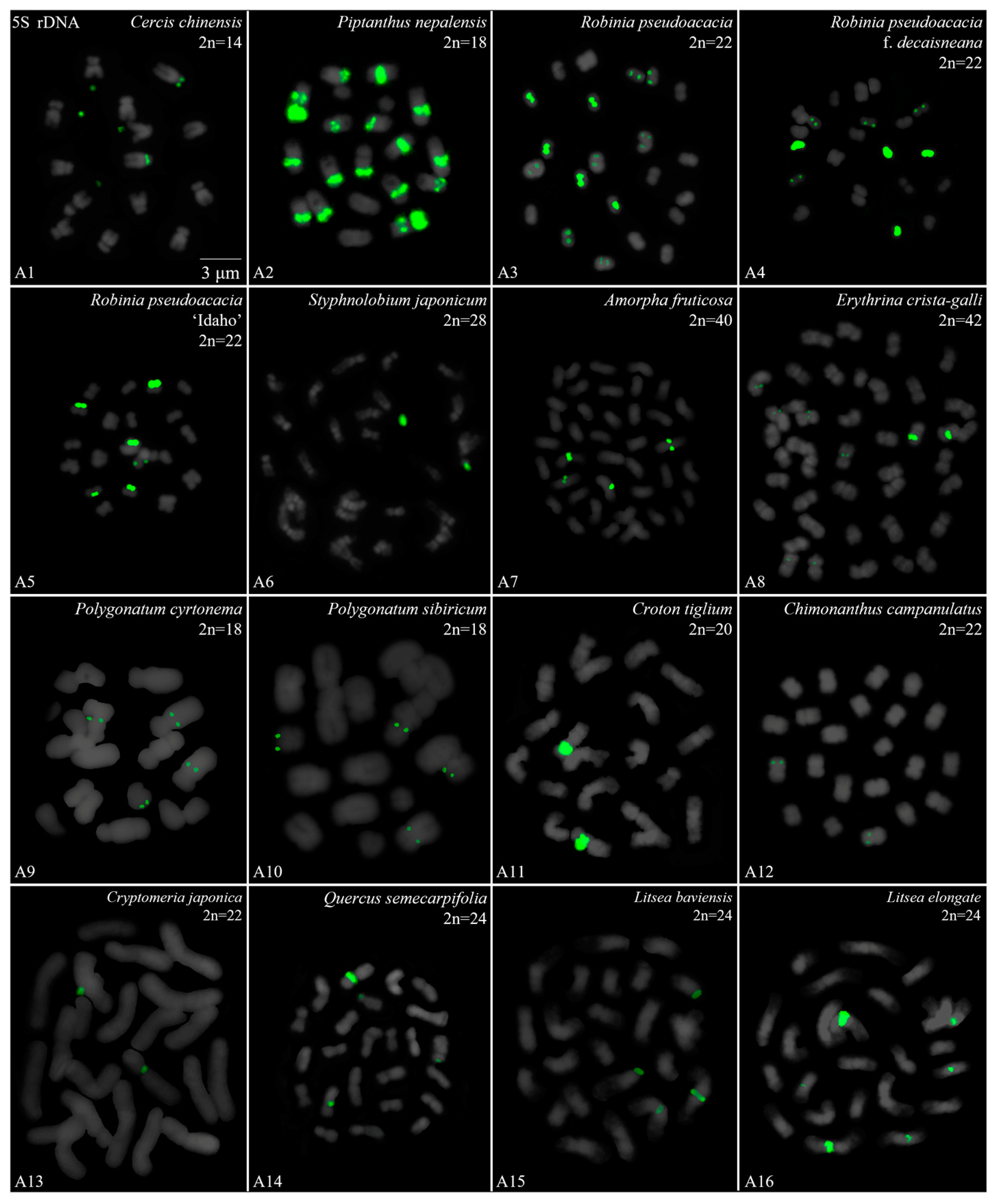
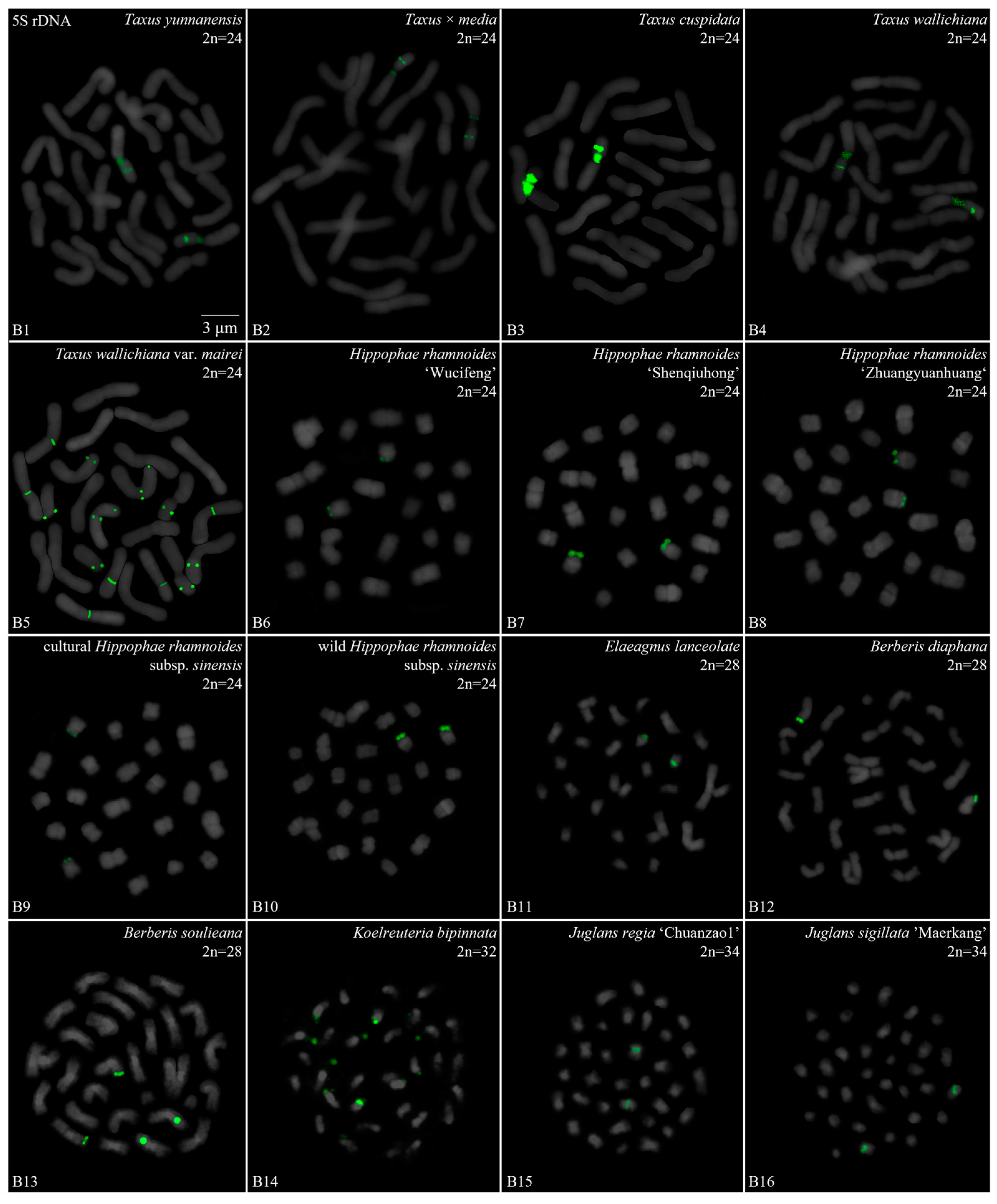

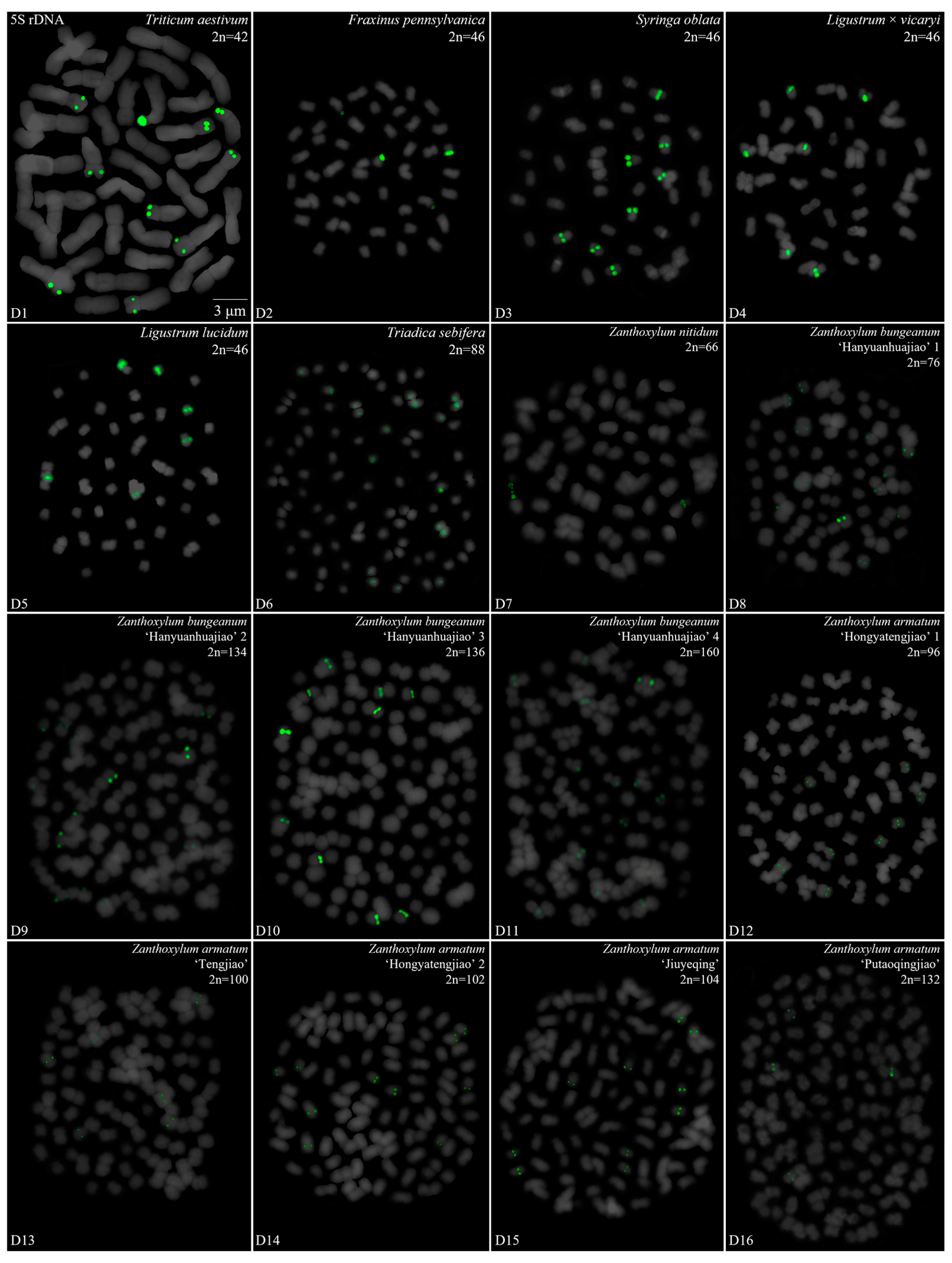
| Family | No. | Species | Collection Location |
|---|---|---|---|
| Rutaceae | 1 | Zanthoxylum nitidum (Roxb.) DC. | Hanyuan, Sichuan |
| 2 | Zanthoxylum bungeanum Maxim. ‘Hanyuanhuajiao’ 1 | Hanyuan, Sichuan | |
| 3 | Zanthoxylum bungeanum Maxim. ‘Hanyuanhuajiao’ 2 | Hanyuan, Sichuan | |
| 4 | Zanthoxylum bungeanum Maxim. ‘Hanyuanhuajiao’ 3 | Hanyuan, Sichuan | |
| 5 | Zanthoxylum bungeanum Maxim. ‘Hanyuanhuajiao’ 4 | Hanyuan, Sichuan | |
| 6 | Zanthoxylum armatum DC. ‘Hongyatengjiao’ 1 | Hongya, Sichuan | |
| 7 | Zanthoxylum armatum DC. ‘Hongyatengjiao’ 2 | Hongya, Sichuan | |
| 8 | Zanthoxylum armatum DC. ‘Jiuyeqing’ | Jiangjin, Chongqing | |
| 9 | Zanthoxylum armatum DC. ‘Putaoqingjiao’ | Hanyuan, Sichuan | |
| 10 | Zanthoxylum armatum DC. ‘Tengjiao’ | Hanyuan, Sichuan | |
| Orchidaceae | 11 | Bletilla striata (Thunb. ex Murray) Rchb. f. ‘Dujiangyan’ | Dujiangyan, Sichuan |
| 12 | Bletilla striata (Thunb. ex Murray) Rchb. f. ‘Neijiang’ | Neijiang, Sichuan | |
| 13 | Bletilla striata (Thunb. ex Murray) Rchb. f. ‘Wenchuan’ | Wenchuan, Sichuan | |
| 14 | Bletilla striata (Thunb. ex Murray) Rchb. f. ‘Baoshan’ | Baoshan, Yunnan | |
| 15 | Bletilla striata (Thunb. ex Murray) Rchb. f. ‘Honghe’ | Honghe, Yunnan | |
| 16 | Bletilla striata (Thunb. ex Murray) Rchb. f. ‘Nanan’ | Nanan, Chongqing | |
