In Search of a Target Gene for a Desirable Phenotype in Aquaculture: Genome Editing of Cyprinidae and Salmonidae Species
Abstract
:1. Introduction
2. Genome Editing in Salmonidae and Cyprinidae Aquaculture Fish Species
2.1. Initial Studies on the Application of Genome Editing in Fish
2.2. Zebrafish as a Model Object in Studies Using Genome Editing
2.3. Genome Editing and Body Development in Fish
2.4. Genome Editing and Growth Traits in Fish
2.5. Genome Editing Affecting Fish Pigmentation
2.6. Genome Editing and Sex Determination in Fishes
2.7. Gene Editing and Disease Resistance in Fishes
3. Overcoming Technical Difficulties in Applying Genetic Engineering Approaches in Aquaculture
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The State of Food and Agriculture 2020; FAO: Rome, Italy, 2020.
- The State of World Fisheries and Aquaculture 2022; FAO: Rome, Italy, 2022.
- Mair, G.; Lucente, D. What are “farmed types” in aquaculture and why do they matter? FAO Aquac. Newsl. 2020, 61, 40–42. [Google Scholar]
- Donaldson, L.R.; Olson, P.R. Development of rainbow trout brood stock by selective breeding. Trans. Am. Fish. Soc. 1957, 85, 93–101. [Google Scholar] [CrossRef]
- Thodesen, J.; Gjedrem, T. Breeding programs on Atlantic salmon in Norway: Lessons learned . RePec 2006, 22–26. Available online: https://digitalarchive.worldfishcenter.org/bitstream/handle/20.500.12348/1861/WF_2455.pdf?sequence=1&isAllowed=y (accessed on 28 May 2024).
- Ponzoni, R.W.; Nguyen, N.H.; Khaw, H.L.; Hamzah, A.; Bakar, K.R.A.; Yee, H.Y. Genetic improvement of Nile tilapia (Oreochromis niloticus) with special reference to the work wonducted by the WorldFish Center with the GIFT srain. Rev. Aquac. 2011, 3, 27–41. [Google Scholar] [CrossRef]
- Zhai, G.; Shu, T.; Chen, K.; Lou, Q.; Jia, J.; Huang, J.; Shi, C.; Jin, X.; He, J.; Jiang, D.; et al. Successful production of an all-female common carp (Cyprinus carpio L.) population using cyp17a1-deficient neomale carp. Engineering 2022, 8, 181–189. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, Y.; Tay, Y.X.; Yue, G.H. Genome editing and its applications in genetic improvement in aquaculture. Rev. Aquac. 2022, 14, 178–191. [Google Scholar] [CrossRef]
- Blix, T.B.; Dalmo, R.A.; Wargelius, A.; Myhr, A.I. Genome editing on finfish: Currents and implications for sustainability. Rev. Aquac. 2021, 13, 2344–2363. [Google Scholar] [CrossRef]
- Chuang, Y.-F.; Phipps, A.J.; Lin, F.-L.; Hecht, V.; Hewitt, A.W.; Wang, P.-Y.; Liu, G.-S. Approach for in vivo delivery of CRISPR/Cas system: A recent update and future prospect. Cell. Mol. Life Sci. 2021, 78, 2683–2708. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fang, W.; Huang, J.; Li, S. The Application of genome editing technology in fish. Mar. Life Sci. Technol. 2021, 3, 326–346. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Kumar, V.; Behera, B.K.; Parhi, J.; Mohapatra, S.; Chakraborty, T.; Das, B.K. CRISPR/Cas Genome Editing—Can It Become a Game Changer in Future Fisheries Sector? Front. Mar. Sci. 2022, 9, 924475. [Google Scholar] [CrossRef]
- Okoli, A.S.; Blix, T.; Myhr, A.I.; Xu, W.; Xu, X. Sustainable use of CRISPR/Cas in fish aquaculture: The biosafety perspective. Transgenic Res. 2022, 31, 1–21. [Google Scholar] [CrossRef]
- Iqbal, G.; Quyoom, N.; Singh, L.S.; Ganpatbhai, A.V.K.; Bhat, N.M.; Gul, S.; Malik, M.A.; Mohanty, A.; Mir, S.A.; Dar, S.A. Genome editing technology in fishes. Curr. Appl. Sci. Technol. 2023, 42, 20–26. [Google Scholar] [CrossRef]
- Gutási, A.; Hammer, S.E.; El-Matbouli, M.; Saleh, M. Review: Recent applications of gene editing in fish species and aquatic medicine. Animals 2023, 13, 1250. [Google Scholar] [CrossRef] [PubMed]
- Mokrani, A.; Liu, S. Harnessing CRISPR/Cas9 system to improve economic traits in aquaculture species. Aquaculture 2024, 579, 740279. [Google Scholar] [CrossRef]
- Yang, L.; Xu, Z.; Zeng, H.; Sun, N.; Wu, B.; Wang, C.; Bo, J.; Li, L.; Dong, Y.; He, S. FishDB: An integrated functional genomics database for fishes. BMC Genom. 2020, 21, 801. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; McCammon, J.M.; Miller, J.C.; Faraji, F.; Ngo, C.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J.; et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Cade, L.; Khayter, C.; Reyon, D.; Peterson, R.T.; Joung, J.K.; Yeh, J.-R.J. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011, 29, 697–698. [Google Scholar] [CrossRef] [PubMed]
- Jao, L.-E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Ge, J.; Li, K.; Xu, Z.; Liang, D.; Li, J.; Li, J.; Jia, W.; Li, Y.; Dong, X.; et al. Heritable targeted inactivation of myostatin gene in yellow catfish (Pelteobagrus fulvidraco) using engineered zinc finger nucleases. PLoS ONE 2011, 6, e28897. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, H.; Zhao, J.; Fang, L.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L.; et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Edvardsen, R.B.; Leininger, S.; Kleppe, L.; Skaftnesmo, K.O.; Wargelius, A. Targeted mutagenesis in Atlantic salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS ONE 2014, 9, e108622. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Niu, P.; Wang, M.; Huang, G.; Xu, S.; Sun, Y.; Xu, X.; Hou, Y.; Sun, X.; Yan, Y.; et al. Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci. Rep. 2016, 6, 22953. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Fan, Y.; Zhou, Y.; Liu, W.; Jiang, N.; Zhang, J.; Zeng, L. Efficient resistance to Grass Carp Reovirus infection in JAM-A knockout cells using CRISPR/Cas9. Fish. Shellfish. Immunol. 2018, 76, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Liu, Y.; Wang, W.; Wang, Q.; Zhang, N.; Lin, F.; Wang, N.; Shao, C.; Dong, Z.; Li, Y.; et al. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 2017, 7, 42213. [Google Scholar] [CrossRef] [PubMed]
- Holtzman, N.G.; Iovine, M.K.; Liang, J.O.; Morris, J. Learning to fish with genetics: A primer on the vertebrate model Danio rerio. Genetics 2016, 203, 1069–1089. [Google Scholar] [CrossRef] [PubMed]
- Teame, T.; Zhang, Z.; Ran, C.; Zhang, H.; Yang, Y.; Ding, Q.; Xie, M.; Gao, C.; Ye, Y.; Duan, M.; et al. The use of zebrafish (Danio rerio) as biomedical models. Anim. Front. 2019, 9, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Chen, X.; Zhang, T.; Wang, R.; Wang, A.; Lan, X.; Zhou, Y.; Ma, S.; Xia, Q. The novel insight into the outcomes of CRISPR/Cas9 editing intra- and inter-species. Int. J. Biol. Macromol. 2020, 163, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, H.A.; Hua, K.; Quigley, H.; Ivare, J.; Tesson, F.; Akimenko, M. A CRISPR/Cas9 zebrafish lamin A/C mutant model of muscular laminopathy. Dev. Dynam 2022, 251, 645–661. [Google Scholar] [CrossRef] [PubMed]
- Naef, V.; Marchese, M.; Ogi, A.; Fichi, G.; Galatolo, D.; Licitra, R.; Doccini, S.; Verri, T.; Argenton, F.; Morani, F.; et al. Efficient neuroprotective rescue of sacsin-related disease phenotypes in zebrafish. Int. J. Mol. Sci. 2021, 22, 8401. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Hao, J.; Yao, Y. CRISPR/Cas9 in zebrafish: An attractive model for FBN1 genetic defects in humans. Mol. Genet. Genomic Med. 2021, 9, e1775. [Google Scholar] [CrossRef] [PubMed]
- Douek, A.M.; Amiri Khabooshan, M.; Henry, J.; Stamatis, S.-A.; Kreuder, F.; Ramm, G.; Änkö, M.-L.; Wlodkowic, D.; Kaslin, J. An engineered sgsh mutant zebrafish recapitulates molecular and behavioural pathobiology of Sanfilippo syndrome A/MPS IIIA. Int. J. Mol. Sci. 2021, 22, 5948. [Google Scholar] [CrossRef] [PubMed]
- Rusterholz, T.D.S.; Hofmann, C.; Bachmann-Gagescu, R. Insights gained from zebrafish models for the ciliopathy Joubert syndrome. Front. Genet. 2022, 13, 939527. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zhou, R.; Meng, P.; Wu, L.; Yang, L.; Liu, W.; Wu, J.; Cheng, Y.; Shi, L.; Zhang, Y. Establishment of a Bernard-Soulier syndrome model in zebrafish. Haematologica 2021, 107, 1655–1668. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, L.M.L.; Branco, E.V.; Sarafian, R.D.; Kobayashi, G.S.; de Araújo, F.T.; Santos Souza, L.; de Paula Moreira, D.; Hsia, G.S.P.; Bertollo, E.M.G.; Buck, C.B.; et al. Establishment of IPSC lines and zebrafish with loss-of-function ahdc1 variants: Models for Xia-Gibbs syndrome. Gene 2023, 871, 147424. [Google Scholar] [CrossRef] [PubMed]
- Haroon, S.; Yoon, H.; Seiler, C.; Osei-Frimpong, B.; He, J.; Nair, R.M.; Mathew, N.D.; Burg, L.; Kose, M.; Venkata, C.R.M.; et al. N-acetylcysteine and cysteamine bitartrate prevent azide-induced neuromuscular decompensation by restoring glutathione balance in two novel surf1−/− zebrafish deletion models of Leigh syndrome. Hum. Mol. Genet. 2023, 32, 1988–2004. [Google Scholar] [CrossRef] [PubMed]
- Heidary, S.; Awasthi, N.; Page, N.; Allnutt, T.; Lewis, R.S.; Liongue, C.; Ward, A.C. A zebrafish model of growth hormone insensitivity syndrome with immune dysregulation 1 (GHISID1). Cell Mol. Life Sci. 2023, 80, 109. [Google Scholar] [CrossRef] [PubMed]
- Withers, S.E.; Rowlands, C.F.; Tapia, V.S.; Hedley, F.; Mosneag, I.-E.; Crilly, S.; Rice, G.I.; Badrock, A.P.; Hayes, A.; Allan, S.M.; et al. Characterization of a mutant samhd1 zebrafish model implicates dysregulation of cholesterol biosynthesis in Aicardi-Goutières syndrome. Front. Immunol. 2023, 14, 1100967. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-S.; Devi, S.; He, J.C.; Zhou, W. A zebrafish model of congenital nephrotic syndrome of the Finnish type. Front. Cell Dev. Biol. 2022, 10, 976043. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Yu, X.; Ping, X.; Shentu, X.; Zou, J. Z nf469 plays a critical role in regulating synthesis of ECM: A zebrafish model of Brittle cornea syndrome. Investig. Ophthalmol. Vis. Sci. 2023, 64, 29. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Shao, W.; Zhong, D.; Ma, C.; Wei, X.; Ahmed, A.; Yu, T.; Jing, W.; Jing, L. Zebrafish mafbb mutants display osteoclast over-activation and bone deformity resembling osteolysis in MCTO patients. Biomolecules 2021, 11, 480. [Google Scholar] [CrossRef]
- Gao, P.; Jia, D.; Li, P.; Huang, Y.; Hu, H.; Sun, K.; Lv, Y.; Chen, X.; Han, Y.; Zhang, Z.; et al. Accumulation of lipid droplets in a novel bietti crystalline dystrophy zebrafish model with impaired PPARα pathway. Investig. Ophthalmol. Vis. Sci. 2022, 63, 32. [Google Scholar] [CrossRef] [PubMed]
- Dalla Barba, F.; Soardi, M.; Mouhib, L.; Risato, G.; Akyürek, E.E.; Lucon-Xiccato, T.; Scano, M.; Benetollo, A.; Sacchetto, R.; Richard, I.; et al. Modeling sarcoglycanopathy in Danio rerio. Int. J. Mol. Sci. 2023, 24, 12707. [Google Scholar] [CrossRef] [PubMed]
- Bögershausen, N.; Krawczyk, H.E.; Jamra, R.A.; Lin, S.; Yigit, G.; Hüning, I.; Polo, A.M.; Vona, B.; Huang, K.; Schmidt, J.; et al. wars1 and sars1: Two tRNA synthetases implicated in autosomal recessive microcephaly. Hum. Mutat. 2022, 43, 1454–1471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Alonzo, I.; Stubben, C.; Geng, Y.; Herdman, C.; Chandler, N.; Doane, K.P.; Pluimer, B.R.; Trauger, S.A.; Peterson, R.T. A Zebrafish model of combined saposin deficiency identifies acid sphingomyelinase as a potential therapeutic target. Dis. Model. Mech. 2023, 16, dmm049995. [Google Scholar] [CrossRef] [PubMed]
- Facchinello, N.; Laquatra, C.; Locatello, L.; Beffagna, G.; Brañas Casas, R.; Fornetto, C.; Dinarello, A.; Martorano, L.; Vettori, A.; Risato, G.; et al. Efficient clofilium tosylate-mediated rescue of POLG-related disease phenotypes in zebrafish. Cell Death Dis. 2021, 12, 100. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Jones, J.L.; Gasperini, R.J.; Charlesworth, J.C.; Liu, G.-S.; Burdon, K.P. Rapid and efficient cataract gene evaluation in F0 zebrafish using CRISPR-Cas9 ribonucleoprotein complexes. Methods 2021, 194, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Jarayseh, T.; Guillemyn, B.; De Saffel, H.; Bek, J.W.; Syx, D.; Symoens, S.; Gansemans, Y.; Van Nieuwerburgh, F.; Jagadeesh, S.; Raja, J.; et al. A tapt1 knock-out zebrafish line with aberrant lens development and impaired vision models human early-onset cataract. Hum. Genet. 2023, 142, 457–476. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, T.; Chen, B.; Liu, Y.; Lu, X.; Li, J. lrpap1 deficiency leads to myopia through TGF-β-induced apoptosis in zebrafish. Cell Commun. Signal 2022, 20, 162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, S.; Wang, L.; Wang, X.; Chen, T.; Chen, Z.; Jiang, Y. Zonular defects in loxl1-deficient zebrafish. Clin. Exp. Ophthalmol. 2022, 50, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Zhang, Z.; Nie, Z.; Wang, C.; Meng, F.; Yi, Q.; Chen, M.; Sun, J.; Zou, J.; Jiang, P.; et al. An animal model for mitochondrial tyrosyl-TRNA synthetase deficiency reveals links between oxidative phosphorylation and retinal function. J. Biol. Chem. 2021, 296, 100437. [Google Scholar] [CrossRef] [PubMed]
