The Analysis, Description, and Examination of the Maize LAC Gene Family’s Reaction to Abiotic and Biotic Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Chromosome Mapping of the LAC Family Genes in Maize
2.2. Analysis of the Physicochemical Characteristics and Conserved Motif of the LAC Gene Family in Maize
2.3. Phylogenetic Analysis of the LAC Family Genes in Maize
2.4. Collinearity Analysis of the LAC Family Genes in Maize
2.5. Functional Analysis of the LAC Family Genes in Maize
2.6. Re-Evaluation of the Large-Scale Data from RNA-Seq for Maize Transcriptome Sequencing
2.7. Analysis Was Conducted on the Expression Patterns of the LAC Gene Family in Maize across Various Tissues
2.8. Analysis of the Expression of LAC Family Genes in Maize under Both Abiotic and Biotic Stress Conditions
3. Results
3.1. An Overview of the Maize LAC Gene Family Members
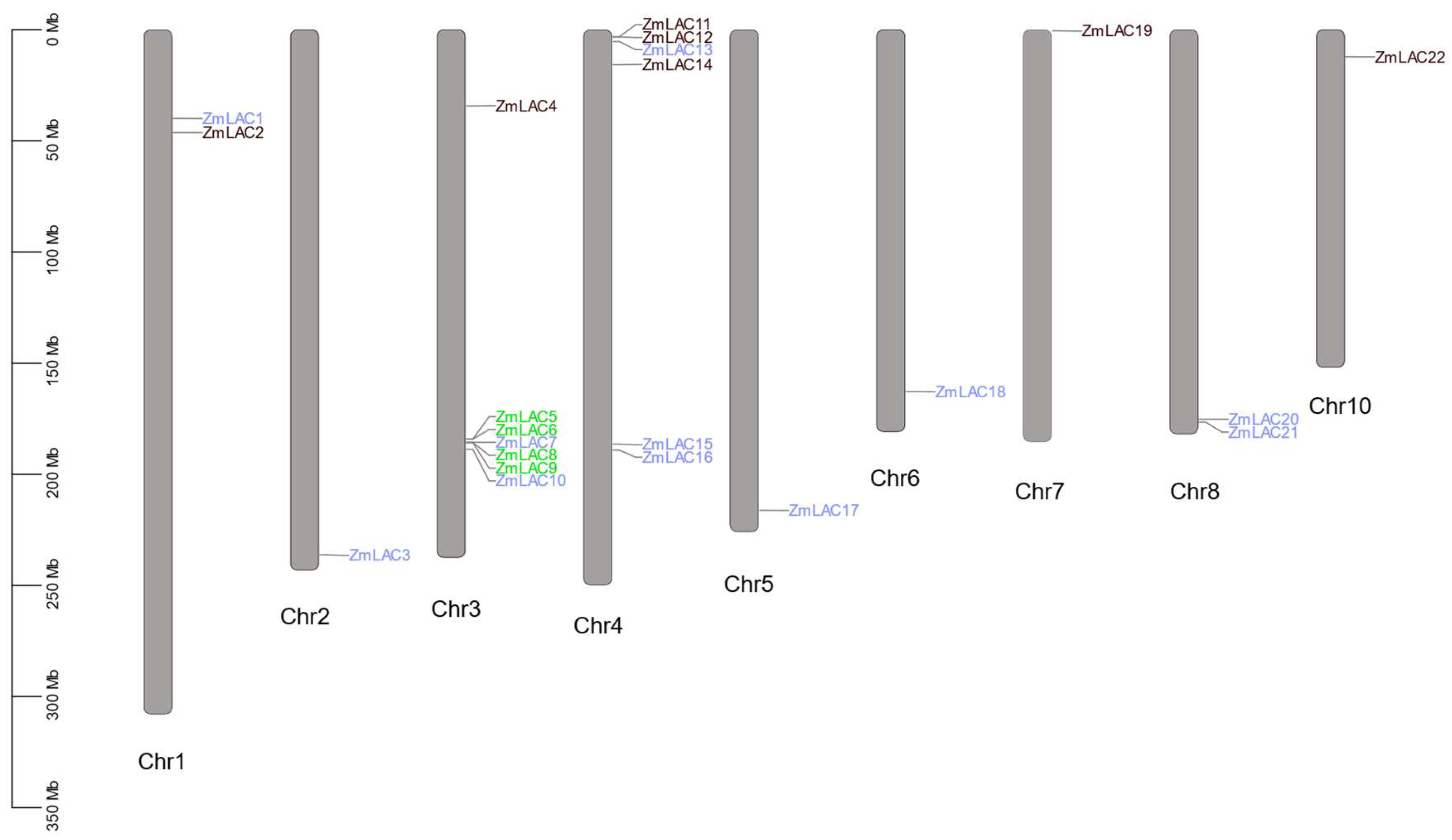
3.2. Mapping the LAC Family Genes in Maize Using Chromosome Analysis
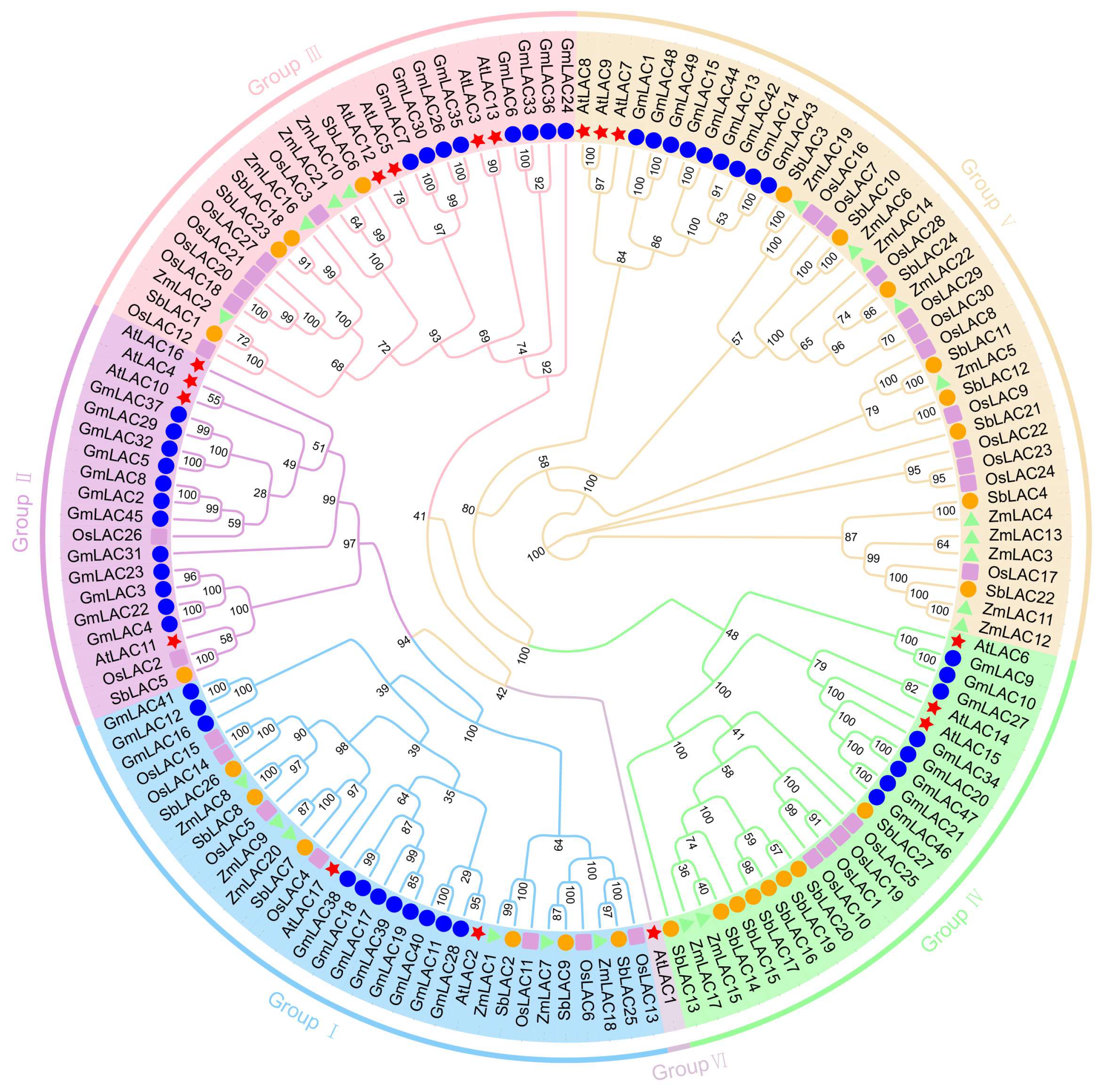
3.3. Phylogenetic Tree Analysis of the LAC Gene Family in Maize
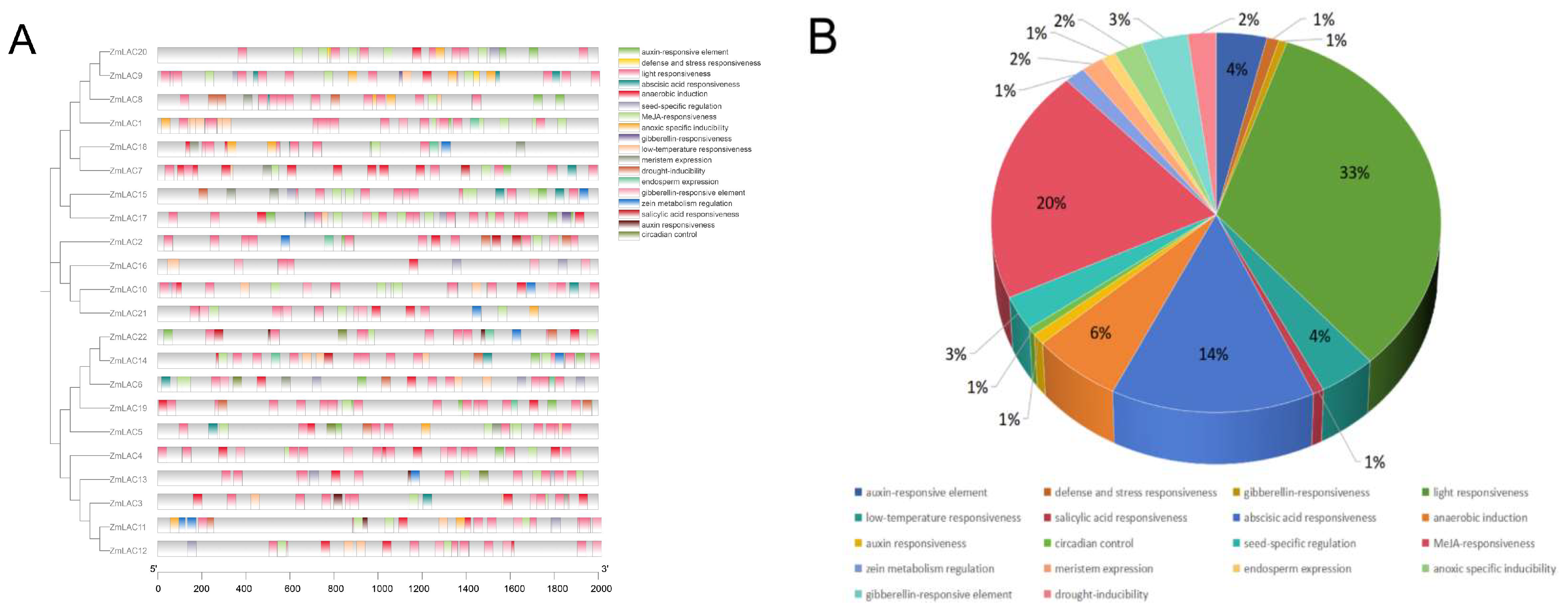
3.4. An Examination of the LAC Gene Family in Maize through Comparing Gene Structure and Conserved Sequences
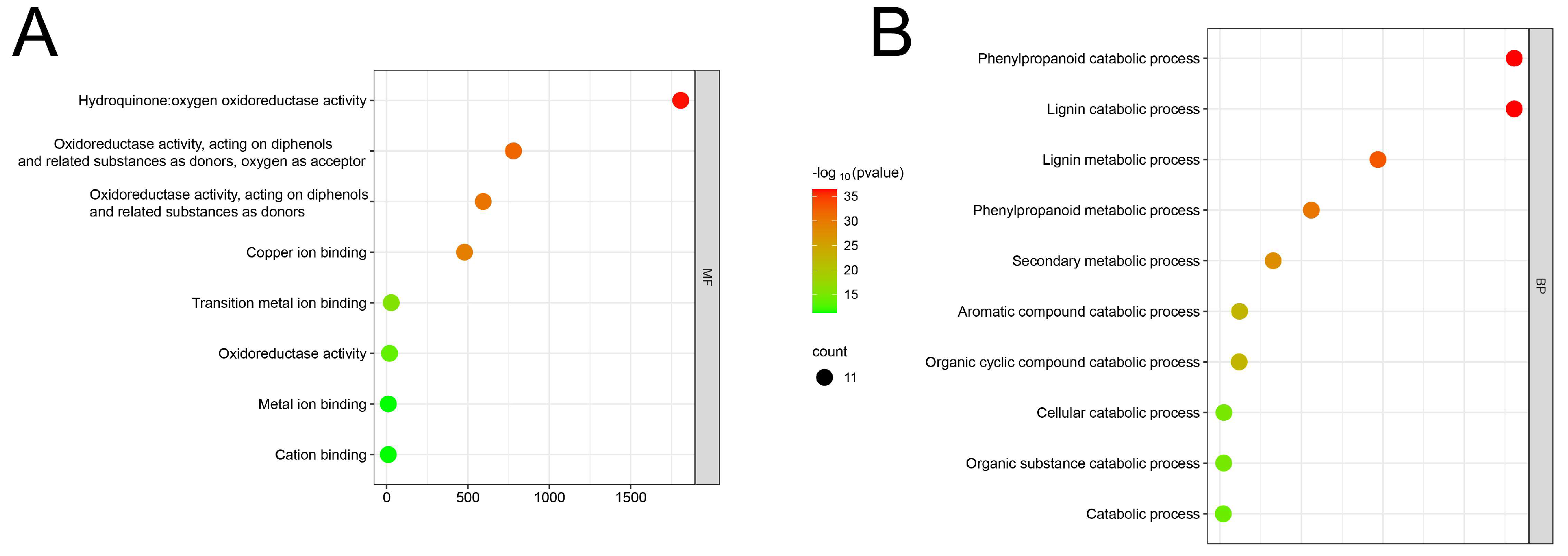
3.5. An Examination of the Functional Aspects of the LAC Gene Family in Maize
3.6. Collinearity Analysis of the LAC Family Genes in Maize
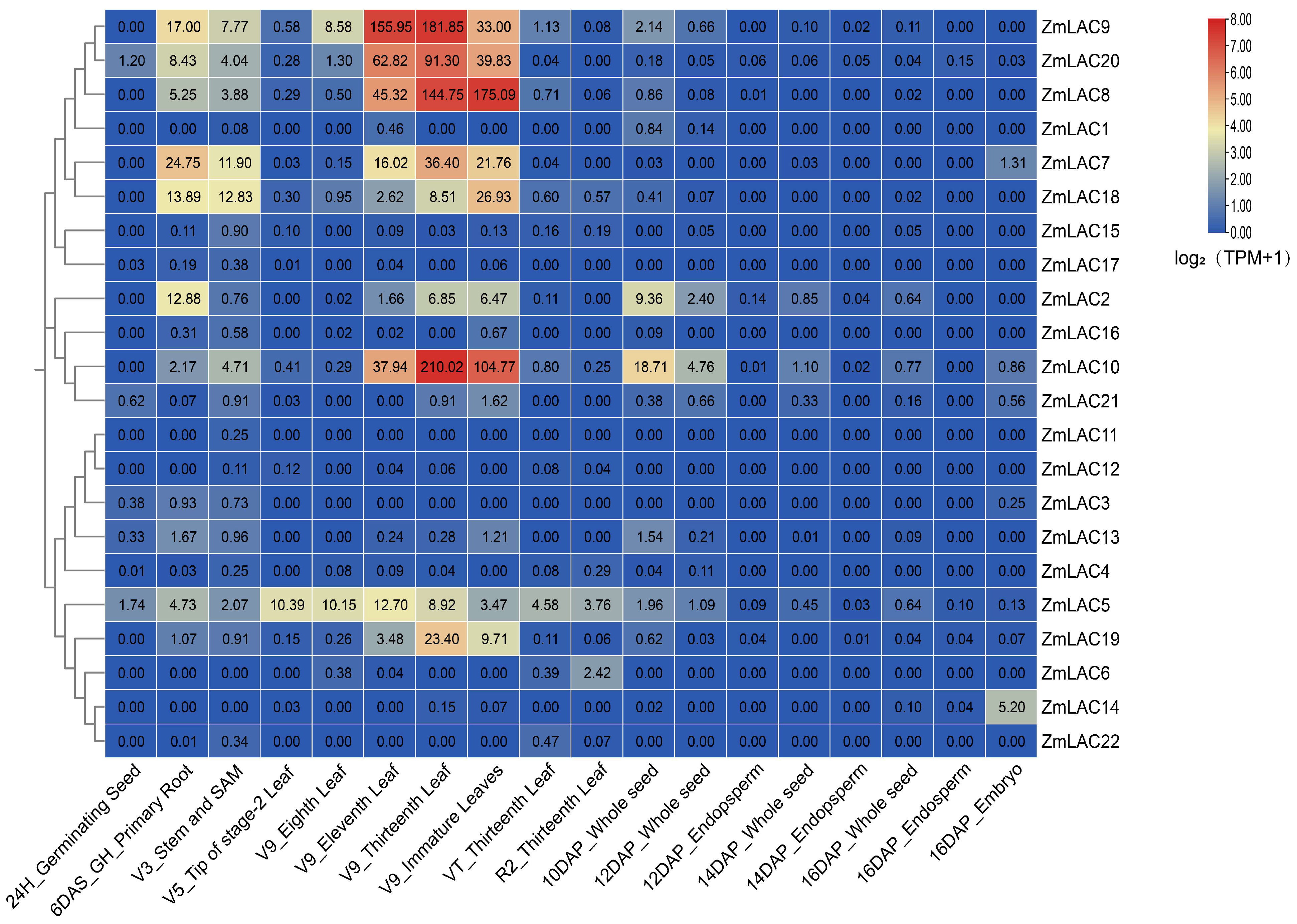
3.7. Analysis Was Conducted on the Expression Patterns of the LAC Family Genes in Maize in Various Tissues
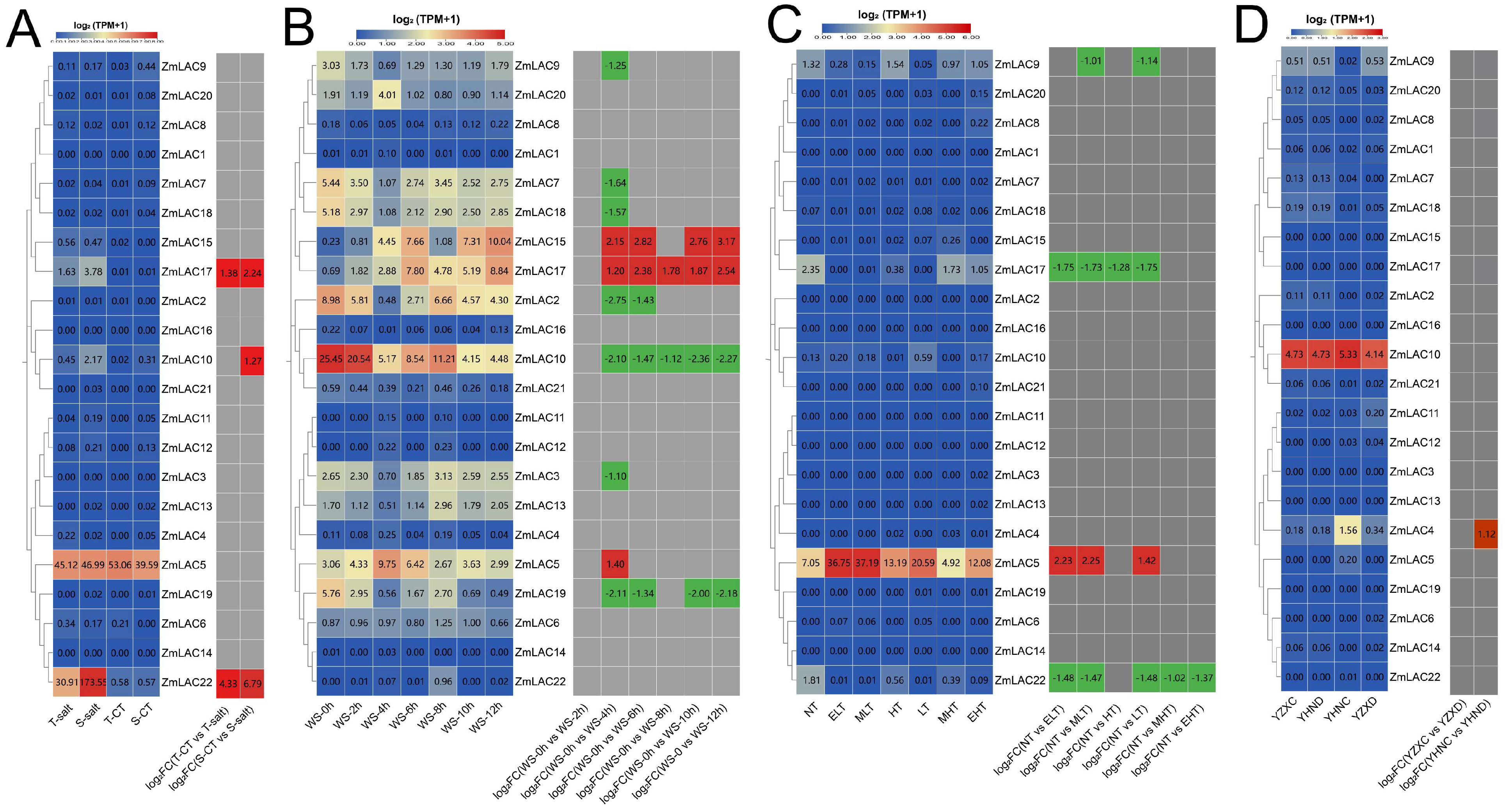
3.8. Analysis of Expression Patterns of Maize LAC Family Genes under Abiotic Stress
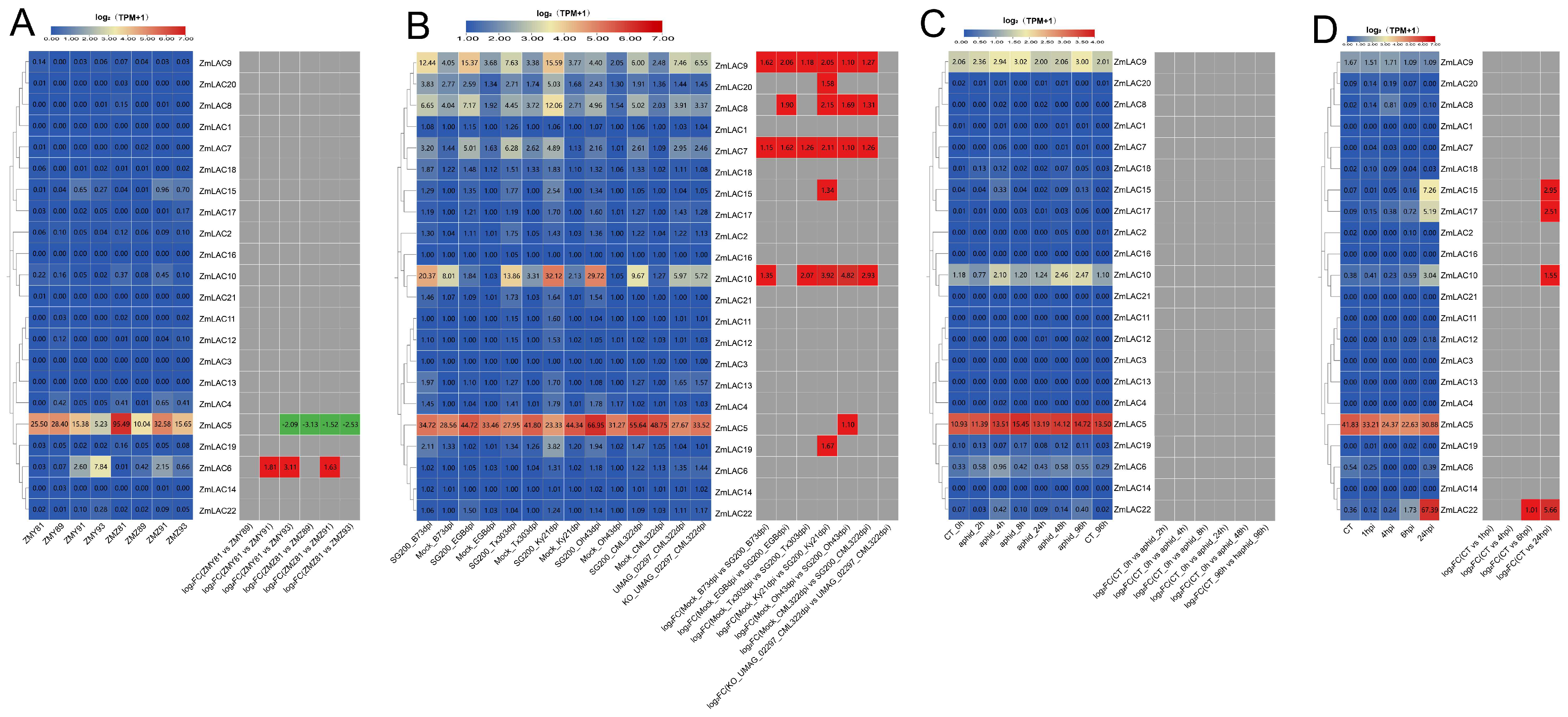
3.9. Analysis on How Maize LAC Family Genes Are Expressed during Biotic Stress
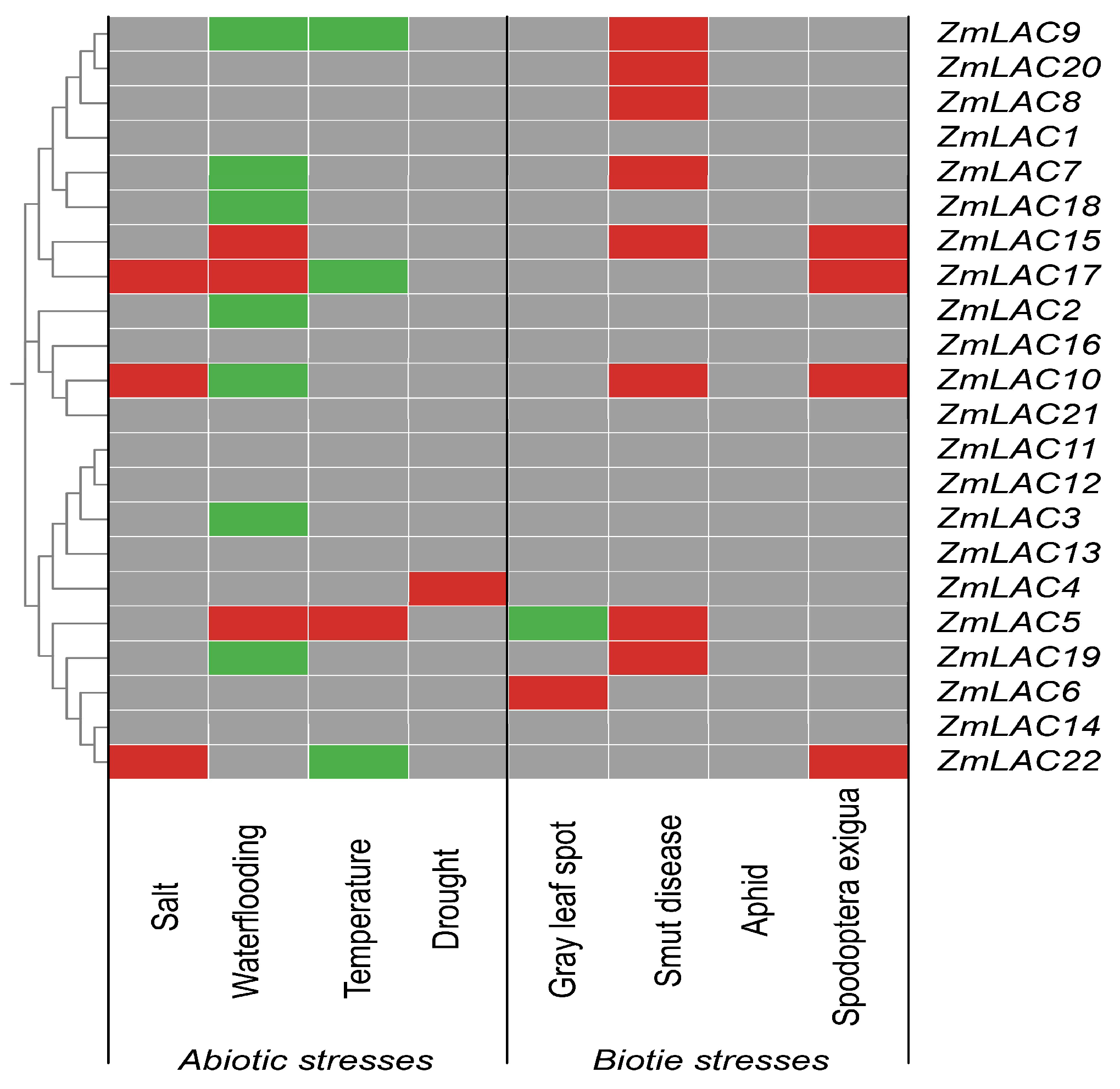
3.10. Analysis of the Regulatory Mode of the Maize LAC Family Genes under Abiotic and Biotic Stressors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yoshida, H. LXIII.—Chemistry of lacquer (Urushi). Part I. Communication from the Chemical Society of Tokio. J. Chem. Soc. Trans. 1883, 43, 472–486. [Google Scholar] [CrossRef]
- Hoegger, P.J.; Kilaru, S.; James, T.Y.; Thacker, J.R.; Kües, U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006, 273, 2308–2326. [Google Scholar] [CrossRef]
- Bao, W.; O’Malley, D.M.; Whetten, R.; Sederoff, R.R. A Laccase Associated with Lignification in Loblolly Pine Xylem. Science 1993, 260, 672–674. [Google Scholar] [CrossRef]