| 17 | Bletilla ochracea Schltr. ‘Dujiangyan’ | Dujiangyan, Sichuan | |
| 18 | Bletilla ochracea Schltr. ‘Zigong’ | Zigong, Sichuan | |
| 19 | Bletilla formosana (Hayata) Schltr. ‘Leshan’ | Leshan, Sichuan | |
| Fabaceae | 20 | Cercis chinensis Bunge | Wenjiang, Sichuan |
| 21 | Piptanthus nepalensis (Hook.) D. Don | Qingbaijiang, Sichuan | |
| 22 | Robinia pseudoacacia L. | Wenjiang, Sichuan | |
| 23 | Robinia pseudoacacia f. decaisneana (Carr.) Voss | Suqian, Jiangsu | |
| 24 | Robinia pseudoacacia ‘Idaho’ | Suqian, Jiangsu | |
| 25 | Styphnolobium japonicum (L.) Schott | Suqian, Jiangsu | |
| 26 | Amorpha fruticosa L. | Suqian, Jiangsu | |
| 27 | Erythrina crista-galli L. | Wenjiang, Sichuan | |
| Elaeagnaceae | 28 | Hippophae rhamnoides L. ‘Shenqiuhong’ | Huai’an, Hebei |
| 29 | Hippophae rhamnoides L. ‘Wucifeng’ | Huai’an, Hebei | |
| 30 | wild Hippophae rhamnoides subsp. sinensis Rousi | Huai’an, Hebei | |
| 31 | cultural Hippophae rhamnoides subsp. sinensis Rousi | Tieling, Liaoning | |
| 32 | Hippophae rhamnoides L. ‘Zhuangyuanhuang’ | Wenchuan, Sichuan | |
| 33 | Elaeagnus lanceolate Warb. apud Diels | Wenchuan, Sichuan | |
| Taxaceae | 34 | Taxus cuspidata Siebold & Zucc. | Suqian, Jiangsu |
| 35 | Taxus × media Rehder | Ya’an, Sichuan | |
| 36 | Taxus wallichiana var. mairei (Lemee & H. Léveillé) L. K. Fu & N. Li | Ya’an, Sichuan | |
| 37 | Taxus wallichiana Zucc. | Xichuang, Sichuan | |
