Lack of T04C9.1, the Homologue of Mammalian APPL2, Leads to Premature Ageing and Shortens Lifespan in Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animals and Treatments
2.3. Cell Culture, Transfection, and SA-β-Gal Staining
2.4. RNA Extraction and qRT-PCR
2.5. Protein Extraction and Western Blot Analysis
2.6. Immunohistochemistry (IHC) Staining
2.7. C. elegans Lifespan and Reproduction Analysis
2.8. Lipofuscin and Lipid Accumulation Assay
2.9. Pharyngeal Pumping and Body Bending Frequency
2.10. Stress Resistance Assay
2.11. ROS Measurement
2.12. Structure Modelling and Molecular Docking
2.13. Protein–Protein Interaction Network
2.14. Data Analysis and Statistics
3. Results
3.1. The Level of APPL2 Decreases in the Major Organs of Aged Mice and Lack of APPL2 Leads to Premature Ageing of HUVECs
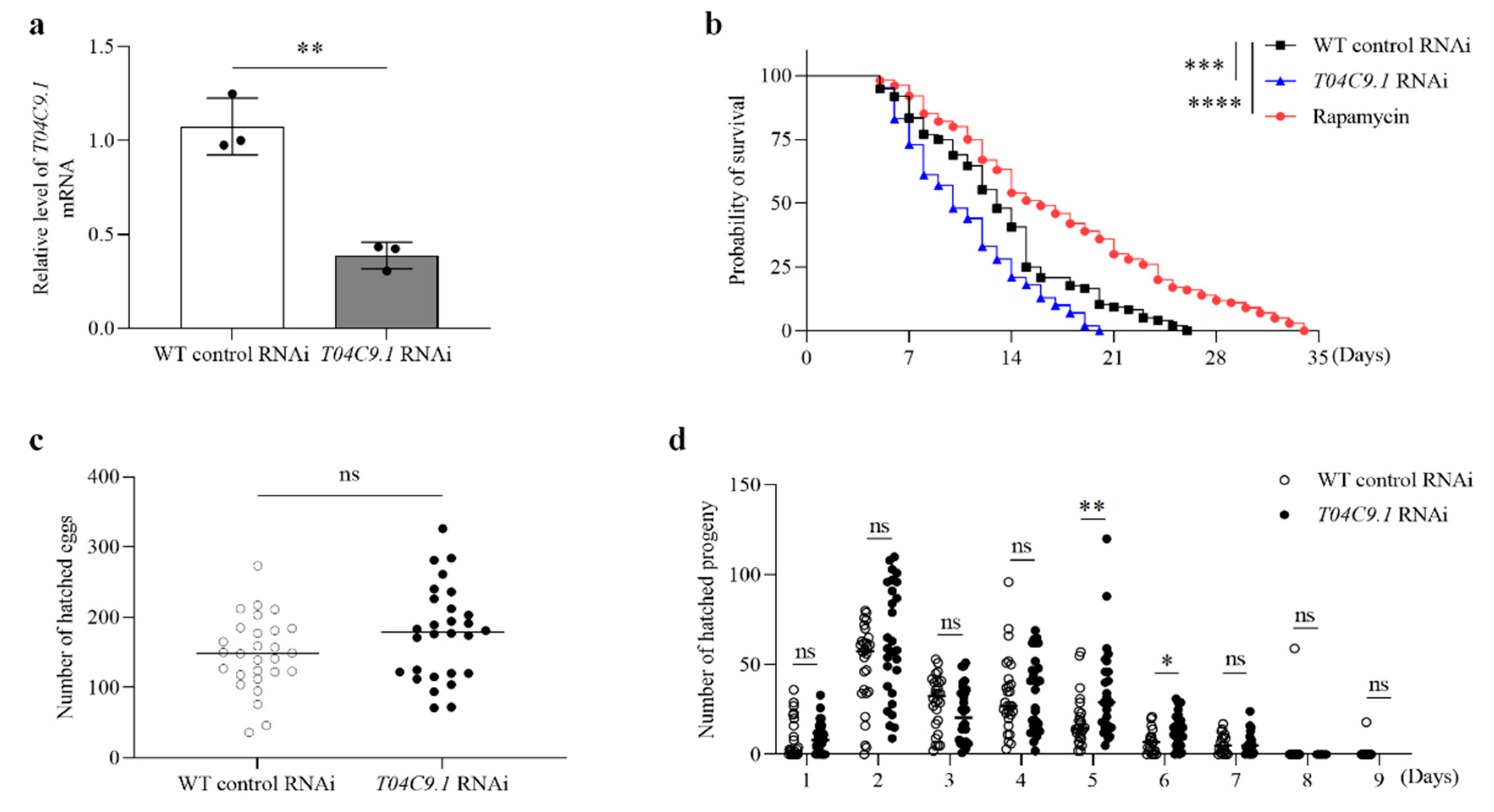
3.2. T04C9.1 Deficiency Reduces Longevity without Altering the Reproduction of C. elegans
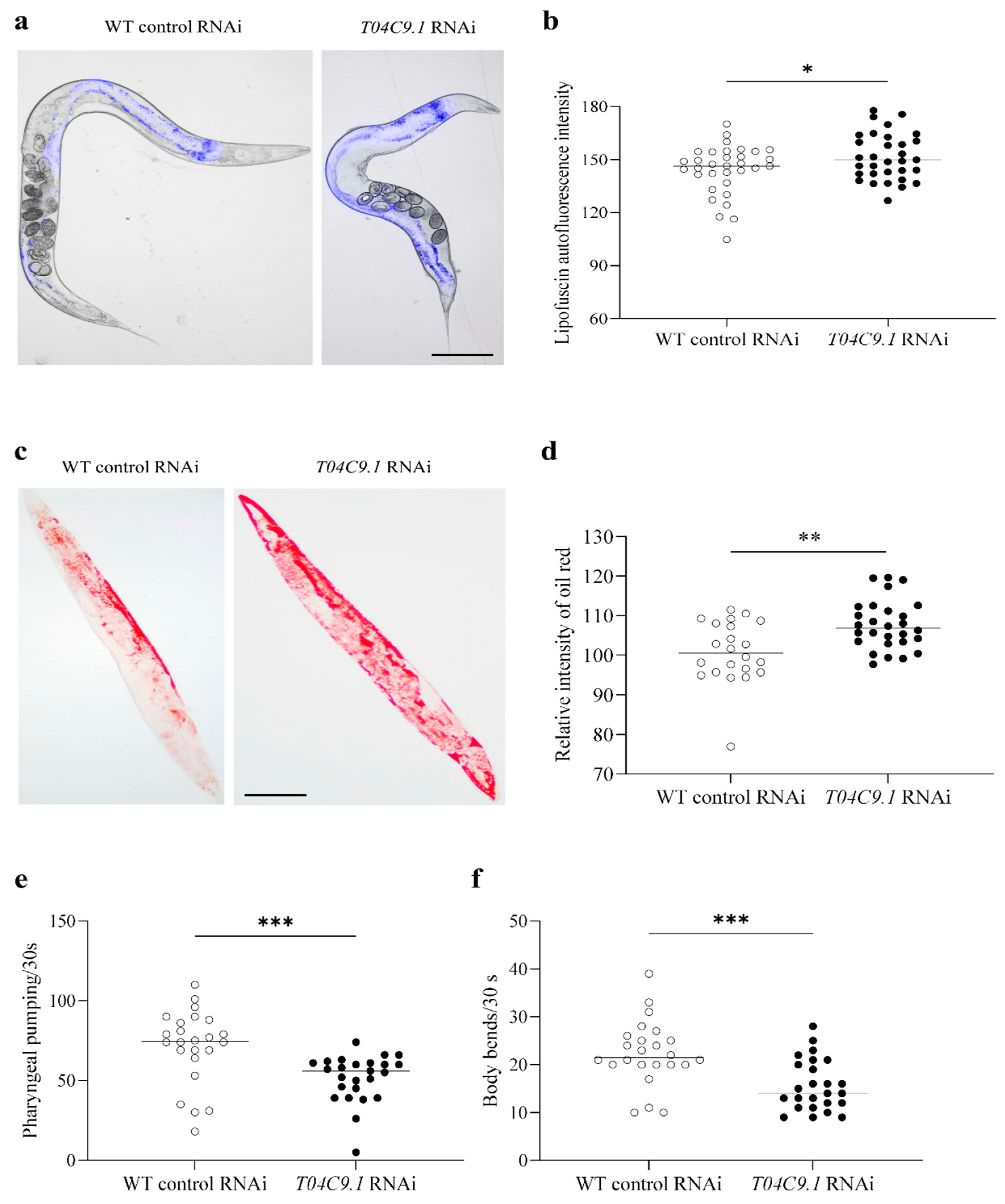
3.3. T04C9.1 Deficiency Leads to Lipofuscin Pigment Accumulation, Lipid Deposition, and Mobility Decline in C. elegans
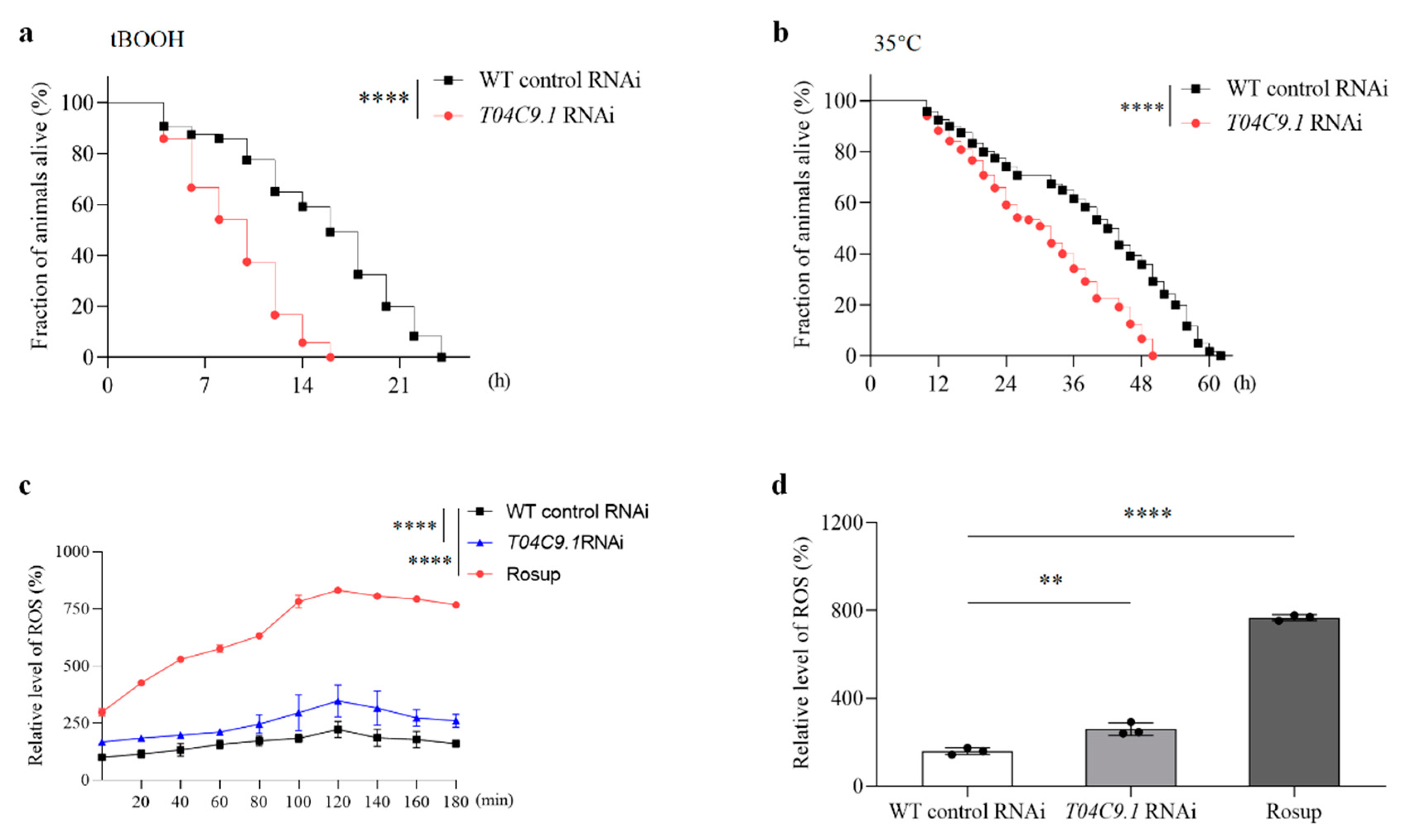
3.4. T04C9.1 Deficiency Increases the Stress Sensitivity and Reactive Oxygen Species (ROS) Levels in C. elegans
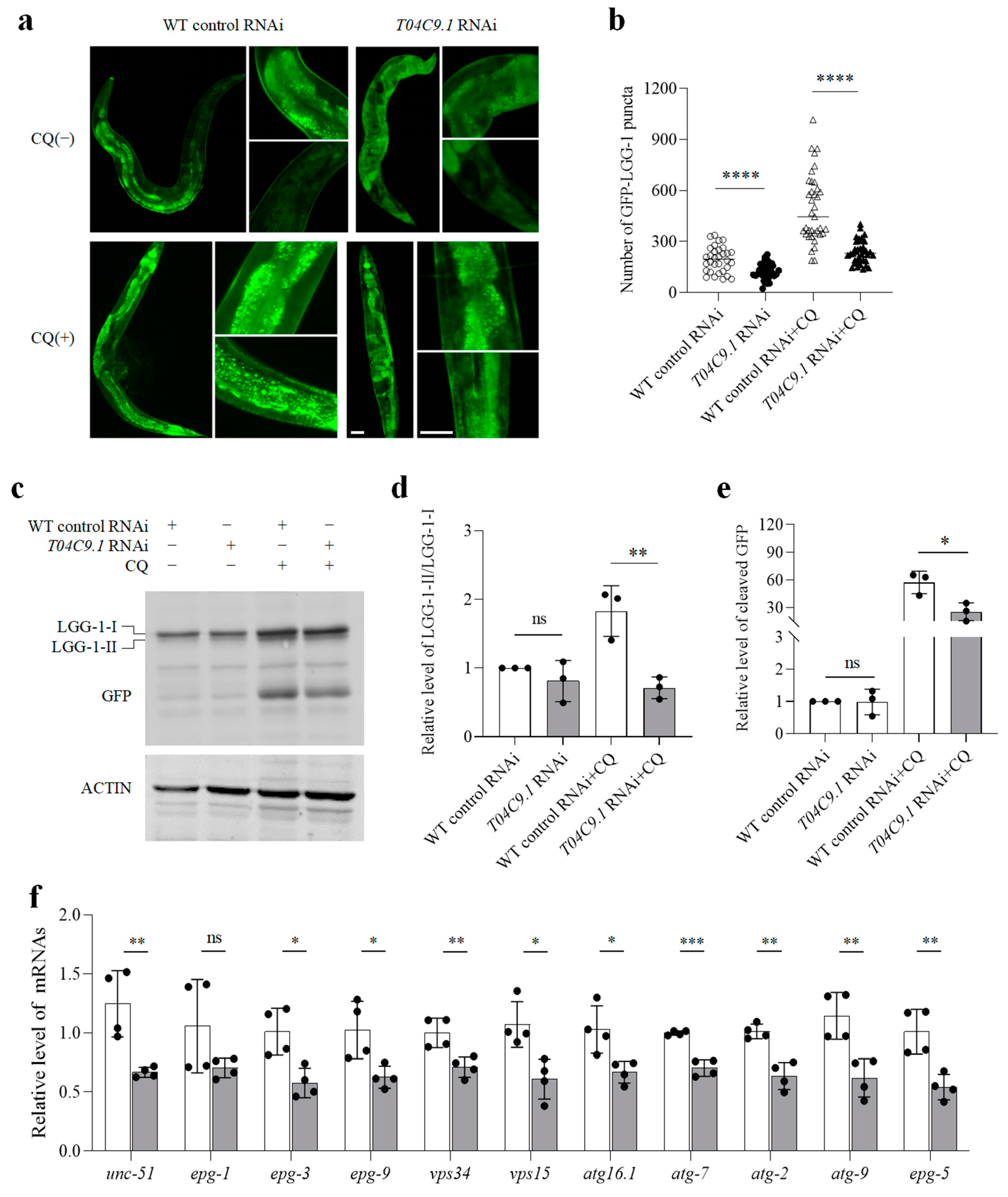
3.5. T04C9.1 Deficiency Decreases Autophagic Activity in C. elegans