- Stemerdink, M.; Broekman, S.; Peters, T.; Kremer, H.; de Vrieze, E.; van Wijk, E. Generation and characterization of a zebrafish model for ADGRV1-associated retinal dysfunction using CRISPR/Cas9 genome editing technology. Cells 2023, 12, 1598. [Google Scholar] [CrossRef] [PubMed]
- Gong, K.; Xie, T.; Yang, Y.; Luo, Y.; Deng, Y.; Chen, K.; Tan, Z.; Guo, H.; Xie, L. Establishment of a dihydrofolate reductase gene knock-in zebrafish strain to aid preliminary analysis of congenital heart disease mechanisms. Front. Cardiovasc. Med. 2021, 8, 763851. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.S.; Wang, D.; Lu, Z.; Zhang, Y.; Li, S.; Lin, Y.; Tan, W. CNPase, a 2′,3′-cyclic-nucleotide 3′-phosphodiesterase, as a therapeutic target to attenuate cardiac hypertrophy by enhancing mitochondrial energy production. Int. J. Mol. Sci. 2021, 22, 10806. [Google Scholar] [CrossRef] [PubMed]
- Chun, Y.W.; Miyamoto, M.; Williams, C.H.; Neitzel, L.R.; Silver-Isenstadt, M.; Cadar, A.G.; Fuller, D.T.; Fong, D.C.; Liu, H.; Lease, R.; et al. Impaired reorganization of centrosome structure underlies human infantile dilated cardiomyopathy. Circulation 2023, 147, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Hayashi, K.; Kobayashi, I.; Hosomichi, K.; Nomura, A.; Teramoto, R.; Usuda, K.; Okada, H.; Deng, Y.; Kobayashi-Sun, J.; et al. The utility of zebrafish cardiac arrhythmia model to predict the pathogenicity of KCNQ1 variants. J. Mol. Cell Cardiol. 2023, 177, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.H.; Seet, S.H.; Maier, M.; Gurel, A.; Traspas, R.M.; Lee, C.; Zhang, S.; Talim, B.; Loh, A.Y.T.; Chia, C.Y.; et al. Loss of c2orf69 defines a fatal autoinflammatory syndrome in humans and zebrafish that evokes a glycogen-storage-associated mitochondriopathy. Am. J. Hum. Genet. 2021, 108, 1301–1317. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.-H.; Yeh, K.-Y.; Hsu, H.-M.; Her, G.M. Deficiency of adipose triglyceride lipase induces metabolic syndrome and cardiomyopathy in zebrafish. Int. J. Mol. Sci. 2022, 24, 117. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Cao, X.; Castro, L.F.C.; Monroig, Ó.; Gao, J. A network-based approach to identify protein kinases critical for regulating srebf1 in lipid deposition causing obesity. Funct. Integr. Genom. 2021, 21, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Ojanen, M.J.T.; Uusi-Mäkelä, M.I.E.; Harjula, S.-K.E.; Saralahti, A.K.; Oksanen, K.E.; Kähkönen, N.; Määttä, J.A.E.; Hytönen, V.P.; Pesu, M.; Rämet, M. Intelectin 3 is dispensable for resistance against a mycobacterial infection in zebrafish (Danio rerio). Sci. Rep. 2019, 9, 995. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, L.; Meng, P.; Zhang, X.; Zhao, D.; Lin, Q.; Zhang, Y. Generation of a thrombopoietin-deficient thrombocytopenia model in zebrafish. J. Thromb. Haemost. 2022, 20, 1900–1909. [Google Scholar] [CrossRef] [PubMed]
- Zada, A.; Kuil, L.E.; de Graaf, B.M.; Kakiailatu, N.; Windster, J.D.; Brooks, A.S.; van Slegtenhorst, M.; de Koning, B.; Wijnen, R.M.H.; Melotte, V.; et al. TFAP2B haploinsufficiency impacts gastrointestinal function and leads to pediatric intestinal pseudo-obstruction. Front. Cell Dev. Biol. 2022, 10, 901824. [Google Scholar] [CrossRef] [PubMed]
- Moreno Traspas, R.; Teoh, T.S.; Wong, P.-M.; Maier, M.; Chia, C.Y.; Lay, K.; Ali, N.A.; Larson, A.; Al Mutairi, F.; Al-Sannaa, N.A.; et al. Loss of FOCAD, operating via the SKI messenger RNA surveillance pathway, causes a pediatric syndrome with liver cirrhosis. Nat. Genet. 2022, 54, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wan, J.-P.; Zhang, R.-J.; Sun, F.; Yang, L.; Zhao, S.-X.; Dong, M.; Song, H.-D. tpo knockout in zebrafish partially recapitulates clinical manifestations of congenital hypothyroidism and reveals the involvement of Th in proper development of glucose homeostasis. Gen. Comp. Endocrinol. 2022, 323–324, 114033. [Google Scholar] [CrossRef] [PubMed]
- Shihana, F.; Cholan, P.M.; Fraser, S.; Oehlers, S.H.; Seth, D. Investigating the role of lipid genes in liver disease using fatty liver models of alcohol and high fat in zebrafish (Danio rerio). Liver Int. 2023, 43, 2455–2468. [Google Scholar] [CrossRef] [PubMed]
- Seda, M.; Crespo, B.; Corcelli, M.; Osborn, D.P.; Jenkins, D. A CRISPR/Cas9-generated mutation in the zebrafish orthologue of PPP2R3B causes idiopathic scoliosis. Sci. Rep. 2023, 13, 6783. [Google Scholar] [CrossRef] [PubMed]
- Rebello, D.; Wohler, E.; Erfani, V.; Li, G.; Aguilera, A.N.; Santiago-Cornier, A.; Zhao, S.; Hwang, S.W.; Steiner, R.D.; Zhang, T.J.; et al. COL11A2 as a candidate gene for vertebral malformations and congenital scoliosis. Hum. Mol. Genet. 2023, 32, 2913–2928. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Lu, C.; Feng, M.; Dai, L.; Wang, M.; Qiu, Y.; Zheng, H.; Liu, Y.; Li, L.; Tang, B.; et al. Cooperation between liver-specific mutations of pten and tp53 genetically induces hepatocarcinogenesis in zebrafish. J. Exp. Clin. Cancer Res. 2021, 40, 262. [Google Scholar] [CrossRef] [PubMed]
- Dona, M.; Waaijers, S.; Richter, S.; Eisenhofer, G.; Korving, J.; Kamel, S.M.; Bakkers, J.; Rapizzi, E.; Rodenburg, R.J.; Zethof, J.; et al. Loss of sdhb in zebrafish larvae recapitulates human paraganglioma characteristics. Endocr. Relat. Cancer 2021, 28, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Moral-Sanz, J.; Fernandez-Rojo, M.A.; Colmenarejo, G.; Kurdyukov, S.; Brust, A.; Ragnarsson, L.; Andersson, Å.; Vila, S.F.; Cabezas-Sainz, P.; Wilhelm, P.; et al. The structural conformation of the tachykinin domain drives the anti-tumoural activity of an octopus peptide in melanoma BRAFV600E. Br. J. Pharmacol. 2022, 179, 4878–4896. [Google Scholar] [CrossRef] [PubMed]
- Oppel, F.; Shao, S.; Gendreizig, S.; Zimmerman, M.W.; Schürmann, M.; Flavian, V.F.; Goon, P.; Chi, S.N.; Aster, J.C.; Sudhoff, H.; et al. P53 pathway inactivation drives SMARCB1-deficient P53-wildtype epithelioid sarcoma onset indicating therapeutic vulnerability through MDM2 inhibition. Mol. Cancer Ther. 2022, 21, 1689–1700. [Google Scholar] [CrossRef] [PubMed]
- Russell, K.L.; Downie, J.M.; Gibson, S.B.; Tsetsou, S.; Keefe, M.D.; Duran, J.A.; Figueroa, K.P.; Bromberg, M.B.; Murtaugh, L.C.; Bonkowsky, J.L.; et al. Pathogenic effect of TP73 gene variants in people with amyotrophic lateral sclerosis. Neurology 2021, 97, e225–e235. [Google Scholar] [CrossRef] [PubMed]
- Griffin, A.; Carpenter, C.; Liu, J.; Paterno, R.; Grone, B.; Hamling, K.; Moog, M.; Dinday, M.T.; Figueroa, F.; Anvar, M.; et al. Phenotypic analysis of catastrophic childhood epilepsy genes. Commun. Biol. 2021, 4, 680. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ortiz, R.; Martínez-Torres, A. Mutants of the zebrafish K+ channel hcn2b exhibit epileptic-like behaviors. Int. J. Mol. Sci. 2021, 22, 11471. [Google Scholar] [CrossRef] [PubMed]
- Miguel Sanz, C.; Martinez Navarro, M.; Caballero Diaz, D.; Sanchez-Elexpuru, G.; Di Donato, V. Toward the use of novel alternative methods in epilepsy modeling and drug discovery. Front. Neurol. 2023, 14, 1213969. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Chen, Y.-H. SCAF4 variants associated with focal epilepsy accompanied by multisystem disorders. Seizure-Eur. J. Epilep 2023, 116, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Dogra, D.; Meza-Santoscoy, P.L.; Gavrilovici, C.; Rehak, R.; de la Hoz, C.L.R.; Ibhazehiebo, K.; Rho, J.M.; Kurrasch, D.M. kcna1a mutant zebrafish model episodic ataxia type 1 (EA1) with epilepsy and show response to first-line therapy carbamazepine. Epilepsia 2023, 64, 2186–2199. [Google Scholar] [CrossRef] [PubMed]
- Kuil, L.E.; MacKenzie, K.C.; Tang, C.S.; Windster, J.D.; Le, T.L.; Karim, A.; de Graaf, B.M.; van der Helm, R.; van Bever, Y.; Sloots, C.E.J.; et al. Size matters: Large copy number losses in hirschsprung disease patients reveal genes involved in enteric nervous system development. PLoS Genet. 2021, 17, e1009698. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Long, F.; Cao, X.; Xiong, B.; Li, Y. Knockout of katnal2 leads to autism-like behaviors and developmental delay in zebrafish. Int. J. Mol. Sci. 2022, 23, 8389. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Long, H.; Guo, J.; Wang, Y.; He, K.; Tao, C.; Li, X.; Jiang, K.; Guo, S.; Pi, Y. Autism-risk gene necab2 regulates psychomotor and social behavior as a neuronal modulator of MGluR1 signaling. Front. Mol. Neurosci. 2022, 15, 901682. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Wang, Y.; Hu, M.; Lin, J.; Li, Q.; Liu, C.; Xu, X. Deleterious variation in BR serine/threonine kinase 2 classified a subtype of autism. Front. Mol. Neurosci. 2022, 15, 904935. [Google Scholar] [CrossRef] [PubMed]
- Sumathipala, S.H.; Khan, S.; Kozol, R.A.; Araki, Y.; Syed, S.; Huganir, R.L.; Dallman, J.E. Context-dependent hyperactivity in syngap1a and syngap1b zebrafish autism models. bioRxiv 2023, 557316. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, C.; Peng, G. ap4s1 truncation leads to axonal defects in a zebrafish model of spastic paraplegia 52. Int. J. Dev. Neurosci. 2023, 83, 753–764. [Google Scholar] [CrossRef]
- Jiang, Y.-J.; Fann, C.S.-J.; Fuh, J.-L.; Chung, M.-Y.; Huang, H.-Y.; Chu, K.-C.; Wang, Y.-F.; Hsu, C.-L.; Kao, L.-S.; Chen, S.-P.; et al. Genome-wide analysis identified novel susceptible genes of restless legs syndrome in migraineurs. J. Headache Pain. 2022, 23, 39. [Google Scholar] [CrossRef] [PubMed]
- Sonti, S.; Littleton, S.H.; Pahl, M.C.; Zimmerman, A.J.; Chesi, A.; Palermo, J.; Lasconi, C.; Brown, E.B.; Pippin, J.A.; Wells, A.D.; et al. Perturbation of the insomnia wdr90 GWAS locus pinpoints Rs3752495 as a causal variant influencing distal expression of neighboring gene, PIG-Q. bioRxiv 2023, 553739. [Google Scholar] [CrossRef] [PubMed]
- Leggieri, A.; García-González, J.; Torres-Perez, J.V.; Havelange, W.; Hosseinian, S.; Mech, A.M.; Keatinge, M.; Busch-Nentwich, E.M.; Brennan, C.H. ankk1 loss of function disrupts dopaminergic pathways in zebrafish. Front. Neurosci. 2022, 16, 794653. [Google Scholar] [CrossRef]
- Baronio, D.; Chen, Y.; Decker, A.R.; Enckell, L.; Fernández-López, B.; Semenova, S.; Puttonen, H.A.J.; Cornell, R.A.; Panula, P. Vesicular monoamine transporter 2 (SLC18A2) regulates monoamine turnover and brain development in zebrafish. Acta Physiol. 2022, 234, e13725. [Google Scholar] [CrossRef]
- Brożko, N.; Baggio, S.; Lipiec, M.A.; Jankowska, M.; Szewczyk, Ł.M.; Gabriel, M.O.; Chakraborty, C.; Ferran, J.L.; Wiśniewska, M.B. Genoarchitecture of the early postmitotic pretectum and the role of wnt signaling in shaping pretectal neurochemical anatomy in zebrafish. Front. Neuroanat. 2022, 16, 838567. [Google Scholar] [CrossRef]
- Yao, Y.; Baronio, D.; Chen, Y.-C.; Jin, C.; Panula, P. The roles of histamine receptor 1 (hrh1) in neurotransmitter system regulation, behavior, and neurogenesis in zebrafish. Mol. Neurobiol. 2023, 60, 6660–6675. [Google Scholar] [CrossRef]
- Maili, L.; Tandon, B.; Yuan, Q.; Menezes, S.; Chiu, F.; Hashmi, S.S.; Letra, A.; Eisenhoffer, G.T.; Hecht, J.T. Disruption of fos causes craniofacial anomalies in developing zebrafish. Front. Cell Dev. Biol. 2023, 11, 1141893. [Google Scholar] [CrossRef] [PubMed]
- Mankiewicz, J.L.; Picklo, M.J.; Idso, J.; Cleveland, B.M. Leptin receptor deficiency results in hyperphagia and increased fatty acid mobilization during fasting in rainbow trout (Oncorhynchus mykiss). Biomolecules 2022, 12, 516. [Google Scholar] [CrossRef] [PubMed]