| 38 | Taxus yunnanensis W.C. Cheng & L.K. Fu | Ya’an, Sichuan | |
| Juglandaceae | 39 | Juglans regia L. ‘Chuanzao1’ | Qingbaijiang, Sichuan |
| 40 | Juglans sigillata Dode ‘Maerkang’ | Maerkang, Sichuan | |
| 41 | Juglans sigillata Dode ‘Muzhilinhe’ | Gulin, Sichuan | |
| 42 | Juglans regia L. ‘Yanyuanzao’ | Yanyuan, Sichuan | |
| Oleaceae | 43 | Fraxinus pennsylvanica Marsh. | Wenjiang, Sichuan |
| 44 | Syringa oblata Lindl. | Wenjiang, Sichuan | |
| 45 | Ligustrum × vicaryi Rehder | Wenjiang, Sichuan | |
| 46 | Ligustrum lucidum Ait. | Wenjiang, Sichuan | |
| Asparagaceae | 47 | Polygonatum cyrtonema Hua | Wenchuan, Sichuan |
| 48 | Polygonatum sibiricum Delar. ex Redoute | Dujiangyan, Sichuan | |
| Berberidaceae | 49 | Berberis diaphana Maxim. | Wenchuan, Sichuan |
| 50 | Berberis soulieana Schneid. | Wenchuan, Sichuan | |
| Lauraceae | 51 | Litsea baviensis Lec. | Jinniu, Sichuan |
| 52 | Litsea elongate (Wall. ex Nees) Benth. et Hook. f. | Jinniu, Sichuan | |
| Malvaceae | 53 | Firmiana simplex (L.) W. Wight | Wenjiang, Sichuan |
| 54 | Hibiscus mutabilis L. | Jinniu, Sichuan | |
| Euphorbiaceae | 55 | Croton tiglium L. | Ya’an, Sichuan |
| 56 | Triadica sebifera (Linnaeus) Small | Jiangyou, Sichuan | |
| Cupressaceae | 57 | Cryptomeria japonica (L.f.) D. Don | Wenjiang, Sichuan |
| Aquifoliaceae | 58 | Ilex chinensis Sims | Dujiangyan, Sichuan |
| Calycanthaceae | 59 | Chimonanthus campanulatus R.H. Chang & C.S. Ding | Jinniu, Sichuan |