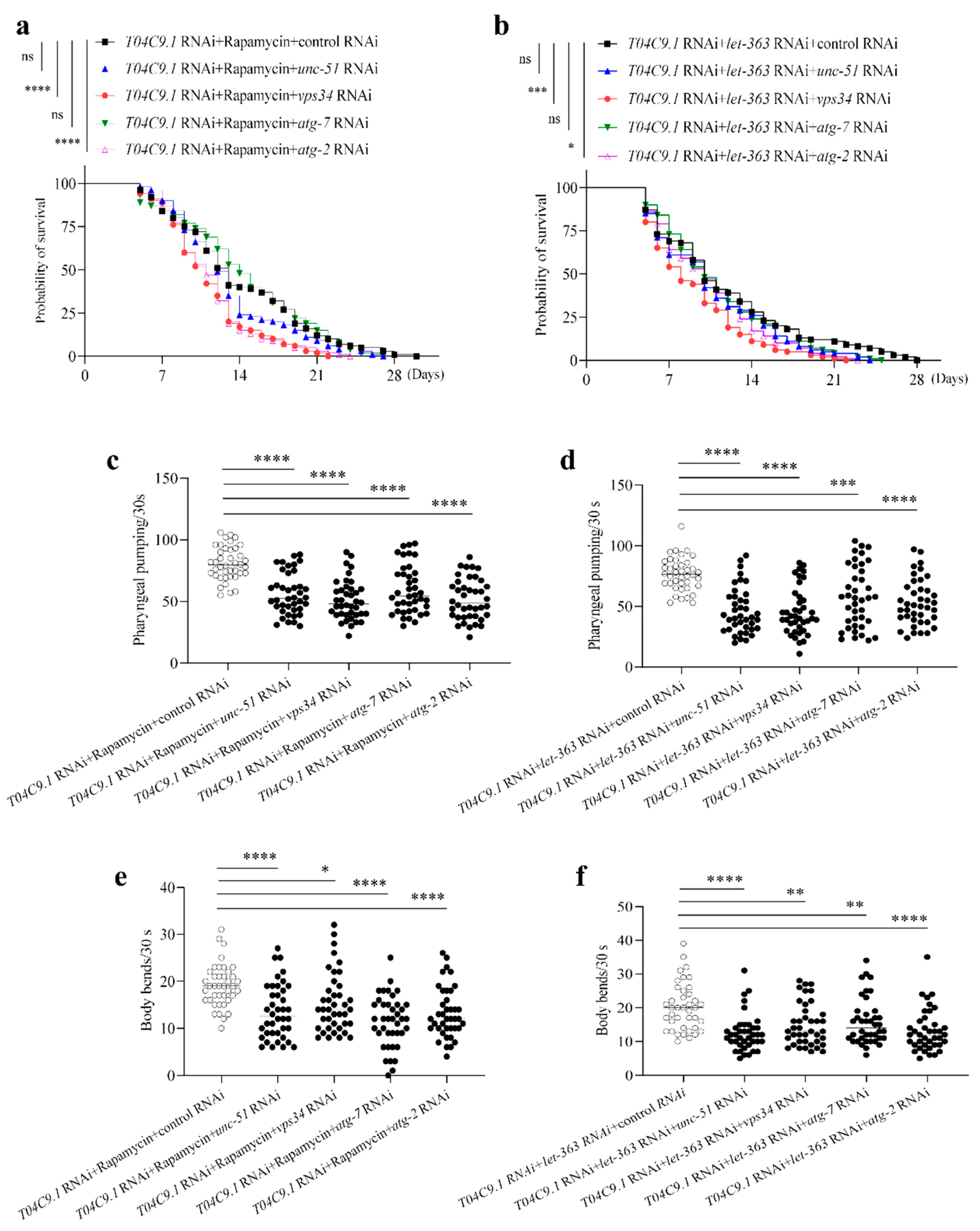
3.6. T04C9.1 Deficiency Leads to Premature Ageing and Shortened Lifespan of C. elegans by Reducing Autophagy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costantino, S.; Paneni, F.; Cosentino, F. Ageing, metabolism and cardiovascular disease. J. Physiol. 2016, 594, 2061–2073. [Google Scholar] [CrossRef]
- Amorim, J.A.; Coppotelli, G.; Rolo, A.P.; Palmeira, C.M.; Ross, J.M.; Sinclair, D.A. Mitochondrial and metabolic dysfunction in ageing and age-related diseases. Nat. Rev. Endocrinol. 2022, 18, 243–258. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Ren, Y.; Wang, Y.; Fang, J.; Yue, H.; Ma, S.; Guan, F. Aging and age-related diseases: From mechanisms to therapeutic strategies. Biogerontology 2021, 22, 165–187. [Google Scholar] [CrossRef]
- Miaczynska, M.; Christoforidis, S.; Giner, A.; Shevchenko, A.; Uttenweiler-Joseph, S.; Habermann, B.; Wilm, M.; Parton, R.G.; Zerial, M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 2004, 116, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Kikani, C.K.; Riojas, R.A.; Langlais, P.; Wang, L.; Ramos, F.J.; Fang, Q.; Christ-Roberts, C.Y.; Hong, J.Y.; Kim, R.Y.; et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat. Cell Biol. 2006, 8, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xiao, T.; Peng, X.; Li, G.; Hu, F. APPLs: More than just adiponectin receptor binding proteins. Cell Signal 2017, 32, 76–84. [Google Scholar] [CrossRef]