- LaFayette, P.R.; Eriksson, K.E.L.; Dean, J.F.D. Nucleotide Sequence of a cDNA Clone Encoding an Acidic Laccase from Sycamore Maple (Acer pseudoplatanus L.). Plant Physiol. 1995, 107, 667–668. [Google Scholar] [CrossRef]
- Driouich, A.; Lainé, A.C.; Vian, B.; Faye, L. Characterization and localization of laccase forms in stem and cell cultures of sycamore. Plant J. 2005, 2, 13–24. [Google Scholar] [CrossRef]
- Morozova, O.V.; Shumakovich, G.P.; Gorbacheva, M.A.; Shleev, S.V.; Yaropolov, A.I. “Blue” laccases. Biochemistry 2007, 72, 1136–1150. [Google Scholar] [CrossRef]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2009, 67, 369–385. [Google Scholar] [CrossRef]
- Dwivedi, U.N.; Singh, P.; Pandey, V.P.; Kumar, A. Structure–function relationship among bacterial, fungal and plant laccases. J. Mol. Catal. B Enzym. 2011, 68, 117–128. [Google Scholar] [CrossRef]
- Yi Chou, E.; Schuetz, M.; Hoffmann, N.; Watanabe, Y.; Sibout, R.; Samuels, A.L. Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J. Exp. Bot. 2018, 69, 1849–1859. [Google Scholar] [CrossRef]
- Diamantidis, G.; Effosse, A.; Potier, P.; Bally, R. Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol. Biochem. 2000, 32, 919–927. [Google Scholar] [CrossRef]
- Jiao, X.; Li, G.; Wang, Y.; Nie, F.; Cheng, X.; Abdullah, M.; Lin, Y.; Cai, Y. Systematic Analysis of the Pleurotus ostreatus Laccase Gene (PoLac) Family and Functional Characterization of PoLac2 Involved in the Degradation of Cotton-Straw Lignin. Molecules 2018, 23, 880. [Google Scholar] [CrossRef]
- Liu, M.; Dong, H.; Wang, M.; Liu, Q. Evolutionary divergence of function and expression of laccase genes in plants. J. Genet. 2020, 99, 1–16. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, J.; Francis, F.; Chen, J. Molecular characterization and gene silencing of Laccase 1 in the grain aphid, Sitobion avenae. Arch. Insect Biochem. Physiol. 2018, 97, e21446. [Google Scholar] [CrossRef]
- Wang, J.P.; Chuang, L.; Loziuk, P.L.; Chen, H.; Lin, Y.-C.; Shi, R.; Qu, G.-Z.; Muddiman, D.C.; Sederoff, R.R.; Chiang, V.L. Phosphorylation is an on/off switch for 5-hydroxyconiferaldehyde O-methyltransferase activity in poplar monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 8481–8486. [Google Scholar] [CrossRef]
- Kishimoto, T.; Hiyama, A.; Toda, H.; Urabe, D. Effect of pH on the Dehydrogenative Polymerization of Monolignols by Laccases from Trametes versicolor and Rhus vernicifera. ACS Omega 2022, 7, 9846–9852. [Google Scholar] [CrossRef]
- McCaig, B.C.; Meagher, R.B.; Dean, J.F.D. Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 2005, 221, 619–636. [Google Scholar] [CrossRef]