| Fagaceae | 60 | Quercus semecarpifolia Smith | Wenchuan, Sichuan |
| Poaceae | 61 | Triticum aestivum L. | Wenjiang, Sichuan |
| Podocarpaceae | 62 | Podocarpus macrophyllus (Thunb.) Sweet | Wenjiang, Sichuan |
| Salicaceae | 63 | Idesia polycarpa Maxim. | Wenjiang, Sichuan |
| Sapindaceae | 64 | Koelreuteria bipinnata Franch. | Wenjiang, Sichuan |
| Accession | Species | Chromosome Number | Chromosome Length | Karyotype Asymmetry |
|---|---|---|---|---|
| A1 | Cercis chinensis | 2n = 14 | 1.58–2.21 μm | 1.40 |
| A2 | Piptanthus nepalensis | 2n = 18 | 1.65–3.02 μm | 1.83 |
| A3 | Robinia pseudoacacia | 2n = 22 | 1.37–1.68 μm | 1.23 |
| A4 | Robinia pseudoacacia f. decaisneana | 2n = 22 | 0.77–1.40 μm | 1.82 |
| A5 | Robinia pseudoacacia ‘Idaho’ | 2n = 22 | 1.09–1.47 μm | 0.74 |
| A6 | Styphnolobium japonicum | 2n = 28 | 0.98–3.19 μm | 3.26 |
| A7 | Amorpha fruticosa | 2n = 40 | 1.44–2.18 μm | 1.51 |
| A8 | Erythrina crista-galli | 2n = 42 | 1.37–3.32 μm | 2.42 |
| A9 | Polygonatum cyrtonema | 2n = 18 | 1.54–4.49 μm | 2.92 |
| A10 * | Polygonatum sibiricum | 2n = 18 | 1.47–6.77 μm | 4.61 |
| A11 | Croton tiglium | 2n = 20 | 1.65–3.61 μm | 2.19 |
| A12 | Chimonanthus campanulatus | 2n = 22 | 1.33–1.93 μm | 1.45 |
| A13 | Cryptomeria japonica | 2n = 22 | 3.09–6.14 μm | 1.99 |
| A14 | Quercus semecarpifolia | 2n = 24 | 1.47–3.10 μm | 3.11 |
| A15 | Litsea baviensis | 2n = 24 | 1.86–4.04 μm | 2.17 |