- Gemmer, A.; Mirkes, K.; Anneser, L.; Eilers, T.; Kibat, C.; Mathuru, A.; Ryu, S.; Schuman, E. Oxytocin receptors influence the development and maintenance of social behavior in zebrafish (Danio rerio). Sci. Rep. 2022, 12, 4322. [Google Scholar] [CrossRef] [PubMed]
- Zoodsma, J.D.; Keegan, E.J.; Moody, G.R.; Bhandiwad, A.A.; Napoli, A.J.; Burgess, H.A.; Wollmuth, L.P.; Sirotkin, H.I. Disruption of grin2b, an ASD-Associated gene, produces social deficits in zebrafish. Mol. Autism 2022, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Barnaby, W.; Dorman Barclay, H.E.; Nagarkar, A.; Perkins, M.; Teicher, G.; Trapani, J.G.; Downes, G.B. GABAA α subunit control of hyperactive behavior in developing zebrafish. Genetics 2022, 220, iyac011. [Google Scholar] [CrossRef] [PubMed]
- Torres-Pérez, J.V.; Anagianni, S.; Mech, A.M.; Havelange, W.; García-González, J.; Fraser, S.E.; Vallortigara, G.; Brennan, C.H. baz1b loss-of-function in zebrafish produces phenotypic alterations consistent with the domestication syndrome. iScience 2023, 26, 105704. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, Y.; Frigato, E.; Noviello, T.M.R.; Toni, M.; Frabetti, F.; Cigliano, L.; Ceccarelli, M.; Sordino, P.; Cerulo, L.; Bertolucci, C.; et al. Loss of circadian rhythmicity in bdnf knockout zebrafish larvae. iScience 2022, 25, 104054. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Vorontsova, I.; Hoshino, M.; Uesugi, K.; Yagi, N.; Hall, J.E.; Schilling, T.F.; Pierscionek, B.K. Aquaporins have regional functions in development of refractive index in the zebrafish eye lens. Investig. Ophthalmol. Vis. Sci. 2021, 62, 23. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Qin, Y.; Huang, Y.; Gao, P.; Li, J.; Yu, S.; Jia, D.; Chen, X.; Lv, Y.; Tu, J.; et al. Rod genesis driven by mafba in an nrl knockout zebrafish model with altered photoreceptor composition and progressive retinal degeneration. PLoS Genet. 2022, 18, e1009841. [Google Scholar] [CrossRef] [PubMed]
- Letelier, J.; Buono, L.; Almuedo-Castillo, M.; Zang, J.; Mounieres, C.; González-Díaz, S.; Polvillo, R.; Sanabria-Reinoso, E.; Corbacho, J.; Sousa-Ortega, A.; et al. Mutation of vsx genes in zebrafish highlights the robustness of the retinal specification network. eLife 2023, 12, e85594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jing, M.; Li, P.; Sun, L.; Pi, X.; Jiang, N.; Zhu, K.; Li, H.; Li, J.; Wang, M.; et al. Knockout of DLIC1 leads to retinal cone degeneration via disturbing Rab8 transport in zebrafish. Biochim. Biophys. Acta 2023, 1869, 166645. [Google Scholar] [CrossRef] [PubMed]
- Davison, C.; Zolessi, F.R. slit2 is necessary for optic axon organization in the zebrafish ventral midline. Cells Dev. 2021, 166, 203677. [Google Scholar] [CrossRef] [PubMed]
- Davison, C.; Bedó, G.; Zolessi, F.R. Zebrafish slit2 and slit3 act together to regulate retinal axon crossing at the midline. J. Dev. Biol. 2022, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Halabi, R.; Watterston, C.; Hehr, C.L.; Mori-Kreiner, R.; Childs, S.J.; McFarlane, S. Semaphorin 3fa controls ocular vascularization from the embryo through to the adult. Investig. Ophthalmol. Vis. Sci. 2021, 62, 21. [Google Scholar] [CrossRef] [PubMed]
- Wohlfart, D.P.; Lou, B.; Middel, C.S.; Morgenstern, J.; Fleming, T.; Sticht, C.; Hausser, I.; Hell, R.; Hammes, H.-P.; Szendrödi, J.; et al. Accumulation of acetaldehyde in aldh2.1 zebrafish causes increased retinal angiogenesis and impaired glucose metabolism. Redox Biol. 2022, 50, 102249. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Silva, R.; Dantas, V.L.G.; Alves, L.U.; Batissoco, A.C.; Oiticica, J.; Lawrence, E.A.; Kawafi, A.; Yang, Y.; Nicastro, F.S.; Novaes, B.C.; et al. ncoa3 identified as a new candidate to explain autosomal dominant progressive hearing loss. Hum. Mol. Genet. 2021, 29, 3691–3705. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Y.; Gao, P.; Lv, Y.; Jia, D.; Sun, K.; Han, Y.; Hu, H.; Tang, Z.; Ren, X.; et al. Knockout of mafba causes inner-ear developmental defects in zebrafish via the impairment of proliferation and differentiation of ionocyte progenitor cells. Biomedicines 2021, 9, 1699. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.L.; Mohanty, S.; Guo, J.; Lekven, A.C.; Riley, B.B. pax2a, sp5a and sp5l act downstream of fgf and wnt to coordinate sensory-neural patterning in the inner ear. Dev. Biol. 2022, 492, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Ezhkova, D.; Schwarzer, S.; Spieß, S.; Geffarth, M.; Machate, A.; Zöller, D.; Stucke, J.; Alexopoulou, D.; Lesche, M.; Dahl, A.; et al. Transcriptome analysis reveals an atoh1b-dependent gene set downstream of dlx3b/4b during early inner ear development in zebrafish. Biol. Open 2023, 12, bio059911. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ping, L.; Gao, R.; Zhang, B.; Chen, X. lmo4a contributes to zebrafish inner ear and vestibular development via regulation of the Bmp pathway. Genes 2023, 14, 1371. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Zeng, C.; Lu, H.; Yue, Y.; Sun, T.; Zhou, X.; Li, G.; Ai, N.; Ge, W. Genetic and epigenetic evidence for nonestrogenic disruption of otolith development by bisphenol a in zebrafish. Environ. Sci. Technol. 2023, 57, 16190–16205. [Google Scholar] [CrossRef] [PubMed]
- Jami, M.S.; Murata, H.; Barnhill, L.M.; Li, S.; Bronstein, J.M. Diesel exhaust exposure alters the expression of networks implicated in neurodegeneration in zebrafish brains. Cell Biol. Toxicol. 2023, 39, 641–655. [Google Scholar] [CrossRef]
- Guo, Y.; Oliveros, C.F.; Ohshima, T. CRMP2 and CRMP4 are required for the formation of commissural tracts in the developing zebrafish forebrain. Dev. Neurobiol. 2022, 82, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-S.; Lu, Y.-F.; Liu, Y.-H.; Huang, C.-J.; Hwang, S.-P.L. Zebrafish cdx1b modulates epithalamic asymmetry by regulating ndr2 and lft1 expression. Dev. Biol. 2021, 470, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Antón-Galindo, E.; Dalla Vecchia, E.; Orlandi, J.G.; Castro, G.; Gualda, E.J.; Young, A.M.J.; Guasch-Piqueras, M.; Arenas, C.; Herrera-Úbeda, C.; Garcia-Fernàndez, J.; et al. Deficiency of the ywhaz gene, involved in neurodevelopmental disorders, alters brain activity and behaviour in zebrafish. Mol. Psychiatry 2022, 27, 3739–3748. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.-Y.; Wu, F.-Y.; Sun, F.; Fang, Y.; Zhang, R.-J.; Zhang, C.-R.; Zhang, C.-X.; Wang, Z.; Yang, R.-M.; Yang, L.; et al. The Isl2a transcription factor regulates pituitary development in zebrafish. Front. Endocrinol. 2023, 14, 920548. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Huang, S.; Zhao, H.; Cao, K.; Gan, J.; Yang, C.; Xu, Z.; Li, S.; Su, B. Zebrafish minichromosome maintenance protein 5 gene regulates the development and migration of facial motor neurons via fibroblast growth factor signaling. Dev. Neurosci. 2021, 43, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Wasserman-Bartov, T.; Admati, I.; Lebenthal-Loinger, I.; Sharabany, J.; Lerer-Goldshtein, T.; Appelbaum, L. tsh induces Agrp1 neuron proliferation in oatp1c1-deficient zebrafish. J. Neurosci. 2022, 42, 8214–8224. [Google Scholar] [CrossRef] [PubMed]
- Poulain, F.E. Analyzing the role of heparan sulfate proteoglycans in axon guidance in vivo in zebrafish. Methods Mol. Biol. 2022, 1229, 469–482. [Google Scholar]
- Soto, X.; Burton, J.; Manning, C.S.; Minchington, T.; Lea, R.; Lee, J.; Kursawe, J.; Rattray, M.; Papalopulu, N. Sequential and additive expression of MIR-9 precursors control timing of neurogenesis. Development 2022, 149, dev200474. [Google Scholar] [CrossRef] [PubMed]
- Limbach, L.E.; Penick, R.L.; Casseday, R.S.; Hyland, M.A.; Pontillo, E.A.; Ayele, A.N.; Pitts, K.M.; Ackerman, S.D.; Harty, B.L.; Herbert, A.L.; et al. Peripheral nerve development in zebrafish requires muscle patterning by tcf15/paraxis. Dev. Biol. 2022, 490, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Vaz, R.; Edwards, S.; Dueñas-Rey, A.; Hofmeister, W.; Lindstrand, A. Loss of ctnnd2b affects neuronal differentiation and behavior in zebrafish. Front. Neurosci. 2023, 17, 1205653. [Google Scholar] [CrossRef] [PubMed]
- Saraswathy, V.M.; Zhou, L.; Mokalled, M.H. Single-cell analysis of innate spinal cord regeneration identifies intersecting modes of neuronal repair. bioRxiv 2023, 541505. [Google Scholar] [CrossRef] [PubMed]
- Altbürger, C.; Holzhauser, J.; Driever, W. CRISPR/Cas9-based QF2 knock-in at the tyrosine hydroxylase (Th) locus reveals novel Th-expressing neuron populations in the zebrafish mid- and hindbrain. Front. Neuroanat. 2023, 17, 1196868. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Xue, R.; Chen, J.; Xu, J. dock8 deficiency attenuates microglia colonization in early zebrafish larvae. Cell Death Discov. 2022, 8, 366. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, Z.; Li, Y.; Song, Z.; Yin, W.; Hu, B. Deletion of the chd7 hinders oligodendrocyte progenitor cell development and myelination in zebrafish. Int. J. Mol. Sci. 2023, 24, 13535. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Sun, Y. The polycomb group gene rnf2 is essential for central and enteric neural system development in zebrafish. Front. Neurosci. 2022, 16, 960149. [Google Scholar] [CrossRef] [PubMed]
- Smits, D.J.; Dekker, J.; Schot, R.; Tabarki, B.; Alhashem, A.; Demmers, J.A.A.; Dekkers, D.H.W.; Romito, A.; van der Spek, P.J.; van Ham, T.J.; et al. CLEC16A interacts with retromer and TRIM27, and its loss impairs endosomal trafficking and neurodevelopment. Hum. Genet. 2023, 142, 379–397. [Google Scholar] [CrossRef] [PubMed]
- Paz, D.; Reyes-Nava, N.G.; Pinales, B.E.; Perez, I.; Gil, C.B.; Gonzales, A.V.; Grajeda, B.; Estevao, I.L.; Ellis, C.C.; Castro, V.L.; et al. Characterization of the zebrafish Gabra1sa43718/Sa43718 germline loss of function allele confirms a function for gabra1 in motility and nervous system development. bioRxiv 2023, 525860. [Google Scholar] [CrossRef] [PubMed]
- Bu, H.; Ding, Y.; Li, J.; Zhu, P.; Shih, Y.-H.; Wang, M.; Zhang, Y.; Lin, X.; Xu, X. Inhibition of mTOR or MAPK ameliorates vmhcl/myh7 cardiomyopathy in zebrafish. JCI Insight 2021, 6, e154215. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Feng, G.; Zhang, Y.; Sun, Y. PRC1 stabilizes cardiac contraction by regulating cardiac sarcomere assembly and cardiac conduction system construction. Int. J. Mol. Sci. 2021, 22, 11368. [Google Scholar] [CrossRef] [PubMed]
- Kamel, S.M.; Koopman, C.D.; Kruse, F.; Willekers, S.; Chocron, S.; Bakkers, J. A heterozygous mutation in cardiac troponin T promotes Ca2+ dysregulation and adult cardiomyopathy in zebrafish. J. Cardiovasc. Dev. Dis. 2021, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Ge, X.; Qian, P.; Lu, X.; Liu, D.; Chen, C. Neuron navigator 3 (NAV3) is required for heart development in zebrafish. Fish. Physiol. Biochem. 2022, 48, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Zink, M.; Seewald, A.; Rohrbach, M.; Brodehl, A.; Liedtke, D.; Williams, T.; Childs, S.J.; Gerull, B. Altered expression of TMEM43 causes abnormal cardiac structure and function in zebrafish. Int. J. Mol. Sci. 2022, 23, 9530. [Google Scholar] [CrossRef] [PubMed]
- Derrick, C.J.; Sánchez-Posada, J.; Hussein, F.; Tessadori, F.; Pollitt, E.J.G.; Savage, A.M.; Wilkinson, R.N.; Chico, T.J.; van Eeden, F.J.; Bakkers, J.; et al. Asymmetric hapln1a drives regionalized cardiac ECM Expansion and promotes heart morphogenesis in zebrafish development. Cardiovasc. Res. 2022, 118, 226–240. [Google Scholar] [CrossRef]
- Huttner, I.G.; Santiago, C.F.; Jacoby, A.; Cheng, D.; Trivedi, G.; Cull, S.; Cvetkovska, J.; Chand, R.; Berger, J.; Currie, P.D.; et al. Loss of Sec-1 family domain-containing 1 (Scfd1) causes severe cardiac defects and endoplasmic reticulum stress in zebrafish. J. Cardiovasc. Dev. Dis. 2023, 10, 408. [Google Scholar] [CrossRef] [PubMed]