- Turlapati, P.V.; Kim, K.-W.; Davin, L.B.; Lewis, N.G. The laccase multigene family in Arabidopsis thaliana: Towards addressing the mystery of their gene function(s). Planta 2010, 233, 439–470. [Google Scholar] [CrossRef]
- Wang, Y.; Bouchabke-Coussa, O.; Lebris, P.; Antelme, S.; Soulhat, C.; Gineau, E.; Dalmais, M.; Bendahmane, A.; Morin, H.; Mouille, G.; et al. LACCASE5 Is Required for Lignification of the Brachypodium distachyon Culm. Plant Physiol. 2015, 168, 192–204. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, K.; Lu, K.; Qu, C.; Liang, Y.; Wang, R.; Chai, Y.; Li, J. Gene Silencing of BnTT10 Family Genes Causes Retarded Pigmentation and Lignin Reduction in the Seed Coat of Brassica napus. PLoS ONE 2013, 8, e61247. [Google Scholar] [CrossRef]
- Wang, G.-D.; Li, Q.-J.; Luo, B.; Chen, X.-Y. Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat. Biotechnol. 2004, 22, 893–897. [Google Scholar] [CrossRef]
- Ranocha, P.; McDougall, G.; Hawkins, S.; Sterjiades, R.; Borderies, G.; Stewart, D.; Cabanes-Macheteau, M.; Boudet, A.M.; Goffner, D. Biochemical characterization, molecular cloning and expression of laccases—A divergent gene family—In poplar. Eur. J. Biochem. 2001, 259, 485–495. [Google Scholar] [CrossRef]
- Liu, Q.; Zheng, L.; He, F.; Zhao, F.-J.; Shen, Z.; Zheng, L. Transcriptional and physiological analyses identify a regulatory role for hydrogen peroxide in the lignin biosynthesis of copper-stressed rice roots. Plant Soil 2014, 387, 323–336. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Wang, X.; Shen, Z.; Zheng, L. Comprehensive Analysis of Rice Laccase Gene (OsLAC) Family and Ectopic Expression of OsLAC10 Enhances Tolerance to Copper Stress in Arabidopsis. Int. J. Mol. Sci. 2017, 18, 209. [Google Scholar] [CrossRef]
- Wang, J.; Feng, J.; Jia, W.; Fan, P.; Bao, H.; Li, S.; Li, Y. Genome-Wide Identification of Sorghum bicolor Laccases Reveals Potential Targets for Lignin Modification. Front. Plant Sci. 2017, 8, 714. [Google Scholar] [CrossRef]
- Gavnholt, B.; Larsen, K.; Rasmussen, S.K. Isolation and characterisation of laccase cDNAs from meristematic and stem tissues of ryegrass (Lolium perenne). Plant Sci. 2002, 162, 873–885. [Google Scholar] [CrossRef]
- Cesarino, I.; Araújo, P.; Sampaio Mayer, J.L.; Vicentini, R.; Berthet, S.; Demedts, B.; Vanholme, B.; Boerjan, W.; Mazzafera, P. Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J. Exp. Bot. 2013, 64, 1769–1781. [Google Scholar] [CrossRef]
- Kiefer-Meyer, M.-C.; Gomord, V.; O’Connell, A.; Halpin, C.; Faye, L. Cloning and sequence analysis of laccase-encoding cDNA clones from tobacco. Gene 1996, 178, 205–207. [Google Scholar] [CrossRef]