- Liu, R.; Meng, J.; Lou, D. Adiponectin inhibits D-gal-induced cardiomyocyte senescence via AdipoR1/APPL1. Mol. Med. Rep. 2021, 24, 1–10. [Google Scholar] [CrossRef]
- Pyrzynska, B.; Banach-Orlowska, M.; Teperek-Tkacz, M.; Miekus, K.; Drabik, G.; Majka, M.; Miaczynska, M. Multifunctional protein APPL2 contributes to survival of human glioma cells. Mol. Oncol. 2013, 7, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lin, H.; Li, X.; Lu, W.; Kim, J.B.; Xu, A.; Cheng, K.K.Y. The adaptor protein APPL2 controls glucose-stimulated insulin secretion via F-actin remodeling in pancreatic β-cells. Proc. Natl. Acad. Sci. USA 2020, 117, 28307–28315. [Google Scholar] [CrossRef]
- Yeo, J.C.; Wall, A.A.; Luo, L.; Condon, N.D.; Stow, J.L. Distinct Roles for APPL1 and APPL2 in Regulating Toll-like Receptor 4 Signaling in Macrophages. Traffic 2016, 17, 1014–1026. [Google Scholar] [CrossRef]
- Son, H.G.; Altintas, O.; Kim, E.J.E.; Kwon, S.; Lee, S.V. Age-dependent changes and biomarkers of aging in Caenorhabditis elegans. Aging Cell 2019, 18, e12853. [Google Scholar] [CrossRef]
- Lee, G.Y.; Sohn, J.; Lee, S.V. Combinatorial Approach Using Caenorhabditis elegans and Mammalian Systems for Aging Research. Mol. Cells 2021, 44, 425–432. [Google Scholar] [CrossRef]