| A16 | Litsea elongate | 2n = 24 | 1.58–4.21 μm | 2.66 |
| B1 | Taxus yunnanensis | 2n = 24 | 2.46–6.28 μm | 2.55 |
| B2 | Taxus × media | 2n = 24 | 2.39–6.88 μm | 2.88 |
| B3 | Taxus cuspidata | 2n = 24 | 2.60–6.32 μm | 2.43 |
| B4 | Taxus wallichiana | 2n = 24 | 2.42–6.42 μm | 2.65 |
| B5 | Taxus wallichiana var. mairei | 2n = 24 | 2.81–6.67 μm | 2.37 |
| B6 | Hippophae rhamnoides ‘Wucifeng’ | 2n = 24 | 1.40–2.60 μm | 1.86 |
| B7 | Hippophae rhamnoides ‘Shenqiuhong’ | 2n = 24 | 1.26–2.46 μm | 1.95 |
| B8 | Hippophae rhamnoides ‘Zhuangyuanhuang’ | 2n = 24 | 1.44–2.81 μm | 1.95 |
| B9 | cultural Hippophae rhamnoides subsp. sinensis | 2n = 24 | 1.30–2.18 μm | 1.68 |
| B10 | wild Hippophae rhamnoides subsp. sinensis | 2n = 24 | 1.05–2.32 μm | 2.21 |
| B11 | Elaeagnus lanceolate | 2n = 28 | 1.02–3.68 μm | 3.61 |
| B12 | Berberis diaphana | 2n = 28 | 1.54–2.84 μm | 1.84 |
| B13 | Berberis soulieana | 2n = 28 | 2.18–3.51 μm | 1.61 |
| B14 | Koelreuteria bipinnata | 2n = 32 | 0.77–1.82 μm | 2.36 |
| B15 | Juglans regia ‘Chuanzao1’ | 2n = 34 | 0.70–1.65 μm | 2.36 |
| B16 | Juglans sigillata ‘Maerkang’ | 2n = 34 | 0.63–1.61 μm | 2.56 |
| C1 | Juglans sigillata ‘Muzhilinhe’ | 2n = 34 | 0.95–2.28 μm | 2.40 |
| C2 | Juglans regia ‘Yanyuanzao’ | 2n = 34 | 0.91–2.19 μm | 2.41 |
| C3 | Bletilla formosana ‘Leshan’ | 2n = 34 | 0.98–3.68 μm | 3.76 |
| C4 | Bletilla striata f. ‘Dujiangyan’ | 2n = 34 | 0.88–3.96 μm | 4.50 |