- Leid, J.; Gray, R.; Rakita, P.; Koenig, A.L.; Tripathy, R.; Fitzpatrick, J.A.J.; Kaufman, C.; Solnica-Krezel, L.; Lavine, K.J. Deletion of taf1 and taf5 in zebrafish capitulate cardiac and craniofacial abnormalities associated with TAFopathies through perturbations in metabolism. Biol. Open 2023, 12, bio059905. [Google Scholar] [CrossRef] [PubMed]
- Singh Angom, R.; Wang, Y.; Wang, E.; Dutta, S.K.; Mukhopadhyay, D. Conditional, Tissue-specific CRISPR/Cas9 vector system in zebrafish reveals the role of nrp1b in heart regeneration. Arterioscler. Thromb. Vasc. Biol. 2023, 43, 1921–1934. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Eghbalian, R.; Boeckel, J.-N.; Frese, K.S.; Haas, J.; Kayvanpour, E.; Sedaghat-Hamedani, F.; Lackner, M.K.; Tugrul, O.F.; Ruppert, T.; et al. NIMA-related kinase 9 regulates the phosphorylation of the essential myosin light chain in the heart. Nat. Commun. 2022, 13, 6209. [Google Scholar] [CrossRef] [PubMed]
- Perl, E.; Ravisankar, P.; Beerens, M.E.; Mulahasanovic, L.; Smallwood, K.; Sasso, M.B.; Wenzel, C.; Ryan, T.D.; Komár, M.; Bove, K.E.; et al. stx4 is required to regulate cardiomyocyte Ca2+ handling during vertebrate cardiac development. Hum. Genet. Genom. Adv. 2022, 3, 100115. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.E.; Ravisankar, P.; Beerens, M.; MacRae, C.A.; Waxman, J.S. nr2f1a maintains atrial nkx2.5 expression to repress pacemaker identity within venous atrial cardiomyocytes of zebrafish. eLife 2023, 12, e77408. [Google Scholar] [CrossRef] [PubMed]
- Vaparanta, K.; Jokilammi, A.; Paatero, I.; Merilahti, J.A.; Heliste, J.; Hemanthakumar, K.A.; Kivelä, R.; Alitalo, K.; Taimen, P.; Elenius, K. STAT5b is a key effector of NRG-1/ERBB4/Scp-mediated Myocardial growth. EMBO Rep. 2023, 24, e56689. [Google Scholar] [CrossRef] [PubMed]
- Hesaraki, M.; Bora, U.; Pahlavan, S.; Salehi, N.; Mousavi, S.A.; Barekat, M.; Rasouli, S.J.; Baharvand, H.; Ozhan, G.; Totonchi, M. A novel missense variant in actin binding domain of MYH7 is associated with left ventricular noncompaction. Front. Cardiovasc. Med. 2022, 9, 839862. [Google Scholar] [CrossRef] [PubMed]
- Joyce, W.; Pan, Y.K.; Garvey, K.; Saxena, V.; Perry, S.F. Regulation of heart rate following genetic deletion of the SS1 adrenergic receptor in larval zebrafish. Acta Physiol. 2022, 235, e13849. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tran, V.; Shaked, I.; Xue, B.; Moore, T.; Lightle, R.; Kleinfeld, D.; Awad, I.A.; Ginsberg, M.H. Abortive intussusceptive angiogenesis causes multi-cavernous vascular malformations. eLife 2021, 10, e62155. [Google Scholar] [CrossRef] [PubMed]
- Khajavi, M.; Zhou, Y.; Schiffer, A.J.; Bazinet, L.; Birsner, A.E.; Zon, L.; D’Amato, R.J. Identification of basp1 as a novel angiogenesis-regulating gene by multi-model system studies. FASEB J. 2021, 35, e21404. [Google Scholar] [CrossRef] [PubMed]
- Parial, R.; Li, H.; Li, J.; Archacki, S.; Yang, Z.; Wang, I.Z.; Chen, Q.; Xu, C.; Wang, Q.K. Role of epigenetic m6A RNA methylation in vascular development: mettl3 regulates vascular development through PHLPP2/MTOR-AKT signaling. FASEB J. 2021, 35, e21465. [Google Scholar] [CrossRef] [PubMed]
- Kempers, L.; Wakayama, Y.; van der Bijl, I.; Furumaya, C.; De Cuyper, I.M.; Jongejan, A.; Kat, M.; van Stalborch, A.-M.D.; van Boxtel, A.L.; Hubert, M.; et al. The endosomal RIN2/Rab5C machinery prevents VEGFR2 degradation to control gene expression and tip cell identity during angiogenesis. Angiogenesis 2021, 24, 695–714. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Wu, C. Functional characterization of stap2b in zebrafish vascular development. FASEB J. 2023, 37, e23053. [Google Scholar] [CrossRef] [PubMed]
- Hopfenmüller, V.L.; Perner, B.; Reuter, H.; Bates, T.J.D.; Große, A.; Englert, C. The Wilms tumor gene wt1a contributes to blood-cerebrospinal fluid barrier function in zebrafish. Front. Cell Dev. Biol. 2022, 9, 809962. [Google Scholar] [CrossRef] [PubMed]
- Pak, B.; Schmitt, C.E.; Oh, S.; Kim, J.-D.; Choi, W.; Han, O.; Kim, M.; Kim, M.-J.; Ham, H.-J.; Kim, S.; et al. pax9 is essential for granulopoiesis but dispensable for erythropoiesis in zebrafish. Biochem. Biophys. Res. Commun. 2021, 534, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Isiaku, A.I.; Zhang, Z.; Pazhakh, V.; Manley, H.R.; Thompson, E.R.; Fox, L.C.; Yerneni, S.; Blombery, P.; Lieschke, G.J. Transient, flexible gene editing in zebrafish neutrophils and macrophages for determination of cell-autonomous functions. Dis. Model. Mech. 2021, 14, dmm047431. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.; Gao, J.; Rivière, T.; Schmid, B.; Walzog, B.; Maier-Begandt, D. Molecular insights into neutrophil biology from the zebrafish perspective: Lessons from CD18 deficiency. Front. Immunol. 2021, 12, 677994. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Castellanos, F.; Ramírez, L.; Lomelí, H. zmiz1a Zebrafish mutants have defective erythropoiesis, altered expression of autophagy genes, and a deficient response to vitamin D. Life Sci. 2021, 284, 119900. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Lin, B.; Lin, Y.; Chang, C.; Tzou, W.; Pei, T.; Hu, C. Meis1, Hi1α, and GATA1 are integrated into a hierarchical regulatory network to mediate primitive erythropoiesis. FASEB J. 2021, 35, e21915. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Ogawa, T.; Fujita, S.; Sone, R.; Kawahara, A. Cooperative contributions of the klf1 and klf17 genes in zebrafish primitive erythropoiesis. Sci. Rep. 2023, 13, 12279. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Ahmed, A.; Wang, W.; Wang, X.; Ma, C.; Jiang, H.; Li, W.; Jing, L. Negative Elongation Factor (NELF) inhibits premature granulocytic development in zebrafish. Int. J. Mol. Sci. 2022, 23, 3833. [Google Scholar] [CrossRef] [PubMed]
- Linehan, J.B.; Lucas Zepeda, J.; Mitchell, T.A.; LeClair, E.E. Follow that cell: Leukocyte migration in L-plastin mutant zebrafish. Cytoskeleton 2022, 79, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Elworthy, S.; Rutherford, H.A.; Prajsnar, T.K.; Hamilton, N.M.; Vogt, K.; Renshaw, S.A.; Condliffe, A.M. Activated PI3K delta syndrome 1 mutations cause neutrophilia in zebrafish larvae. Dis. Model. Mech. 2023, 16, dmm049841. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Sun, Y.; Yang, S.; Wu, Y.; Wang, L.; Zou, W.; Jiang, N.; Chen, J.; Han, Y.; Huang, C.; et al. Novel Chemical-sructure TPOR agonist, TMEA, promotes megakaryocytes differentiation and thrombopoiesis via MTOR and ERK signalings. Phytomedicine 2023, 110, 154637. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, G.; Zhao, Y.; Ren, J.; Li, Q.; Cui, Z. Functions of SMC2 in the development of zebrafish liver. Biomedicines 2021, 9, 1240. [Google Scholar] [CrossRef] [PubMed]
- Serifi, I.; Besta, S.; Karetsou, Z.; Giardoglou, P.; Beis, D.; Niewiadomski, P.; Papamarcaki, T. Targeting of SET/I2PP2A oncoprotein inhibits gli1 transcription revealing a new modulator of hedgehog signaling. Sci. Rep. 2021, 11, 13940. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Han, X.; Chen, H.; Zhao, C.; Chen, Y.; Zhou, J.; de The, H.; Zhu, J.; Yuan, H. ypel5 regulates liver development and function in zebrafish. J. Mol. Cell Biol. 2023, 15, mjad019. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.Q.; Meyer-Miner, A.; David-Rachel, M.; Lee, F.J.H.; Wilkins, B.J.; Karpen, S.J.; Ciruna, B.; Ghanekar, A.; Kamath, B.M. Loss of zebrafish pkd1l1 causes biliary defects that have implications for biliary atresia splenic malformation. Dis. Model. Mech. 2023, 16, dmm049326. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Yang, D.; Ni, R. tmed10 deficiency results in impaired exocrine pancreatic differentiation in zebrafish larvae. Dev. Biol. 2023, 503, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Elsaid, H.O.A.; Furriol, J.; Blomqvist, M.; Diswall, M.; Leh, S.; Gharbi, N.; Anonsen, J.H.; Babickova, J.; Tøndel, C.; Svarstad, E.; et al. Reduced α-galactosidase A activity in zebrafish (Danio rerio) mirrors distinct features of fabry nephropathy phenotype. Mol. Genet. Metab. Rep. 2022, 31, 100851. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Qin, Y.; Sheng, J.; Duan, X.; Shen, L.; Liu, D. Aquaporin 8ab is required in zebrafish embryonic intestine development. Acta Biochim. Biophys. Sin. 2022, 54, 952–960. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.A.; Rabahi, S.; Diaz, O.E.; Salloum, Y.; Kern, B.C.; Westling, M.; Luo, X.; Parigi, S.M.; Monasterio, G.; Das, S.; et al. Interleukin-10 regulates goblet cell numbers through notch signaling in the developing zebrafish intestine. Mucosal Immunol. 2022, 15, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Wiweger, M.; Majewski, L.; Adamek-Urbanska, D.; Wasilewska, I.; Kuznicki, J. npc2-deficient zebrafish reproduce neurological and inflammatory symptoms of Niemann-pick type C disease. Front. Cell Neurosci. 2021, 15, 647860. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, X.; Li, Y.; Yu, L.; Zhang, B.; Zhang, J.; Cho, W.J.; Venkatarangan, V.; Chen, L.; Burugula, B.B.; et al. GCAF(TMEM251) regulates lysosome biogenesis by activating the mannose-6-phosphate pathway. Nat. Commun. 2022, 13, 5351. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jia, J.; Zhao, Q.; Zhang, Y.; Huang, B.; Wang, L.; Tian, J.; Huang, C.; Li, M.; Li, X. Novel loss-of-function variant in HNF1a induces β-Cell dysfunction through endoplasmic reticulum stress. Int. J. Mol. Sci. 2022, 23, 13022. [Google Scholar] [CrossRef] [PubMed]
- Naylor, R.W.; Watson, E.; Williamson, S.; Preston, R.; Davenport, J.B.; Thornton, N.; Lowe, M.; Williams, M.; Lennon, R. Basement membrane defects in CD151-associated glomerular disease. Pediatr. Nephrol. 2022, 37, 3105–3115. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, R.; Fatehi, M.; Wang, Y.; Long, W.; Tian, R.; Deng, X.; Weng, Z.; Xu, Q.; Light, P.E.; et al. Regulation of PKD2 channel function by TACAN. J. Physiol. 2023, 601, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, L.; Leyhr, J.; Zhang, H.; Öhman-Mägi, C.; Allalou, A.; Haitina, T. The broad role of nkx3.2 in the development of the zebrafish axial skeleton. PLoS ONE 2021, 16, e0255953. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Wang, X.; Jin, M.; Li, L.; Zhu, J.; Kang, Y.; Chen, Z.; Sun, Y.; Zhao, C. Cilia regulate meiotic recombination in zebrafish. J. Mol. Cell Biol. 2022, 14, mjac049. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, H.; Schulz, E.T.; Azuma, Y.; Azuma, M. EWSR1 prevents the induction of aneuploidy through direct regulation of Aurora B. Front. Cell Dev. Biol. 2023, 11, 987153. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Kambakam, S.; Almeida, M.P.; Ming, Z.; Welker, J.M.; Wierson, W.A.; Schultz-Rogers, L.E.; Ekker, S.C.; Clark, K.J.; Essner, J.J.; et al. Cre/Lox regulated conditional rescue and inactivation with zebrafish UFlip alleles generated by CRISPR-Cas9 targeted integration. eLife 2022, 11, e71478. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.A.; Oyarbide, U. Neuronal expression of ndst3 in early zebrafish development is responsive to wnt signaling manipulation. Gene Expr. Patterns 2023, 47, 119300. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, G.; Chi, Y.-Y.; Kou, Y.-X.; Li, Y. Expression profiling and functional characterization of the duplicated oxr1b gene in zebrafish. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2021, 39, 100857. [Google Scholar] [CrossRef] [PubMed]
- Lane, B.M.; Chryst-Stangl, M.; Wu, G.; Shalaby, M.; El Desoky, S.; Middleton, C.C.; Huggins, K.; Sood, A.; Ochoa, A.; Malone, A.F.; et al. Steroid-sensitive nephrotic syndrome candidate gene CLVS1 regulates podocyte oxidative stress and endocytosis. JCI Insight 2022, 7, e152102. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Muraki, K.; Tamaoki, J.; Kobayashi, M. Soy-derived equol induces antioxidant activity in zebrafish in an nrf2-independent manner. Int. J. Mol. Sci. 2022, 23, 5243. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Peng, T.; Zhou, Q.; Zhu, J.; Liao, G.; Zou, F.; Meng, X. Evaluation of the oxidative toxicity induced by lead, manganese, and cadmium using genetically modified nrf2a-mutant zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2023, 266, 109550. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, R.; Eberhart, J.K. Loss of nicotinamide nucleotide transhydrogenase sensitizes embryos to ethanol-induced neural crest and neural apoptosis via generation of reactive oxygen species. Front. Neurosci. 2023, 17, 1154621. [Google Scholar] [CrossRef] [PubMed]