- Hsieh, P.N.; Zhou, G.; Yuan, Y.; Zhang, R.; Prosdocimo, D.A.; Sangwung, P.; Borton, A.H.; Boriushkin, E.; Hamik, A.; Fujioka, H.; et al. A conserved KLF-autophagy pathway modulates nematode lifespan and mammalian age-associated vascular dysfunction. Nat. Commun. 2017, 8, 914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, P.; Chen, J.; Chen, Z.; Liu, Z.; Feng, G.; Sha, F.; Li, Z.; Xu, Z.; Huang, Y.; et al. CD44 connects autophagy decline and ageing in the vascular endothelium. Nat. Commun. 2023, 14, 5524. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Hadley, J.T.; Li, Z.; Dong, F.; Xu, H.; Xin, X.; Zhang, Y.; Chen, C.; Li, S.; Guo, X.; et al. Adiponectin Alleviates Diet-Induced Inflammation in the Liver by Suppressing MCP-1 Expression and Macrophage Infiltration. Diabetes 2021, 70, 1303–1316. [Google Scholar] [CrossRef] [PubMed]
- Possik, E.; Klein, L.L.; Sanjab, P.; Zhu, R.; Côté, L.; Bai, Y.; Zhang, D.; Sun, H.; Al-Mass, A.; Oppong, A.; et al. Glycerol 3-phosphate phosphatase/PGPH-2 counters metabolic stress and promotes healthy aging via a glycogen sensing-AMPK-HLH-30-autophagy axis in C. elegans. Nat. Commun. 2023, 14, 5214. [Google Scholar] [CrossRef]