| C5 | Bletilla striata f. ‘Wenchuan’ | 2n = 34 | 1.33–4.74 μm | 3.56 |
| C6 | Bletilla striata f. ‘Neijiang’ | 2n = 34 | 1.40–3.16 μm | 2.26 |
| C7 | Bletilla striata f. ‘Baoshan’ | 2n = 34 | 1.26–3.61 μm | 2.87 |
| C8 | Bletilla striata f. ‘Nanan’ | 2n = 34 | 1.12–3.86 μm | 3.45 |
| C9 | Bletilla striata f. ‘Honghe’ | 2n = 34 | 1.23–4.04 μm | 3.28 |
| C10 | Bletilla ochracea ‘Zigong’ | 2n = 34 | 1.12–2.63 μm | 2.35 |
| C11 | Bletilla ochracea ‘Dujiangyan’ | 2n = 34 | 1.19–3.02 μm | 2.63 |
| C12 | Firmiana simplex | 2n = 36 | 0.98–2.21 μm | 2.26 |
| C13 | Hibiscus mutabilis | 2n = 90 | 1.09–2.98 μm | 2.73 |
| C14 | Podocarpus macrophyllus | 2n = 36 | 1.93–5.26 μm | 2.73 |
| C15 | Idesia polycarpa | 2n = 38 | 0.77–2.46 μm | 3.19 |
| C16 * | Ilex chinensis | 2n = 40 | 0.63–1.58 μm | 2.51 |
| D1 | Triticum aestivum | 2n = 42 | 3.85–6.88 μm | 1.79 |
| D2 | Fraxinus pennsylvanica | 2n = 46 | 0.88–1.96 μm | 2.23 |
| D3 | Syringa oblata | 2n = 46 | 0.98–1.93 μm | 1.97 |
| D4 | Ligustrum × vicaryi | 2n = 46 | 0.95–2.14 μm | 2.25 |
| D5 | Ligustrum lucidum | 2n = 46 | 0.70–2.11 μm | 3.01 |
| D6 * | Triadica sebifera | 2n = 88 | 0.60–1.23 μm | 2.05 |
| D7 | Zanthoxylum nitidum | 2n = 66 | 1.40–2.56 μm | 1.83 |
| D8 | Zanthoxylum bungeanum ‘Hanyuanhuajiao’ 1 | 2n = 76 | 0.84–1.72 μm | 2.05 |
| D9 | Zanthoxylum bungeanum ‘Hanyuanhuajiao’ 2 | 2n = 134 | 0.88–1.82 μm | 2.07 |