- Kevin Pan, Y.; Julian, T.; Garvey, K.; Perry, S.F. Catecholamines modulate the hypoxic ventilatory response of larval zebrafish (Danio rerio). J. Exp. Biol. 2023, 226, jeb245051. [Google Scholar] [CrossRef] [PubMed]
- Lelièvre, E.; Bureau, C.; Bordat, Y.; Frétaud, M.; Langevin, C.; Jopling, C.; Kissa, K. Deficiency in hereditary hemorrhagic telangiectasia-associated endoglin elicits hypoxia-driven heart failure in zebrafish. Dis. Model. Mech. 2023, 16, dmm049488. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, A.; Betto, R.M.; Diamante, L.; Tesoriere, A.; Ghirardo, R.; Cioccarelli, C.; Meneghetti, G.; Peron, M.; Laquatra, C.; Tiso, N.; et al. STAT3 and HIF1α cooperatively mediate the transcriptional and physiological responses to hypoxia. Cell Death Discov. 2023, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.M.; Perry, S.F. The Rhesus glycoprotein Rhcgb is expendable for ammonia excretion and Na+ uptake in zebrafish (Danio rerio). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2020, 247, 110722. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.M.; Shir-Mohammadi, K.; Kwong, R.W.M.; Perry, S.F. Reassessing the contribution of the Na+/H+ exchanger Nhe3b to Na+ uptake in zebrafish (Danio rerio) using CRISPR/Cas9 gene editing. J. Exp. Biol. 2019, 223 Pt 2, jeb215111. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.M.; Mandic, M.; Yew, H.M.; Kunert, E.; Pan, Y.K.; Ha, J.; Kwong, R.W.M.; Gilmour, K.M.; Perry, S.F. Use of a carbonic anhydrase ca17a knockout to investigate mechanisms of ion uptake in zebrafish (Danio rerio). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2021, 320, R55–R68. [Google Scholar] [CrossRef] [PubMed]
- Linnerz, T.; Sung, Y.J.; Rolland, L.; Astin, J.W.; Dalbeth, N.; Hall, C.J. Uricase-deficient larval zebrafish with elevated urate levels demonstrate suppressed acute inflammatory response to monosodium urate crystals and prolonged crystal persistence. Genes 2022, 13, 2179. [Google Scholar] [CrossRef] [PubMed]
- Solman, M.; Blokzijl-Franke, S.; Piques, F.; Yan, C.; Yang, Q.; Strullu, M.; Kamel, S.M.; Ak, P.; Bakkers, J.; Langenau, D.M.; et al. Inflammatory response in hematopoietic stem and progenitor cells triggered by activating SHP2 mutations evokes blood defects. eLife 2022, 11, e73040. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, B.; Peng, Z.; Yang, T.; Qin, B.; Ao, J.; Yang, Y.; Wang, J.; Zheng, L.; Xie, H. Transcriptomics and phenotypic analysis of gpr56 knockout in zebrafish. Int. J. Mol. Sci. 2023, 24, 7740. [Google Scholar] [CrossRef] [PubMed]
- Sobah, M.L.; Scott, A.C.; Laird, M.; Koole, C.; Liongue, C.; Ward, A.C. socs3b regulates the development and function of innate immune cells in zebrafish. Front. Immunol. 2023, 14, 1119727. [Google Scholar] [CrossRef] [PubMed]
- Bian, W.-P.; Xie, S.-L.; Wang, C.; Martinovich, G.G.; Ma, Y.-B.; Jia, P.-P.; Pei, D.-S. mitfa deficiency promotes immune vigor and potentiates antitumor effects in zebrafish. Fish. Shellfish. Immunol. 2023, 142, 109130. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, N.; Ward, A.C.; Liongue, C. Analysis of potential non-canonical or alternate STAT5 functions in immune development and growth. Front. Biosci.-Landmark 2023, 28, 187. [Google Scholar] [CrossRef]
- Shanaka, K.A.S.N.; Jung, S.; Madushani, K.P.; Wijerathna, H.M.S.M.; Neranjan Tharuka, M.D.; Kim, M.-J.; Lee, J. Generation of viperin-knockout zebrafish by CRISPR/Cas9-mediated genome engineering and the effect of this mutation under VHSV infection. Fish. Shellfish. Immunol. 2022, 131, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Streiff, C.; He, B.; Morvan, L.; Zhang, H.; Delrez, N.; Fourrier, M.; Manfroid, I.; Suárez, N.M.; Betoulle, S.; Davison, A.J.; et al. Susceptibility and permissivity of zebrafish (Danio rerio) larvae to Cypriniviruses. Viruses 2023, 15, 768. [Google Scholar] [CrossRef] [PubMed]
- Sellaththurai, S.; Jung, S.; Kim, M.-J.; Nadarajapillai, K.; Ganeshalingam, S.; Jeong, J.B.; Lee, J. CRISPR/Cas9-induced knockout of sting increases susceptibility of zebrafish to bacterial infection. Biomolecules 2023, 13, 324. [Google Scholar] [CrossRef] [PubMed]
- Saralahti, A.K.; Harjula, S.-K.E.; Rantapero, T.; Uusi-Mäkelä, M.I.E.; Kaasinen, M.; Junno, M.; Piippo, H.; Nykter, M.; Lohi, O.; Rounioja, S.; et al. Characterization of the innate immune response to streptococcus pneumoniae infection in zebrafish. PLoS Genet. 2023, 19, e1010586. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.; de Silva, K.; Plain, K.M.; Purdie, A.C.; Blair, T.A.; Duggin, I.G.; Britton, W.J.; Oehlers, S.H. Mycobacterial infection-induced MiR-206 inhibits protective neutrophil recruitment via the CXCL12/CXCR4 signalling axis. PLoS Pathog. 2021, 17, e1009186. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Casini, A.; Olivieri, M.; Petris, G.; Montagna, C.; Reginato, G.; Maule, G.; Lorenzin, F.; Prandi, D.; Romanel, A.; Demichelis, F.; et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018, 36, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. FDA, Briefing Packet AquAdvantage Salmon; FDA Center for Veterinary Medicine: Laurel, MD, USA, 2010; p. 61. [Google Scholar]
- Prykhozhij, S.V.; Cordeiro-Santanach, A.; Caceres, L.; Berman, J.N. Genome editing in zebrafish using high-fidelity Cas9 nucleases: Choosing the right nuclease for the task. Methods Mol. Biol. 2020, 2115, 385–405. [Google Scholar] [PubMed]
- Liu, P.; Luk, K.; Shin, M.; Idrizi, F.; Kwok, S.; Roscoe, B.; Mintzer, E.; Suresh, S.; Morrison, K.; Frazão, J.B.; et al. Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res. 2019, 47, 4169–4180. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, M.M.; Biayna, J.; Supek, F. TP53-dependent toxicity of CRISPR/Cas9 cuts is differential across genomic loci and can confound genetic screening. Nat. Commun. 2022, 13, 4520. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.J.; Carvalho, L.; Logarinho, E.; Bessa, J. foxm1 modulates cell non-autonomous response in zebrafish skeletal muscle homeostasis. Cells 2021, 10, 1241. [Google Scholar] [CrossRef] [PubMed]
- Ah-Fong, A.M.V.; Boyd, A.M.; Matson, M.E.H.; Judelson, H.S. A Cas12a-based gene editing system for Phytophthora Infestans reveals monoallelic expression of an elicitor. Mol. Plant Pathol. 2021, 22, 737–752. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.S.; Lam, I.I.; Clay, H.; Duong, D.N.; Deo, R.C.; Coughlin, S.R. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev. Cell 2018, 46, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Uusi-Mäkelä, M.I.E.; Barker, H.R.; Bäuerlein, C.A.; Häkkinen, T.; Nykter, M.; Rämet, M. Chromatin accessibility is associated with CRISPR-Cas9 efficiency in the zebrafish (Danio rerio). PLoS ONE 2018, 13, e0196238. [Google Scholar] [CrossRef] [PubMed]
- Zhi, L.I.; Zhou, L.; Zhang, J.; Wang, Y.; Gui, J.F. A fast and efficient microinjection method in the fertilized eggs of teleost fish with adhesive eggs. Acta Hydrobiol. Sin. 2016, 40, 76–82. [Google Scholar]
- Goto, R.; Saito, T.; Matsubara, T.; Yamaha, E. Microinjection of marine fish eggs. Methods Mol. Biol. 2019, 1874, 475–487. [Google Scholar] [PubMed]
- JoVE. Science Education Database Biology II: Mouse, Zebrafish, and Chick. Zebrafish Microinjection Techniques. Available online: https://www.jove.com/v/5130/introduction-to-microinjection-of-early-zebrafish-embryos (accessed on 29 May 2024).
- Putri, R.R.; Chen, L. Spatiotemporal control of zebrafish (Danio rerio) gene expression using a light-activated CRISPR activation system. Gene 2018, 677, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Shang, M.; Liu, T.; Wang, K. High-throughput methods for genome editing: The more the better. Plant Physiol. 2022, 188, 1731–1745. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Davey, C.F.; Whitebirch, A.C.; Miller, A.C.; Moens, C.B. Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods 2015, 12, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Datlinger, P.; Rendeiro, A.F.; Schmidl, C.; Krausgruber, T.; Traxler, P.; Klughammer, J.; Schuster, L.C.; Kuchler, A.; Alpar, D.; Bock, C. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 2017, 14, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Dalgin, G.; Prince, V.E. Midline Morphogenesis of zebrafish foregut endoderm is dependent on hoxb5b. Dev. Biol. 2021, 471, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chrystal, P.W.; French, C.R.; Jean, F.; Havrylov, S.; van Baarle, S.; Peturson, A.-M.; Xu, P.; Crump, J.G.; Pilgrim, D.B.; Lehmann, O.J.; et al. The axenfeld–rieger syndrome gene foxc1 contributes to left–right patterning. Genes 2021, 12, 170. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.-R.; Wan, S.-M.; Zhou, J.-J.; Nie, C.-H.; Chen, Y.-L.; Diao, J.-H.; Gao, Z.-X. Functional differentiation of bmp7 genes in zebrafish: bmp7a for dorsal-ventral pattern and bmp7b for melanin synthesis and eye development. Front. Cell Dev. Biol. 2022, 10, 10:838721. [Google Scholar] [CrossRef] [PubMed]
- Livne, H.; Avital, T.; Ruppo, S.; Harazi, A.; Mitrani-Rosenbaum, S.; Daya, A. Generation and characterization of a novel gne knockout model in zebrafish. Front. Cell Dev. Biol. 2022, 24, 976111. [Google Scholar] [CrossRef] [PubMed]
- Seni-Silva, A.C.; Maleski, A.L.A.; Souza, M.M.; Falcao, M.A.P.; Disner, G.R.; Lopes-Ferreira, M.; Lima, C. Natterin-like depletion by CRISPR/Cas9 impairs zebrafish (Danio rerio) embryonic development. BMC Genom. 2022, 23, 123. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Han, F.; Huang, Y.; Liu, J.; Liao, X.; Cao, Z.; Li, W. sphk1 deficiency induces apoptosis and developmental defects and premature death in zebrafish. Fish. Physiol. Biochem. 2023, 49, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Wen, D.; Wang, L.; Meng, C.; Wang, Y.; Zhao, Z.; Wu, C. CRISPR/Cas9-induced asap1a and asap1b co-knockout mutant zebrafish displayed abnormal embryonic development and impaired neutrophil migration. Gene Expr. Patterns 2023, 49, 119331. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.; Bishop, K.; Broadbridge, E.; Yu, K.; Carrington, B.; Elkahloun, A.; Zhen, T.; Pei, W.; Burgess, S.M.; Liu, P.; et al. zrsr2 is essential for the embryonic development and splicing of minor introns in RNA and protein processing genes in zebrafish. Int. J. Mol. Sci. 2022, 23, 10668. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; LaDu, J.K.; Garcia, G.R.; Li, S.; Tomono-Duval, K.; Rericha, Y.; Huang, L.; Tanguay, R.L. A CRISPR-Cas9 mutation in sox9b long intergenic noncoding RNA (slincR) affects zebrafish development, behavior, and regeneration. Toxicol. Sci. 2023, 194, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Zhang, F.; Ping, L.; Wang, Y.; Zhang, B.; Wang, J.; Chen, X. vwa1 knockout in zebrafish causes abnormal craniofacial chondrogenesis by regulating FGF pathway. Genes 2023, 14, 838. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, F.; Leoni, L.; Daponte, V.; Gioia, R.; Cotti, S.; Fiedler, I.A.K.; Larianova, D.; Willaert, A.; Coucke, P.J.; Villani, S.; et al. Zebrafish tric-b is required for skeletal development and bone cells differentiation. Front. Endocrinol. 2023, 14, 1002914. [Google Scholar] [CrossRef] [PubMed]