- Escorcia, W.; Ruter, D.L.; Nhan, J.; Curran, S.P. Quantification of Lipid Abundance and Evaluation of Lipid Distribution in Caenorhabditis elegans by Nile Red and Oil Red O Staining. J. Vis. Exp. 2018, 133, e57352. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Tan, Y.; Xin, X.; Coffey, F.J.; Wiest, D.L.; Dong, L.Q.; Testa, J.R. Appl1 and Appl2 are Expendable for Mouse Development But Are Essential for HGF-Induced Akt Activation and Migration in Mouse Embryonic Fibroblasts. J. Cell Physiol. 2016, 231, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Berdichevsky, A.; Nedelcu, S.; Boulias, K.; Bishop, N.A.; Guarente, L.; Horvitz, H.R. 3-Ketoacyl thiolase delays aging of Caenorhabditis elegans and is required for lifespan extension mediated by sir-2.1. Proc. Natl. Acad. Sci. USA 2010, 107, 18927–18932. [Google Scholar] [CrossRef] [PubMed]
- Soo, S.K.; Rudich, Z.D.; Ko, B.; Moldakozhayev, A.; AlOkda, A.; Van Raamsdonk, J.M. Biological resilience and aging: Activation of stress response pathways contributes to lifespan extension. Ageing Res. Rev. 2023, 88, 101941. [Google Scholar] [CrossRef]
- Gusarov, I.; Shamovsky, I.; Pani, B.; Gautier, L.; Eremina, S.; Katkova-Zhukotskaya, O.; Mironov, A.; Makarov, A.; Nudler, E. Dietary thiols accelerate aging of C. elegans. Nat. Commun. 2021, 12, 4336. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Fang, Q.; Yu, W.; Zhang, R.; Hu, C.; Dong, K.; Bao, Y.; Wang, C.; Xiang, K.; Jia, W. Genetic variations in APPL2 are associated with overweight and obesity in a Chinese population with normal glucose tolerance. BMC Med. Genet. 2012, 13, 22. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.W.; Ding, S.; Ma, X.D.; Gu, N.; Guo, X.H. Genetic variability in adapter proteins with APPL1/2 is associated with the risk of coronary artery disease in type 2 diabetes mellitus in Chinese Han population. Chin. Med. J. 2011, 124, 3618–3621. [Google Scholar] [PubMed]
- Barbieri, M.; Esposito, A.; Angellotti, E.; Rizzo, M.R.; Marfella, R.; Paolisso, G. Association of genetic variation in adaptor protein APPL1/APPL2 loci with non-alcoholic fatty liver disease. PLoS ONE 2013, 8, e71391. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Lin, W.; Nie, T.; Hui, X.; Gao, X.; Li, K.; Ding, M.; Tang, X.; Li, P.; Wang, Y.; et al. Absence of Appl2 sensitizes endotoxin shock through activation of PI3K/Akt pathway. Cell Biosci. 2014, 4, 60. [Google Scholar] [CrossRef]
- Shen, P.; Yue, Y.; Park, Y. A living model for obesity and aging research: Caenorhabditis elegans. Crit. Rev. Food Sci. Nutr. 2018, 58, 741–754. [Google Scholar] [CrossRef]