- LaFayette, P.R. Characterization and heterologous expression of laccase cDNAs from xylem tissues of yellow-poplar (Liriodendron tulipifera). Plant Mol. Biol. 1999, 40, 23–35. [Google Scholar] [CrossRef]
- Baldrian, P. Fungal laccases—Occurrence and properties. FEMS Microbiol. Rev. 2006, 30, 215–242. [Google Scholar] [CrossRef]
- Courty, P.E.; Hoegger, P.J.; Kilaru, S.; Kohler, A.; Buée, M.; Garbaye, J.; Martin, F.; Kües, U. Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol. 2009, 182, 736–750. [Google Scholar] [CrossRef]
- Han, K.-H.; Sapmak, A.; Boyce, K.J.; Andrianopoulos, A.; Vanittanakom, N. The pbrB Gene Encodes a Laccase Required for DHN-Melanin Synthesis in Conidia of Talaromyces (Penicillium) marneffei. PLoS ONE 2015, 10, e0122728. [Google Scholar]
- Cai, X.; Davis, E.J.; Ballif, J.; Liang, M.; Bushman, E.; Haroldsen, V.; Torabinejad, J.; Wu, Y. Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 2006, 57, 2563–2569. [Google Scholar] [CrossRef]
- Liang, M.; Davis, E.; Gardner, D.; Cai, X.; Wu, Y. Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 2006, 224, 1185–1196. [Google Scholar] [CrossRef]
- Balasubramanian, V.K.; Rai, K.M.; Thu, S.W.; Hii, M.M.; Mendu, V. Genome-wide identification of multifunctional laccase gene family in cotton (Gossypium spp.); expression and biochemical analysis during fiber development. Sci. Rep. 2016, 6, 34309. [Google Scholar] [CrossRef]
- Yu, Y.; Li, Q.-F.; Zhang, J.-P.; Zhang, F.; Zhou, Y.-F.; Feng, Y.-Z.; Chen, Y.-Q.; Zhang, Y.-C. Laccase-13 Regulates Seed Setting Rate by Affecting Hydrogen Peroxide Dynamics and Mitochondrial Integrity in Rice. Front. Plant Sci. 2017, 8, 1324. [Google Scholar] [CrossRef]
- Swetha, C.; Basu, D.; Pachamuthu, K.; Tirumalai, V.; Nair, A.; Prasad, M.; Shivaprasad, P.V. Major Domestication-Related Phenotypes inIndicaRice Are Due to Loss of miRNA-Mediated Laccase Silencing. Plant Cell 2018, 30, 2649–2662. [Google Scholar] [CrossRef]
- Davis, K.R.; Cheng, X.; Li, G.; Ma, C.; Abdullah, M.; Zhang, J.; Zhao, H.; Jin, Q.; Cai, Y.; Lin, Y. Comprehensive genome-wide analysis of the pear (Pyrus bretschneideri) laccase gene (PbLAC) family and functional identification of PbLAC1 involved in lignin biosynthesis. PLoS ONE 2019, 14, e0210892. [Google Scholar]
- Hu, Q.; Min, L.; Yang, X.; Jin, S.; Zhang, L.; Li, Y.; Ma, Y.; Qi, X.; Li, D.; Liu, H.; et al. Laccase GhLac1 Modulates Broad-Spectrum Biotic Stress Tolerance via Manipulating Phenylpropanoid Pathway and Jasmonic Acid Synthesis. Plant Physiol. 2018, 176, 1808–1823. [Google Scholar] [CrossRef]