- McGowan, L.M.; Kague, E.; Vorster, A.; Newham, E.; Cross, S.; Hammond, C.L. wnt16 elicits a protective effect against fractures and supports bone repair in zebrafish. JBMR Plus 2021, 5, e10461. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Liao, M.; Liu, W.; Cai, Y.; Yi, Q.; Long, J.; Tan, L.; Deng, Y.; Deng, H.; Chen, X. Loss of wnt16 leads to skeletal deformities and downregulation of bone developmental pathway in zebrafish. Int. J. Mol. Sci. 2021, 22, 6673. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Maeno, A.; Araki, S.; Kikuchi, M.; Suzuki, M.; Ishizaka, M.; Satoh, K.; Akama, K.; Kawabe, Y.; Suzuki, K.; et al. An atlas of seven zebrafish HOX cluster mutants provides insights into sub/neofunctionalization of vertebrate HOX clusters. Development 2021, 148, dev198325. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, A.; Nie, C.; Gao, Z.; Wan, S.-M. Functional differentiation of bmp2a and bmp2b genes in zebrafish. Gene Expr. Patterns 2022, 46, 119288. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Macorra, C.; Sintes, M.; Tabanera, N.; Grasa, I.; Bovolenta, P.; Cardozo, M.J. mosmo is required for zebrafish craniofacial formation. Front. Cell Dev. Biol. 2021, 22, 767048. [Google Scholar] [CrossRef] [PubMed]
- Kague, E.; Hughes, S.M.; Lawrence, E.A.; Cross, S.; Martin-Silverstone, E.; Hammond, C.L.; Hinits, Y. Scleraxis genes are required for normal musculoskeletal development and for rib growth and mineralization in zebrafish. FASEB J. 2019, 33, 9116–9130. [Google Scholar] [CrossRef] [PubMed]
- Alexandre-Moreno, S.; Bonet-Fernández, J.-M.; Atienzar-Aroca, R.; Aroca-Aguilar, J.-D.; Escribano, J. Null cyp1b1 activity in zebrafish leads to variable craniofacial defects associated with altered expression of extracellular matrix and lipid metabolism genes. Int. J. Mol. Sci. 2021, 22, 6430. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, X.; Wang, X.; Zhai, Y.; Li, M.; Pan, J.; Bai, Y.; Rong, X.; Zhou, J. Genetic deletion of hspa8 leads to selective tissue malformations in zebrafish embryonic development. J. Cell Sci. 2022, 135, jcs259734. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Wu, J.; Jing, J.; Huang, P.; Li, Z.; Mei, J.; Gui, J.-F. Loss of stat3 function leads to spine malformation and immune disorder in zebrafish. Sci. Bull. 2017, 62, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Terhune, E.A.; Cuevas, M.T.; Monley, A.M.; Wethey, C.I.; Chen, X.; Cattell, M.V.; Bayrak, M.N.; Bland, M.R.; Sutphin, B.; Trahan, G.D.; et al. Mutations in kif7 implicated in idiopathic scoliosis in humans and axial curvatures in zebrafish. Hum. Mutat. 2021, 42, 392–407. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, L.; Leyhr, J.; Zhang, H.; Allalou, A.; Öhman-Mägi, C.; Haitina, T. The role of gdf5 in the development of the zebrafish fin endoskeleton. Dev. Dyn. 2022, 251, 1535–1549. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.-H.; Wan, S.-M.; Chen, Y.-L.; Huysseune, A.; Wu, Y.-M.; Zhou, J.-J.; Hilsdorf, A.W.S.; Wang, W.-M.; Witten, P.E.; Lin, Q.; et al. Single-cell transcriptomes and runx2b−/− mutants reveal the genetic signatures of intermuscular bone formation in zebrafish. Natl. Sci. Rev. 2022, 9, nwac152. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.-H.; Li, Z.; Wang, Z.-W.; Li, X.-Y.; Wang, Y.; Zhang, X.-J.; Tong, J.-F.; Wu, Y.; Xia, L.-Y.; Gao, Z.-X.; et al. Creation of intermuscular bone-free mutants in amphitriploid gibel carp by editing two duplicated runx2b homeologs. Aquaculture 2023, 567, 739300. [Google Scholar] [CrossRef]
- Ka, H.I.; Seo, H.; Choi, Y.; Kim, J.; Cho, M.; Choi, S.-Y.; Park, S.; Han, S.; An, J.; Chung, H.S.; et al. Loss of splicing factor IK impairs normal skeletal muscle development. BMC Biol. 2021, 19, 44. [Google Scholar] [CrossRef] [PubMed]
- Bhat, N.; Narayanan, A.; Fathzadeh, M.; Shah, K.; Dianatpour, M.; Abou Ziki, M.D.; Mani, A. dyrk1b promotes autophagy during skeletal muscle differentiation by upregulating 4e-Bp1. Cell Signal 2022, 90, 110186. [Google Scholar] [CrossRef] [PubMed]
- Voisard, P.; Diofano, F.; Glazier, A.A.; Rottbauer, W.; Just, S. CRISPR/Cas9-mediated constitutive loss of VCP (valosin-containing protein) impairs proteostasis and leads to defective striated muscle structure and function in vivo. Int. J. Mol. Sci. 2022, 23, 6722. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.L.; Webb, S.E.; Miller, A.L. Localized TPC1-mediated Ca2+ release from endolysosomes contributes to myoseptal junction development in zebrafish. J. Cell Sci. 2022, 135, jcs259564. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Shi, Y.; Zhang, C.; Peng, Y.; Wan, Y.; Xu, Y.; Liu, X.; Han, B.; Zhao, S.; Kuang, Y.; et al. In-frame deletion of SMC5 related with the phenotype of primordial dwarfism, chromosomal instability and insulin resistance. Clin. Transl. Med. 2023, 13, e1007. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Mei, J.; Huang, P.; Jing, J.; Li, Z.; Kang, J.; Gui, J.-F. Essential roles of stat5.1/stat5b in controlling fish somatic growth. J. Genet. Genom. 2017, 44, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, B.M.; Yamaguchi, G.; Radler, L.M.; Shimizu, M. Editing the duplicated insulin-like growth factor binding protein-2b gene in rainbow trout (Oncorhynchus mykiss). Sci. Rep. 2018, 8, 16054. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, B.M.; Habara, S.; Oikawa, J.; Radler, L.M.; Shimizu, M. Compensatory response of the somatotropic axis from IGFBP-2b gene editing in rainbow trout (Oncorhynchus mykiss). Genes 2020, 11, 1488. [Google Scholar] [CrossRef] [PubMed]
- Shahi, N.; Mallik, S.K.; Sarma, D. Muscle growth in targeted knockout common carp (Cyprinus carpio) mstn gene with low off-target effects. Aquaculture 2022, 547, 737423. [Google Scholar] [CrossRef]
- Dong, Z.; Ge, J.; Xu, Z.; Dong, X.; Cao, S.; Pan, J.; Zhao, Q. Generation of myostatin b knockout yellow catfish (Tachysurus fulvidraco) using transcription activator-like effector nucleases. Zebrafish 2014, 11, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Khalil, K.; Elayat, M.; Khalifa, E.; Daghash, S.; Elaswad, A.; Miller, M.; Abdelrahman, H.; Ye, Z.; Odin, R.; Drescher, D.; et al. Generation of myostatin gene-edited channel catfish (Ictalurus punctatus) via zygote injection of CRISPR/Cas9 system. Sci. Rep. 2017, 7, 7301. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Cho, J.Y.; Kim, J.-W.; Kim, H.-C.; Noh, J.K.; Kim, Y.-O.; Hwang, H.-K.; Kim, W.-J.; Yeo, S.-Y.; An, C.M.; et al. CRISPR/Cas9-mediated myostatin disruption enhances muscle mass in the olive flounder (Paralichthys olivaceus). Aquaculture 2019, 512, 734336. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Kinoshita, M.; Ng, T.H.; Chang, Y.-H.; Maekawa, S.; Chiang, Y.-A.; Aoki, T.; Wang, H.-C. Using CRISPR/Cas9-mediated gene editing to further explore growth and trade-off effects in myostatin-mutated F4 medaka (Oryzias latipes). Sci. Rep. 2017, 7, 11435. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wu, T.; Yang, L.; Su, Y.; Zhao, C.; Li, L.; Cai, J.; Dai, X.; Wang, D.; Zhou, L. Generation of fast growth Nile tilapia (Oreochromis niloticus) by myostatin gene mutation. Aquaculture 2023, 562, 738762. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, J.; Wang, L.; Sun, F.; Yue, G.H. pomc knockout increases growth in zebrafish. Aquaculture 2023, 574, 739707. [Google Scholar] [CrossRef]
- Che, J.; Hu, C.; Wang, Q.; Fan, C.; Si, Y.; Gong, X.; Bao, B. The double mutations of acvr2aa and acvr2ba leads to muscle hypertrophy in zebrafish. Aquac. Fish. 2023, 8, 706–712. [Google Scholar] [CrossRef]
- Li, S.-Z.; Liu, W.; Li, Z.; Li, W.-H.; Wang, Y.; Zhou, L.; Gui, J.-F. greb1 regulates convergent extension movement and pituitary development in zebrafish. Gene 2017, 627, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Lu, Y.; Cheng, X.; Shi, C.; Lou, Q.; Jin, X.; He, J.; Zhai, G.; Yin, Z. Functions of the thyroid-stimulating hormone on key developmental features revealed in a series of zebrafish dyshormonogenesis models. Cells 2021, 10, 1984. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Fang, Y.; Zhang, M.-M.; Zhang, R.-J.; Wu, F.-Y.; Yang, R.-M.; Tu, P.-H.; Dong, M.; Zhao, S.-X.; Song, H.-D. Genetic manipulation on zebrafish duox recapitulate the clinical manifestations of congenital hypothyroidism. Endocrinology 2021, 162, bqab101. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, G.; Murashita, K.; Verri, T.; Gomes, A.S.; Rønnestad, I. Leptin receptor-deficient (knockout) zebrafish: Effects on nutrient acquisition. Gen. Comp. Endocrinol. 2021, 310, 113832. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Ding, Y.; Nowik, N.; Jager, C.; Eeza, M.N.H.; Alia, A.; Baelde, H.J.; Spaink, H.P. Leptin deficiency affects glucose homeostasis and results in adiposity in zebrafish. J. Endocrinol. 2021, 249, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ai, N.; Chen, W.; Wong, Q.W.-L.; Ge, W. Leptin and its signaling are not involved in zebrafish puberty onset. Biol. Reprod. 2022, 106, 928–942. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Cheng, C.; Cheng, Z.-C.; Wu, Q.; Shen, H.; Yuan, M.; Zhang, B.; Yang, J.-K. The dual role of rfx6 in directing β cell development and insulin production. J. Mol. Endocrinol. 2021, 66, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Schmöhl, F.; Li, X.; Qian, X.; Tabler, C.T.; Bennewitz, K.; Sticht, C.; Morgenstern, J.; Fleming, T.; Volk, N.; et al. Reduced acrolein detoxification in akr1a1a zebrafish mutants causes impaired insulin receptor signaling and microvascular alterations. Adv. Sci. 2021, 8, e2101281. [Google Scholar] [CrossRef] [PubMed]
- Zeng, N.; Bao, J.; Shu, T.; Shi, C.; Zhai, G.; Jin, X.; He, J.; Lou, Q.; Yin, Z. Sexual dimorphic effects of igf1 deficiency on metabolism in zebrafish. Front. Endocrinol. 2022, 13, 879962. [Google Scholar] [CrossRef] [PubMed]
- Mattis, K.K.; Krentz, N.A.J.; Metzendorf, C.; Abaitua, F.; Spigelman, A.F.; Sun, H.; Ikle, J.M.; Thaman, S.; Rottner, A.K.; Bautista, A.; et al. Loss of RREB1 in pancreatic β cells reduces cellular insulin content and affects endocrine cell gene expression. Diabetologia 2023, 66, 674–694. [Google Scholar] [CrossRef] [PubMed]
- Tabler, C.T.; Lodd, E.; Bennewitz, K.; Middel, C.S.; Erben, V.; Ott, H.; Poth, T.; Fleming, T.; Morgenstern, J.; Hausser, I.; et al. Loss of glyoxalase 2 alters the glucose metabolism in zebrafish. Redox Biol. 2023, 59, 102576. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tian, J.; Kang, Q.; Yuan, H.; Liu, C.; Li, Z.; Liu, J.; Li, M. Knockout of nur77 leads to amino acid, lipid, and glucose metabolism disorders in zebrafish. Front. Endocrinol. 2022, 13, 864631. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Castro, L.F.C.; Monroig, Ó.; Cao, X.; Sun, Y.; Gao, J. A zebrafish pparγ gene deletion reveals a protein kinase network associated with defective lipid metabolism. Funct. Integr. Genom. 2022, 22, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, K.; Medishetti, R.; Kotha, J.; Behera, P.; Chandra, K.; Mavuduru, V.A.; Joshi, M.B.; Samineni, R.; Katika, M.R.; Ball, W.B.; et al. PHLPP1 promotes neutral lipid accumulation through AMPK/ChREBP-dependent lipid uptake and fatty acid synthesis pathways. iScience 2022, 25, 103766. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, A.; Helfenrath, K.; Wisniewsky, M.; Hinrichs, K.; Burmester, T.; Fabrizius, A. The knockout of cytoglobin 1 in zebrafish (Danio rerio) alters lipid metabolism, iron homeostasis and oxidative stress response. Biochim. Biophys. Acta 2023, 1870, 119558. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, Y.; Goh, P.-T.; Lopes-Marques, M.; Castro, L.F.C.; Monroig, Ó.; Kuah, M.-K.; Cao, X.; Shu-Chien, A.C.; Gao, J. Evolution and functional characteristics of the novel elovl8 that play pivotal roles in fatty acid biosynthesis. Genes 2021, 12, 1287. [Google Scholar] [CrossRef] [PubMed]
- Bláhová, Z.; Franěk, R.; Let, M.; Bláha, M.; Pšenička, M.; Mráz, J. Partial fads2 gene knockout diverts LC-PUFA Biosynthesis via an alternative δ8 pathway with an impact on the reproduction of female zebrafish (Danio rerio). Genes 2022, 13, 700. [Google Scholar] [CrossRef] [PubMed]
- Datsomor, A.K.; Olsen, R.E.; Zic, N.; Madaro, A.; Bones, A.M.; Edvardsen, R.B.; Wargelius, A.; Winge, P. CRISPR/Cas9-mediated editing of δ5 and δ6 desaturases impairs δ8-desaturation and docosahexaenoic acid synthesis in Atlantic salmon (Salmo salar L.). Sci. Rep. 2019, 9, 16888. [Google Scholar] [CrossRef] [PubMed]