| D10 | Zanthoxylum bungeanum ‘Hanyuanhuajiao’ 3 | 2n = 136 | 0.91–2.14 μm | 2.35 |
| D11 | Zanthoxylum bungeanum ‘Hanyuanhuajiao’ 4 | 2n = 160 | 0.74–1.68 μm | 2.27 |
| D12 | Zanthoxylum armatum ‘Hongyatengjiao’ 1 | 2n = 96 | 0.70–1.47 μm | 3.01 |
| D13 | Zanthoxylum armatum ‘Tengjiao’ | 2n = 100 | 0.88–2.00 μm | 2.27 |
| D14 | Zanthoxylum armatum ‘Hongyatengjiao’ 2 | 2n = 102 | 0.95–2.04 μm | 2.14 |
| D15 | Zanthoxylum armatum ‘Jiuyeqing’ | 2n = 104 | 0.88–2.00 μm | 2.27 |
| D16 | Zanthoxylum armatum ‘Putaoqingjiao’ | 2n = 132 | 0.74–1.79 μm | 2.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Liu, Y.; Gong, X.; Ye, M.; Xiao, Q.; Zeng, Z. Karyotype Description and Comparative Chromosomal Mapping of 5S rDNA in 42 Species. Genes 2024, 15, 647. https://doi.org/10.3390/genes15050647
Luo X, Liu Y, Gong X, Ye M, Xiao Q, Zeng Z. Karyotype Description and Comparative Chromosomal Mapping of 5S rDNA in 42 Species. Genes. 2024; 15(5):647. https://doi.org/10.3390/genes15050647
Chicago/Turabian StyleLuo, Xiaomei, Yunke Liu, Xiao Gong, Meng Ye, Qiangang Xiao, and Zhen Zeng. 2024. "Karyotype Description and Comparative Chromosomal Mapping of 5S rDNA in 42 Species" Genes 15, no. 5: 647. https://doi.org/10.3390/genes15050647
APA StyleLuo, X., Liu, Y., Gong, X., Ye, M., Xiao, Q., & Zeng, Z. (2024). Karyotype Description and Comparative Chromosomal Mapping of 5S rDNA in 42 Species. Genes, 15(5), 647. https://doi.org/10.3390/genes15050647