- Hashemipetroudi, S.H.; Arab, M.; Heidari, P.; Kuhlmann, M. Genome-wide analysis of the laccase (LAC) gene family in Aeluropus littoralis: A focus on identification, evolution and expression patterns in response to abiotic stresses and ABA treatment. Front. Plant Sci. 2023, 14, 1112354. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, L.; Wang, X.; Chen, B.; Zhao, J.; Cui, J.; Li, Z.; Yang, J.; Wu, L.; Wu, J.; et al. The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 2018, 20, 309–322. [Google Scholar] [CrossRef]
- Wei, T.; Tang, Y.; Jia, P.; Zeng, Y.; Wang, B.; Wu, P.; Quan, Y.; Chen, A.; Li, Y.; Wu, J. A Cotton Lignin Biosynthesis Gene, GhLAC4, Fine-Tuned by ghr-miR397 Modulates Plant Resistance against Verticillium dahliae. Front. Plant Sci. 2021, 12, 743795. [Google Scholar] [CrossRef]
- Nitta, K.; Kataoka, K.; Sakurai, T. Primary structure of a Japanese lacquer tree laccase as a prototype enzyme of multicopper oxidases. J. Inorg. Biochem. 2002, 91, 125–131. [Google Scholar] [CrossRef]
- Liang, M.; Haroldsen, V.; Cai, X.; Wu, Y. Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress*. Plant Cell Environ. 2006, 29, 746–753. [Google Scholar] [CrossRef]
- Pei, L.; Gao, X.; Tian, X.; Liu, N.; Chen, M.; Fernie, A.R.; Li, H. A microRNA528-ZmLac3 module regulates low phosphate tolerance in maize. Plant J. 2024. [Google Scholar] [CrossRef]
- Benz, B.F. Archaeological evidence of teosinte domestication from Guilá Naquitz, Oaxaca. Proc. Natl. Acad. Sci. USA 2001, 98, 2104–2106. [Google Scholar] [CrossRef]
- Ray, L.I.P.; Jyothi, K.S.; Singh, A.K.; Bharti, V.; Pandey, P.K. Strategies for water productivity enhancement in maize—A comprehensive review. Irrig. Drain. 2023, 73, 359–374. [Google Scholar] [CrossRef]
- Barošević, T.; Bagi, F.; Savić, Z.; Ljubičić, N.; Ivanović, I. Assessment of Maize Hybrids Resistance to Aspergillus Ear Rot and Aflatoxin Production in Environmental Conditions in Serbia. Toxins 2022, 14, 887. [Google Scholar] [CrossRef]
- Gabaldn, T.; Huynen, M.A. Prediction of protein function and pathways in the genome era. Cell. Mol. Life Sci. 2004, 61, 930–944. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef]
- Li, J.; Yang, S.; Yang, X.; Wu, H.; Tang, H.; Yang, L. PlantGF: An analysis and annotation platform for plant gene families. Database 2022, 2022, baab088. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Cai, Q.; Li, X.; Sun, Y.; Yu, T.; Yang, J.; Zhang, J. Analysis of the C2H2 Gene Family in Maize (Zea mays L.) under Cold Stress: Identification and Expression. Life 2022, 13, 122. [Google Scholar] [CrossRef]