- Datsomor, A.K.; Zic, N.; Li, K.; Olsen, R.E.; Jin, Y.; Vik, J.O.; Edvardsen, R.B.; Grammes, F.; Wargelius, A.; Winge, P. CRISPR/Cas9-mediated ablation of elovl2 in Atlantic salmon (Salmo salar L.) inhibits elongation of polyunsaturated fatty acids and induces srebp-1 and target genes. Sci. Rep. 2019, 9, 7533. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.-J.; Li, J.; Li, L.; Chen, X.; Wei, J.-R.; Yin, Z.; He, S.; Liang, X.-F. Knockout of t1r1 gene in zebrafish (Danio rerio) by CRISPR/Cas9 reveals its roles in regulating feeding behavior. Aquaculture 2021, 545, 737189. [Google Scholar] [CrossRef]
- Wargelius, A.; Leininger, S.; Skaftnesmo, K.O.; Kleppe, L.; Andersson, E.; Taranger, G.L.; Schulz, R.W.; Edvardsen, R.B. dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci. Rep. 2016, 6, 21284. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.S.; Hsu, J.Y.; Rossi, C.C.; Artinger, K.B. Requirement of zebrafish pcdh10a and pcdh10b in melanocyte precursor migration. Dev. Biol. 2018, 444, S274–S286. [Google Scholar] [CrossRef] [PubMed]
- Saunders, L.M.; Mishra, A.K.; Aman, A.J.; Lewis, V.M.; Toomey, M.B.; Packer, J.S.; Qiu, X.; McFaline-Figueroa, J.L.; Corbo, J.C.; Trapnell, C.; et al. Thyroid hormone regulates distinct paths to maturation in pigment cell lineages. eLife 2019, 8, e45181. [Google Scholar] [CrossRef] [PubMed]
- Nagao, Y.; Takada, H.; Miyadai, M.; Adachi, T.; Seki, R.; Kamei, Y.; Hara, I.; Taniguchi, Y.; Naruse, K.; Hibi, M.; et al. Distinct interactions of sox5 and sox10 in fate specification of pigment cells in medaka and zebrafish. PLoS Genet. 2018, 14, e1007260. [Google Scholar] [CrossRef] [PubMed]
- Miyadai, M.; Takada, H.; Shiraishi, A.; Kimura, T.; Watakabe, I.; Kobayashi, H.; Nagao, Y.; Naruse, K.; Higashijima, S.; Shimizu, T.; et al. A gene regulatory network combining pax3/7, sox10 and mitf generates diverse pigment cell types in medaka and zebrafish. Development 2023, 150, dev202114. [Google Scholar] [CrossRef]
- Granneman, J.G.; Kimler, V.A.; Zhang, H.; Ye, X.; Luo, X.; Postlethwait, J.H.; Thummel, R. Lipid droplet biology and evolution illuminated by the characterization of a novel perilipin in teleost fish. eLife 2017, 6, e21771. [Google Scholar] [CrossRef] [PubMed]
- Verwilligen, R.A.F.; Mulder, L.; Araújo, P.M.; Carneiro, M.; Bussmann, J.; Hoekstra, M.; Van Eck, M. Zebrafish as outgroup model to study evolution of scavenger receptor class B type I functions. Biochim. Biophys. Acta 2023, 1868, 159308. [Google Scholar] [CrossRef] [PubMed]
- Mo, E.S.; Cheng, Q.; Reshetnyak, A.V.; Schlessinger, J.; Nicoli, S. Alk and Ltk ligands are essential for iridophore development in zebrafish mediated by the receptor tyrosine kinase LTK. Proc. Natl. Acad. Sci. USA 2017, 114, 12027–12032. [Google Scholar] [CrossRef] [PubMed]
- Spiewak, J.E.; Bain, E.J.; Liu, J.; Kou, K.; Sturiale, S.L.; Patterson, L.B.; Diba, P.; Eisen, J.S.; Braasch, I.; Ganz, J.; et al. Evolution of endothelin signaling and diversification of adult pigment pattern in Danio fishes. PLoS Genet. 2018, 14, e1007538. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, J.; Du, J.; Si, Z.; Yang, H.; Xu, X.; Wang, C. ASIP disruption via CRISPR/Cas9 system induces black patches dispersion in Oujiang color common carp. Aquaculture 2019, 498, 230–235. [Google Scholar] [CrossRef]
- Mandal, B.K.; Chen, H.; Si, Z.; Hou, X.; Yang, H.; Xu, X.; Wang, J.; Wang, C. Shrunk and scattered black spots turn out due to MC1R knockout in a white-black Oujiang color common carp (Cyprinus carpio Var. color). Aquaculture 2020, 518, 734822. [Google Scholar] [CrossRef]
- Chen, H.; Wang, J.; Du, J.; Mandal, B.K.; Si, Z.; Xu, X.; Yang, H.; Wang, C. Analysis of recently duplicated tyrp1 genes and their effect on the formation of black patches in Oujiang-color common carp (Cyprinus carpio var. color). Anim. Genet. 2021, 52, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Qi, Y.; Liang, Q.; Song, J.; Liu, J.; Li, W.; Shu, Y.; Tao, M.; Zhang, C.; Qin, Q.; et al. Targeted disruption of tyrosinase causes melanin reduction in Carassius auratus cuvieri and its hybrid progeny. Sci. China Life Sci. 2019, 62, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.E.; Castranova, D.; Weinstein, B.M. Rapid generation of pigment free, immobile zebrafish embryos and larvae in any genetic background using CRISPR-Cas9 DgRNPs. Zebrafish 2021, 18, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Cal, L.; Suarez-Bregua, P.; Braasch, I.; Irion, U.; Kelsh, R.; Cerdá-Reverter, J.M.; Rotllant, J. Loss-of-function mutations in the melanocortin 1 receptor cause disruption of dorso-ventral countershading in teleost fish. Pigment. Cell Melanoma Res. 2019, 32, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Cal, L.; Suarez-Bregua, P.; Comesaña, P.; Owen, J.; Braasch, I.; Kelsh, R.; Cerdá-Reverter, J.M.; Rotllant, J. Countershading in zebrafish results from an asip1 controlled dorsoventral gradient of pigment cell differentiation. Sci. Rep. 2019, 9, 3449. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Gui, J.-F. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Kichigin, I.G.; Andreyushkova, D.A.; Pobedintseva, M.A.; Trifonov, V.A. Diversity of sex determining mechanisms in ray-finned fishes (Actinopterygii). Tsitologiya 2016, 58, 405–411. [Google Scholar]
- Lau, E.S.-W.; Zhang, Z.; Qin, M.; Ge, W. Knockout of zebrafish ovarian aromatase gene (cyp19a1a) by TALEN and CRISPR/Cas9 leads to all-male offspring due to failed ovarian differentiation. Sci. Rep. 2016, 6, 37357. [Google Scholar] [CrossRef] [PubMed]
- Dranow, D.B.; Hu, K.; Bird, A.M.; Lawry, S.T.; Adams, M.T.; Sanchez, A.; Amatruda, J.F.; Draper, B.W. bmp15 is an oocyte-produced signal required for maintenance of the adult female sexual phenotype in zebrafish. PLoS Genet. 2016, 12, e1006323. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Tang, H.; Liu, Y.; Chen, Y.; Li, G.; Liu, X.; Lin, H. Targeted disruption of aromatase reveals dual functions of cyp19a1a during sex differentiation in zebrafish. Endocrinology 2017, 158, 3030–3041. [Google Scholar] [CrossRef] [PubMed]
- WU, K.; Song, W.; Zhang, Z.; Ge, W. Disruption of dmrt1 rescues the all-male phenotype of cyp19a1a mutant in zebrafish—A novel insight into the roles of aromatase/estrogens in gonadal differentiation and early folliculogenesis. Development 2020, 147, dev182758. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, B.; Ge, W. Genetic analysis of zebrafish gonadotropin (FSH and LH) functions by TALEN-mediated gene disruption. Mol. Endocrinol. 2015, 29, 76–98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lau, S.-W.; Zhang, L.; Ge, W. Disruption of zebrafish follicle-stimulating hormone receptor (fshr) but not luteinizing hormone receptor (lhcgr) gene by TALEN leads to failed follicle activation in females followed by sexual reversal to males. Endocrinology 2015, 156, 3747–3762. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, D.; Shaner, Z.C.; Chen, S.; Hong, W.; Stellwag, E.J. Nuclear progestin receptor (pgr) knockouts in zebrafish demonstrate role for pgr in ovulation but not in rapid non-genomic steroid mediated meiosis resumption. Front. Endocrinol. 2015, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Desvignes, T.; Bremiller, R.; Wilson, C.; Dillon, D.; High, S.; Draper, B.; Buck, C.L.; Postlethwait, J. gonadal soma controls ovarian follicle proliferation through gsdf in zebrafish. Dev. Dyn. 2017, 246, 925–945. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Cui, Y.; Jiang, L.; Ge, W. Functional analysis of nuclear estrogen receptors in zebrafish reproduction by genome editing approach. Endocrinology 2017, 158, 2292–2308. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Li, C.; Ying, R.; Lin, H.; Li, J.; Zhao, Y.; Cheng, H.; Zhou, R. Loss-of-function of sox3 causes follicle development retardation and reduces fecundity in zebrafish. Protein Cell 2019, 10, 347–364. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhai, Y.; Geng, R.; Wu, K.; Song, W.; Ai, N.; Ge, W. Genetic analysis of activin/inhibin β subunits in zebrafish development and reproduction. PLoS Genet. 2022, 18, e1010523. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Zhang, X.; Zhao, C.; Geng, R.; Wu, K.; Yuan, M.; Ai, N.; Ge, W. Rescue of bmp15 deficiency in zebrafish by mutation of inha reveals mechanisms of BMP15 regulation of folliculogenesis. PLoS Genet. 2023, 19, e1010954. [Google Scholar] [CrossRef] [PubMed]
- Zhai, G.; Shu, T.; Xia, Y.; Jin, X.; He, J.; Yin, Z. Androgen signaling regulates the transcription of anti-müllerian hormone via synergy with SRY-related protein SOX9A. Sci. Bull. 2017, 62, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Shu, T.; Zhai, G.; Pradhan, A.; Olsson, P.-E.; Yin, Z. Zebrafish cyp17a1 knockout reveals that androgen-mediated signaling is important for male brain sex differentiation. Gen. Comp. Endocrinol. 2020, 295, 113490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ye, D.; Wang, H.; Wang, Y.; Hu, W.; Sun, Y. Zebrafish cyp11c1 knockout reveals the roles of 11-ketotestosterone and cortisol in sexual development and reproduction. Endocrinology 2020, 161, bqaa048s. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Zhang, D.; Liu, W.; Wang, J.; Liu, X.; Zhou, C.; Gui, J.; Xiao, W. Zebrafish androgen receptor is required for spermatogenesis and maintenance of ovarian function. Oncotarget 2018, 9, 24320–24334. [Google Scholar] [CrossRef] [PubMed]
- Crowder, C.M.; Lassiter, C.S.; Gorelick, D.A. Nuclear androgen receptor regulates testes organization and oocyte maturation in zebrafish. Endocrinology 2018, 159, 980–993. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Chen, Y.; Wang, L.; Yin, Y.; Li, G.; Guo, Y.; Liu, Y.; Lin, H.; Cheng, C.H.K.; Liu, X. Fertility impairment with defective spermatogenesis and steroidogenesis in male zebrafish lacking androgen receptor. Biol. Reprod. 2018, 98, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, K.; Ren, Z.; Ge, W. Genetic evidence for amh modulation of gonadotropin actions to control gonadal homeostasis and gametogenesis in zebrafish and its noncanonical signalling through Bmpr2a receptor. Development 2020, 147, dev189811. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Mei, J.; Li, Z.; Zhang, X.; Zhou, L.; Gui, J.-F. Distinct and cooperative roles of amh and dmrt1 in self-renewal and differentiation of male germ cells in zebrafish. Genetics 2017, 207, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Kossack, M.E.; High, S.K.; Hopton, R.E.; Yan, Y.; Postlethwait, J.H.; Draper, B.W. Female sex development and reproductive duct formation depend on wnt4a in zebrafish. Genetics 2019, 211, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Atienzar-Aroca, R.; Aroca-Aguilar, J.-D.; Alexandre-Moreno, S.; Ferre-Fernández, J.-J.; Bonet-Fernández, J.-M.; Cabañero-Varela, M.-J.; Escribano, J. Knockout of myoc provides evidence for the role of myocilin in zebrafish sex determination associated with wnt signalling downregulation. Biology 2021, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chen, W.; Zhu, B.; Ge, W. Disruption of Epidermal growth factor receptor but not EGF blocks follicle activation in zebrafish ovary. Front. Cell Dev. Biol. 2022, 9, 750888. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Xie, Q.; Wu, K.; Zhou, X.; Ge, W. Loss of nobox prevents ovarian differentiation from juvenile ovaries in zebrafish. Biol. Reprod. 2022, 106, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Nanjappa, D.P.; De Saffel, H.; Kalladka, K.; Arjuna, S.; Babu, N.; Prasad, K.; Sips, P.; Chakraborty, A. Poly (A)-specific ribonuclease deficiency impacts oogenesis in zebrafish. Sci. Rep. 2023, 13, 10026. [Google Scholar] [CrossRef] [PubMed]