- Jacob, K.D.; Noren Hooten, N.; Trzeciak, A.R.; Evans, M.K. Markers of oxidant stress that are clinically relevant in aging and age-related disease. Mech. Ageing Dev. 2013, 134, 139–157. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Yoshimori, T. Autophagy and Longevity. Mol. Cells 2018, 41, 65–72. [Google Scholar] [PubMed]
- Rubinsztein, D.C.; Mariño, G.; Kroemer, G. Autophagy and aging. Cell 2011, 146, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Aman, Y.; Schmauck-Medina, T.; Hansen, M.; Morimoto, R.I.; Simon, A.K.; Bjedov, I.; Palikaras, K.; Simonsen, A.; Johansen, T.; Tavernarakis, N.; et al. Autophagy in healthy aging and disease. Nat. Aging 2021, 1, 634–650. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Ookuma, S.; Nishida, E. Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans. Genes. Cells 2009, 14, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Canciglieri, P.H.; Kuga, G.K.; Muñoz, V.R.; Gaspar, R.C.; da Rocha, A.L.; Breda, L.; Anaruma, C.P.; Minuzzi, L.G.; da Silva, A.S.R.; Cintra, D.E.; et al. The reversal effect of physical exercise on aging-related increases in APPL2 content in skeletal muscle. Life Sci. 2018, 210, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Christoforidis, S.; Miaczynska, M.; Ashman, K.; Wilm, M.; Zhao, L.; Yip, S.C.; Waterfield, M.D.; Backer, J.M.; Zerial, M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1999, 1, 249–252. [Google Scholar] [CrossRef]