- Nawaz, A.F.; Zia, M.A.; Shoukat, S.; Arif, M.; Ali, S. Genome-wide identification and expression analysis of the Glutamine synthetase family genes in Zea mays under drought stress. Plant Stress 2023, 9, 100180. [Google Scholar] [CrossRef]
- Wang, X.; Tian, X.; Zhang, H.; Li, H.; Zhang, S.; Li, H.; Zhu, J. Genome-wide analysis of the maize LACS gene family and functional characterization of the ZmLACS9 responses to heat stress. Plant Stress 2023, 10, 100271. [Google Scholar] [CrossRef]
- Tang, H.; Jing, D.; Liu, C.; Xie, X.; Zhang, L.; Chen, X.; Li, C. Genome-Wide Identification and Expression Analyses of the FAR1/FHY3 Gene Family Provide Insight into Inflorescence Development in Maize. Curr. Issues Mol. Biol. 2024, 46, 430–449. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, L.; Li, T.; Ruan, Y.; Zhang, A.; Dong, X.; Zhu, Y.; Li, C.; Fan, J. Genome-Wide Investigation and Characterization of SWEET Gene Family with Focus on Their Evolution and Expression during Hormone and Abiotic Stress Response in Maize. Genes 2022, 13, 1682. [Google Scholar] [CrossRef]
- Sun, Q.; Liu, X.; Yang, J.; Liu, W.; Du, Q.; Wang, H.; Fu, C.; Li, W.-X. MicroRNA528 Affects Lodging Resistance of Maize by Regulating Lignin Biosynthesis under Nitrogen-Luxury Conditions. Mol. Plant 2018, 11, 806–814. [Google Scholar] [CrossRef]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2007, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2011, 40, D290–D301. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Brown, G.R.; Maglott, D.R. NCBI Reference Sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Res. 2011, 40, D130–D135. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Poon, A.F.Y.; Zhang, Y.-J.; Ma, P.-F.; Li, D.-Z. High-Throughput Sequencing of Six Bamboo Chloroplast Genomes: Phylogenetic Implications for Temperate Woody Bamboos (Poaceae: Bambusoideae). PLoS ONE 2011, 6, e20596. [Google Scholar]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Feng, Q.; Gao, Z.; Lu, H.; Hu, T.; Yao, N.; Liu, K.; et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R.; Valencia, A. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Yin, Y.; Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A.; Wren, J. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef]
- Sun, M.-x.; Sekhon, R.S.; Briskine, R.; Hirsch, C.N.; Myers, C.L.; Springer, N.M.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Maize Gene Atlas Developed by RNA Sequencing and Comparative Evaluation of Transcriptomes Based on RNA Sequencing and Microarrays. PLoS ONE 2013, 8, e61005. [Google Scholar]