- Fontana, C.M.; Terrin, F.; Facchinello, N.; Meneghetti, G.; Dinarello, A.; Gambarotto, L.; Zuccarotto, A.; Caichiolo, M.; Brocca, G.; Verin, R.; et al. Zebrafish ambra1b knockout reveals a novel role for ambra1 in primordial germ cells survival, sex differentiation and reproduction. Biol. Res. 2023, 56, 19. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.; Mikwar, M.; Da Fonte, D.; Lu, C.; Tao, B.; Peng, D.; Erandani, W.K.C.U.; Hu, W.; Trudeau, V.L. Secretoneurin is a secretogranin-2 derived hormonal peptide in vertebrate neuroendocrine systems. Gen. Comp. Endocrinol. 2020, 299, 113588. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, D.; Sharma, K.; Saxena, V.; Nipu, N.; Rajapaksha, D.C.; Mennigen, J.A. Knock-out of vasotocin reduces reproductive success in female zebrafish (Danio rerio). Front. Endocrinol. 2023, 14, 1151299. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.-H.; Wang, Y.; Li, Z.; Yu, Z.-X.; Li, X.-Y.; Tong, J.-F.; Wang, Z.-W.; Zhang, X.-J.; Zhou, L.; Gui, J.-F. Functional divergence of multiple duplicated foxl2 homeologs and alleles in a recurrent polyploid fish. Mol. Biol. Evol. 2021, 38, 1995–2013. [Google Scholar] [CrossRef] [PubMed]
- Webster, K.A.; Schach, U.; Ordaz, A.; Steinfeld, J.S.; Draper, B.W.; Siegfried, K.R. dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 2017, 422, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Yano, A.; Guyomard, R.; Nicol, B.; Jouanno, E.; Quillet, E.; Klopp, C.; Cabau, C.; Bouchez, O.; Fostier, A.; Guiguen, Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout (Oncorhynchus mykiss). Curr. Biol. 2012, 22, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
- Yano, A.; Nicol, B.; Jouanno, E.; Guiguen, Y. Heritable targeted inactivation of the rainbow trout (Oncorhynchus mykiss) master sex-determining gene using zinc-finger nucleases. Mar. Biotechnol. 2014, 16, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Zhong, C.; Wu, X.; Chen, J.; Tao, B.; Xia, X.; Shi, M.; Zhu, Z.; Trudeau, V.L.; Hu, W. mettl3 mutation disrupts gamete maturation and reduces fertility in zebrafish. Genetics 2018, 208, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.-K.; Huang, S.-C.; Chang, C.-F.; Wu, J.-L.; Gong, H.-Y. Infertility control of transgenic fluorescent zebrafish with targeted mutagenesis of the dnd1 gene by CRISPR/Cas9 genome editing. Front. Genet. 2023, 14, 1029200. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, R.; Katayama, N.; Sadaie, S.; Miwa, M.; Sanchez Matias, G.A.; Ichida, K.; Fujii, W.; Naito, K.; Hayashi, M.; Yoshizaki, G. Production of germ cell-less rainbow trout by dead end gene knockout and their use as recipients for germ cell transplantation. Mar. Biotechnol. 2022, 24, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, O.; Patinote, A.; Nguyen, T.; Com, E.; Pineau, C.; Bobe, J. Genome editing reveals reproductive and developmental dependencies on specific types of vitellogenin in zebrafish (Danio rerio). Mol. Reprod. Dev. 2019, 86, 1168–1188s. [Google Scholar] [CrossRef] [PubMed]
- Pachoensuk, T.; Fukuyo, T.; Rezanujjaman, M.; Wanlada, K.; Yamamoto, C.; Maeno, A.; Rahaman, M.M.; Ali, M.H.; Tokumoto, T. Zebrafish stm is involved in the development of otoliths and of the fertilization envelope. J. Reprod. Infertil. 2021, 2, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yue, G.H.; Wong, S.-M. VNN disease and status of breeding for resistance to NNV in aquaculture. Aquac. Fish. 2022, 7, 147–157. [Google Scholar] [CrossRef]
- Zhu, J.; Yang, J.; Wen, H.; Wang, M.; Zheng, X.; Zhao, J.; Sun, X.; Yang, P.; Mao, Q.; Li, Y.; et al. Expression and functional analysis of fam76b in zebrafish. Fish. Shellfish. Immunol. 2023, 142, 109161. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wong, S.M.; Yue, G.H. Effects of rrm1 on NNV resistance revealed by RNA-seq and gene editing. Mar. Biotechnol. 2021, 23, 854–869. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liu, X.; Cai, X.; Ouyang, G.; Fan, S.; Wang, J.; Xiao, W. Zebrafish prmt7 negatively regulates antiviral responses by suppressing the retinoic acid-inducible gene-I-like receptor signaling. FASEB J. 2020, 34, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yu, Y.; Wang, L.; Wong, S.-M.; Yue, G.H. Silencing Asian seabass gab3 inhibits Nervous Necrosis Virus replication. Mar. Biotechnol. 2022, 24, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- Nadarajapillai, K.; Jung, S.; Sellaththurai, S.; Ganeshalingam, S.; Kim, M.-J.; Lee, J. CRISPR/Cas9-mediated knockout of tnf-α1 in zebrafish reduces disease resistance after Edwardsiella piscicida bacterial infection. Fish. Shellfish. Immunol. 2024, 144, 109249. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Torraca, V.; Kamel, S.M.; Lombardi, A.; Meijer, A.H. Frontline science: Antagonism between regular and atypical Cxcr3 receptors regulates macrophage migration during infection and injury in zebrafish. J. Leukoc. Biol. 2020, 107, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Coogan, M.; Alston, V.; Su, B.; Khalil, K.; Elaswad, A.; Khan, M.; Simora, R.M.C.; Johnson, A.; Xing, D.; Li, S.; et al. CRISPR/Cas-9 induced knockout of myostatin gene improves growth and disease resistance in channel catfish (Ictalurus punctatus). Aquaculture 2022, 557, 738290. [Google Scholar] [CrossRef]
- Lu, G.; Luo, M. Genomes of major fishes in world fisheries and aquaculture: Status, application and perspective. Aquac. Fish. 2020, 5, 163–173. [Google Scholar] [CrossRef]
- Aparicio, S.; Chapman, J.; Stupka, E.; Putnam, N.; Chia, J.; Dehal, P.; Christoffels, A.; Rash, S.; Hoon, S.; Smit, A.; et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 2002, 297, 1301–1310. [Google Scholar] [CrossRef]
- Houston, R.D.; Bean, T.P.; Macqueen, D.J.; Gundappa, M.K.; Jin, Y.H.; Jenkins, T.L.; Selly, S.L.C.; Martin, S.A.M.; Stevens, J.R.; Santos, E.M.; et al. Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 2020, 21, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Xiong, L.; Wang, Y.; He, L.; Huang, R.; Liao, L.; Zhu, Z.; Wang, Y. ITGB1b-deficient rare minnows delay Grass Carp Reovirus (GCRV) entry and attenuate GCRV-triggered apoptosis. Int. J. Mol. Sci. 2018, 19, 3175. [Google Scholar] [CrossRef] [PubMed]
- Gratacap, R.L.; Regan, T.; Dehler, C.E.; Martin, S.A.M.; Boudinot, P.; Collet, B.; Houston, R.D. Efficient CRISPR/Cas9 genome editing in a salmonid fish cell line using a lentivirus delivery system. BMC Biotechnol. 2020, 20, 35. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Robledo, D.; Hickey, J.M.; McGrew, M.J.; Houston, R.D. Surrogate broodstock to enhance biotechnology research and applications in aquaculture. Biotechnol. Adv. 2021, 49, 107756. [Google Scholar] [CrossRef] [PubMed]
- Zohar, Y. Endocrinology and fish farming: Aspects in reproduction, growth, and smoltification. Fish. Physiol. Biochem. 1989, 7, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Muir, W.M.; Howard, R.D. Possible ecological risks of transgenic organism release when transgenes affect mating success: Sexual selection and the trojan gene hypothesis. Proc. Natl. Acad. Sci. USA 1999, 96, 13853–13856. [Google Scholar] [CrossRef] [PubMed]
- Stigebrandt, A.; Aure, J.; Ervik, A.; Hansen, P.K. Regulating the local environmental impact of intensive marine fish farming. Aquaculture 2004, 234, 239–261. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, H.; Liu, L.; Yang, P.; Shao, L.; Tang, L. Progress in infertility control technology of fish. Isr. J. Aquacult.-Bamid. 2022, 74, 1–9. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, M.; Ryu, J.H.; Hayman, E.S.; Fairgrieve, W.T.; Zohar, Y.; Luckenbach, J.A.; Wong, T. Reproductive sterility in aquaculture: A review of induction methods and an emerging approach with application to pacific northwest finfish species. Rev. Aquac. 2023, 15, 220–241. [Google Scholar] [CrossRef]
- Benfey, T.J.; Sutterlin, A.M. Triploidy induced by heat shock and hydrostatic pressure in landlocked Atlantic salmon (Salmo salar L.). Aquaculture 1984, 36, 359–367. [Google Scholar] [CrossRef]
- Bazaz, A.I.; Ahmad, I.; Nafath-ul-Arab, I.; Shah, T.H.; Asimi, O.; Bhat, B.A.; Yousuf, Z.; Baba, S.H.; Razak, N.; Fatima, A. A Review on induction of triploidy in fish using heat, pressure and cold shock treatments. J. Entomol. Zool. Stud. 2020, 8, 381–385. [Google Scholar]
- Chourrout, D.; Nakayama, I. Chromosome studies of progenies of tetraploid female rainbow trout. Theor. Appl. Genet. 1987, 74, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Madaro, A.; Kjøglum, S.; Hansen, T.; Fjelldal, P.G.; Stien, L.H. A Comparison of triploid and diploid Atlantic salmon (Salmo salar) performance and welfare under commercial farming conditions in Norway. J. Appl. Aquac. 2022, 34, 1021–1035. [Google Scholar] [CrossRef]
- Chevassus, B. Hybridization in fish. Aquaculture 1983, 33, 245–262. [Google Scholar] [CrossRef]
- Hulata, G. A Review of genetic improvement of the common carp (Cyprinus carpio L.) and other cyprinids by crossbreeding, hybridization and selection. Aquaculture 1995, 129, 143–155. [Google Scholar] [CrossRef]
- Smitherman, R.O.; Dunham, R.A. Genetics and Breeding. In Channel Catfish Culture; Tucker, C.S., Ed.; Elsevier Science Ltd.: Amsterdam, The Netherlands, 1985. [Google Scholar]
- Hulata, G. Genetic manipulations in aquaculture: A review of stock improvement by classical and modern technologies. Genetica 2001, 111, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Chen, J.; Peng, W.; Wang, Y.; Liao, L.; Li, Y.; Trudeau, V.L.; Zhu, Z.; Hu, W. Effects of Growth hormone over-expression on reproduction in the common carp (Cyprinus carpio L.). Gen. Comp. Endocrinol. 2014, 195, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.-T.; Zohar, Y. Production of reproductively sterile fish by a non-transgenic gene silencing technology. Sci. Rep. 2015, 5, 15822. [Google Scholar] [CrossRef] [PubMed]
- Osterwalder, T.; Yoon, K.S.; White, B.H.; Keshishian, H. A Conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 2001, 98, 12596–12601. [Google Scholar] [CrossRef] [PubMed]
- Distel, M.; Wullimann, M.F.; Köster, R.W. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. USA 2009, 106, 13365–13370. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.-Y.; Lin, P.-Y.; Liao, C.-H.; Gong, H.-Y.; Lin, G.-H.; Kawakami, K.; Wu, J.-L. Nitroreductase-mediated gonadal dysgenesis for infertility control of genetically modified zebrafish. Mar. Biotechnol. 2010, 12, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-C.; Hou, M.-F.; Hong, J.-R.; Wu, J.-L.; Her, G.M. Inducible male infertility by targeted cell ablation in zebrafish testis. Mar. Biotechnol. 2010, 12, 466–478. [Google Scholar] [CrossRef]
- Wong, A.C.; Van Eenennaam, A.L. Transgenic approaches for the reproductive containment of genetically engineered fish. Aquaculture 2008, 275, 1–12. [Google Scholar] [CrossRef]
- Ahmed, N.; Thompson, S.; Glaser, M. Global aquaculture productivity, environmental sustainability, and climate change adaptability. Environ. Manag. 2019, 63, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Güralp, H.; Skaftnesmo, K.O.; Kjærner-Semb, E.; Straume, A.H.; Kleppe, L.; Schulz, R.W.; Edvardsen, R.B.; Wargelius, A. Rescue of germ cells in dnd crispant embryos opens the possibility to produce inherited sterility in Atlantic salmon. Sci. Rep. 2020, 10, 18042. [Google Scholar] [CrossRef] [PubMed]
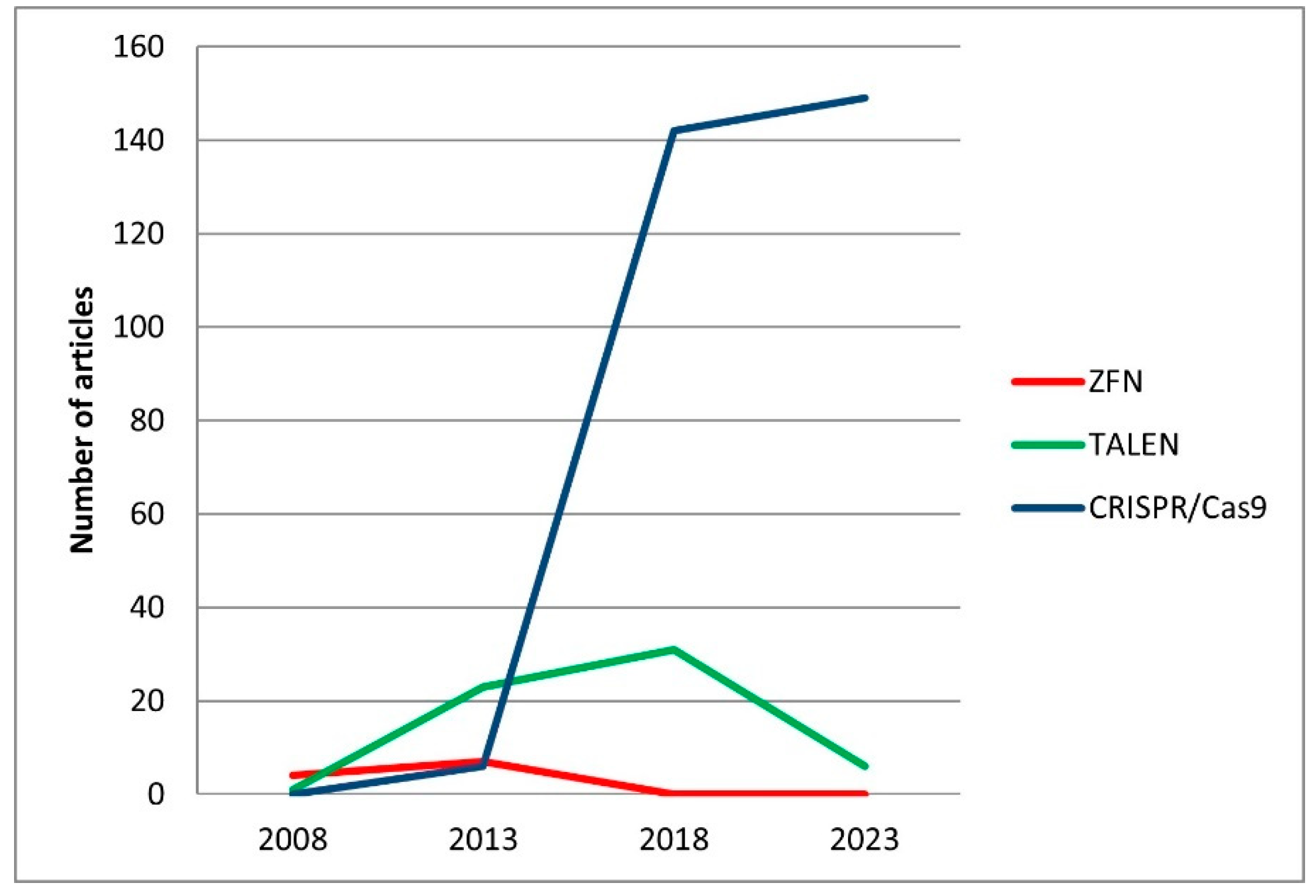
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orlova, S.Y.; Ruzina, M.N.; Emelianova, O.R.; Sergeev, A.A.; Chikurova, E.A.; Orlov, A.M.; Mugue, N.S. In Search of a Target Gene for a Desirable Phenotype in Aquaculture: Genome Editing of Cyprinidae and Salmonidae Species. Genes 2024, 15, 726. https://doi.org/10.3390/genes15060726
Orlova SY, Ruzina MN, Emelianova OR, Sergeev AA, Chikurova EA, Orlov AM, Mugue NS. In Search of a Target Gene for a Desirable Phenotype in Aquaculture: Genome Editing of Cyprinidae and Salmonidae Species. Genes. 2024; 15(6):726. https://doi.org/10.3390/genes15060726
Chicago/Turabian StyleOrlova, Svetlana Yu., Maria N. Ruzina, Olga R. Emelianova, Alexey A. Sergeev, Evgeniya A. Chikurova, Alexei M. Orlov, and Nikolai S. Mugue. 2024. "In Search of a Target Gene for a Desirable Phenotype in Aquaculture: Genome Editing of Cyprinidae and Salmonidae Species" Genes 15, no. 6: 726. https://doi.org/10.3390/genes15060726
APA StyleOrlova, S. Y., Ruzina, M. N., Emelianova, O. R., Sergeev, A. A., Chikurova, E. A., Orlov, A. M., & Mugue, N. S. (2024). In Search of a Target Gene for a Desirable Phenotype in Aquaculture: Genome Editing of Cyprinidae and Salmonidae Species. Genes, 15(6), 726. https://doi.org/10.3390/genes15060726