- Ravikumar, B.; Imarisio, S.; Sarkar, S.; O’Kane, C.J.; Rubinsztein, D.C. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J. Cell Sci. 2008, 121, 1649–1660. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Chen, Z.; Zhao, L.; Sun, J.; Yin, L.; Jiang, Y.; Shi, X.; Song, Z.; Zhang, L. Lack of T04C9.1, the Homologue of Mammalian APPL2, Leads to Premature Ageing and Shortens Lifespan in Caenorhabditis elegans. Genes 2024, 15, 659. https://doi.org/10.3390/genes15060659
Li Z, Chen Z, Zhao L, Sun J, Yin L, Jiang Y, Shi X, Song Z, Zhang L. Lack of T04C9.1, the Homologue of Mammalian APPL2, Leads to Premature Ageing and Shortens Lifespan in Caenorhabditis elegans. Genes. 2024; 15(6):659. https://doi.org/10.3390/genes15060659
Chicago/Turabian StyleLi, Zirui, Zhiqiang Chen, Lianghao Zhao, Jiaqi Sun, Lin Yin, Yuwei Jiang, Xiaotong Shi, Ziye Song, and Lu Zhang. 2024. "Lack of T04C9.1, the Homologue of Mammalian APPL2, Leads to Premature Ageing and Shortens Lifespan in Caenorhabditis elegans" Genes 15, no. 6: 659. https://doi.org/10.3390/genes15060659
APA StyleLi, Z., Chen, Z., Zhao, L., Sun, J., Yin, L., Jiang, Y., Shi, X., Song, Z., & Zhang, L. (2024). Lack of T04C9.1, the Homologue of Mammalian APPL2, Leads to Premature Ageing and Shortens Lifespan in Caenorhabditis elegans. Genes, 15(6), 659. https://doi.org/10.3390/genes15060659