- Wang, M.; Wang, Y.; Zhang, Y.; Li, C.; Gong, S.; Yan, S.; Li, G.; Hu, G.; Ren, H.; Yang, J.; et al. Comparative transcriptome analysis of salt-sensitive and salt-tolerant maize reveals potential mechanisms to enhance salt resistance. Genes Genom. 2019, 41, 781–801. [Google Scholar] [CrossRef]
- Yu, F.; Tan, Z.; Fang, T.; Tang, K.; Liang, K.; Qiu, F. A Comprehensive Transcriptomics Analysis Reveals Long Non-Coding RNA to Be Involved in the Key Metabolic Pathway in Response to Waterlogging Stress in Maize. Genes 2020, 11, 267. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Li, Y.; Zhang, Y.; Gou, Z.; Qi, X.; Zhang, J. Transcriptomic Analysis Revealed the Common and Divergent Responses of Maize Seedling Leaves to Cold and Heat Stresses. Genes 2020, 11, 881. [Google Scholar] [CrossRef]
- Prasad, M.; Jin, H.; Liu, S.; Zenda, T.; Wang, X.; Liu, G.; Duan, H. Maize leaves drought-responsive genes revealed by comparative transcriptome of two cultivars during the filling stage. PLoS ONE 2019, 14, e0223786. [Google Scholar]
- Yu, Y.; Shi, J.; Li, X.; Liu, J.; Geng, Q.; Shi, H.; Ke, Y.; Sun, Q. Transcriptome analysis reveals the molecular mechanisms of the defense response to gray leaf spot disease in maize. BMC Genom. 2018, 19, 1–17. [Google Scholar] [CrossRef]
- Schurack, S.; Depotter, J.R.L.; Gupta, D.; Thines, M.; Doehlemann, G. Comparative transcriptome profiling identifies maize line specificity of fungal effectors in the maize–Ustilago maydis interaction. Plant J. 2021, 106, 733–752. [Google Scholar] [CrossRef]
- Tzin, V.; Fernandez-Pozo, N.; Richter, A.; Schmelz, E.A.; Schoettner, M.; Schäfer, M.; Ahern, K.R.; Meihls, L.N.; Kaur, H.; Huffaker, A.; et al. Dynamic maize responses to aphid feeding are revealed by a time series of transcriptomic and metabolomic assays. Plant Physiol. 2015, 169, 1727–1743. [Google Scholar] [CrossRef]
- Tzin, V.; Hojo, Y.; Strickler, S.R.; Bartsch, L.J.; Archer, C.M.; Ahern, K.R.; Zhou, S.; Christensen, S.A.; Galis, I.; Mueller, L.A.; et al. Rapid defense responses in maize leaves induced by Spodoptera exigua caterpillar feeding. J. Exp. Bot. 2017, 68, 4709–4723. [Google Scholar] [CrossRef]
- Hamilton, J.P.; Robin Buell, C. Advances in plant genome sequencing. Plant J. 2012, 70, 177–190. [Google Scholar] [CrossRef]
- Schuetz, M.; Benske, A.; Smith, R.A.; Watanabe, Y.; Tobimatsu, Y.; Ralph, J.; Demura, T.; Ellis, B.; Samuels, A.L. Laccases Direct Lignification in the Discrete Secondary Cell Wall Domains of Protoxylem. Plant Physiol. 2014, 166, 798–807. [Google Scholar] [CrossRef]
- Wang, J.; Feng, J.; Jia, W.; Chang, S.; Li, S.; Li, Y. Lignin engineering through laccase modification: A promising field for energy plant improvement. Biotechnol. Biofuels 2015, 8, 1–11. [Google Scholar] [CrossRef]
- Bai, Y.; Ali, S.; Liu, S.; Zhou, J.; Tang, Y. Characterization of plant laccase genes and their functions. Gene 2023, 852, 147060. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 1–19. [Google Scholar] [CrossRef]
- Clark, J.W.; Donoghue, P.C.J. Constraining the timing of whole genome duplication in plant evolutionary history. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170912. [Google Scholar] [CrossRef]
- Luo, Y.; Liao, X.; Wu, F.-X.; Wang, J. Computational Approaches for Transcriptome Assembly Based on Sequencing Technologies. Curr. Bioinform. 2020, 15, 2–16. [Google Scholar] [CrossRef]
- Ahlawat, Y.K.; Biswal, A.K.; Harun, S.; Harman-Ware, A.E.; Doeppke, C.; Sharma, N.; Joshi, C.P.; Hankoua, B.B. Heterologous expression of Arabidopsis laccase2, laccase4 and peroxidase52 driven under developing xylem specific promoter DX15 improves saccharification in populus. Biotechnol. Biofuels Bioprod. 2024, 17, 1–14. [Google Scholar] [CrossRef]
- Li, L.; Steffens, J. Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta 2002, 215, 239–247. [Google Scholar] [CrossRef]
- Rains, M.K.; Gardiyehewa de Silva, N.D.; Molina, I. Reconstructing the suberin pathway in poplar by chemical and transcriptomic analysis of bark tissues. Tree Physiol. 2018, 38, 340–361. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Liu, Y.; Zou, K.; Guan, M.; Wu, Y.; Hu, Y.; Yu, H.; Du, J.; Wu, D. The Analysis, Description, and Examination of the Maize LAC Gene Family’s Reaction to Abiotic and Biotic Stress. Genes 2024, 15, 749. https://doi.org/10.3390/genes15060749
Wang T, Liu Y, Zou K, Guan M, Wu Y, Hu Y, Yu H, Du J, Wu D. The Analysis, Description, and Examination of the Maize LAC Gene Family’s Reaction to Abiotic and Biotic Stress. Genes. 2024; 15(6):749. https://doi.org/10.3390/genes15060749
Chicago/Turabian StyleWang, Tonghan, Yang Liu, Kunliang Zou, Minhui Guan, Yutong Wu, Ying Hu, Haibing Yu, Junli Du, and Degong Wu. 2024. "The Analysis, Description, and Examination of the Maize LAC Gene Family’s Reaction to Abiotic and Biotic Stress" Genes 15, no. 6: 749. https://doi.org/10.3390/genes15060749
APA StyleWang, T., Liu, Y., Zou, K., Guan, M., Wu, Y., Hu, Y., Yu, H., Du, J., & Wu, D. (2024). The Analysis, Description, and Examination of the Maize LAC Gene Family’s Reaction to Abiotic and Biotic Stress. Genes, 15(6), 749. https://doi.org/10.3390/genes15060749




