Analysis of Hyperosmotic Tolerance Mechanisms in Gracilariopsis lemaneiformis Based on Weighted Co-Expression Network Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Material Handling and Sampling
2.3. Total RNA Extraction and Library Construction
2.4. Quality Control and Analysis of Sequencing Data
2.5. Weighted Correlation Network Analysis (WGCNA)
3. Results
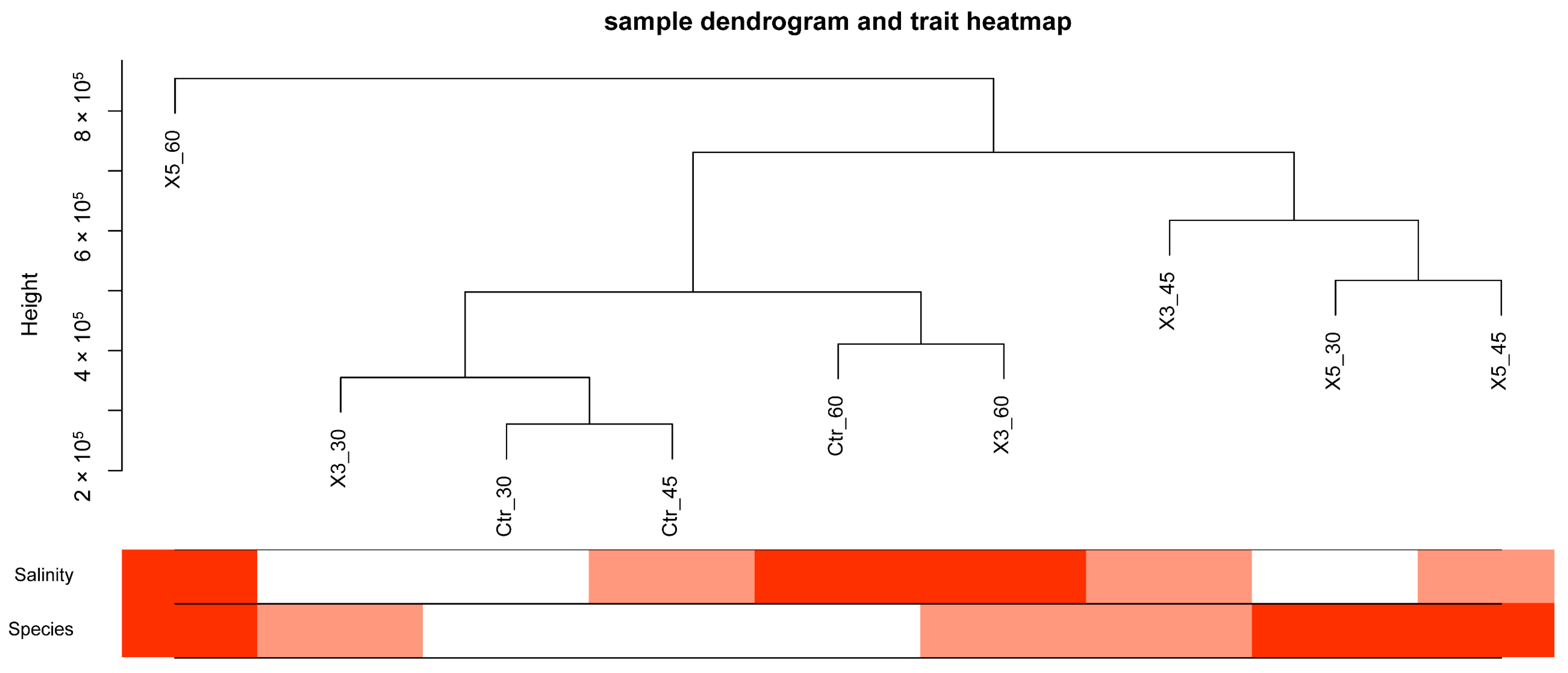
3.1. Correlation Analysis of Samples and Traits
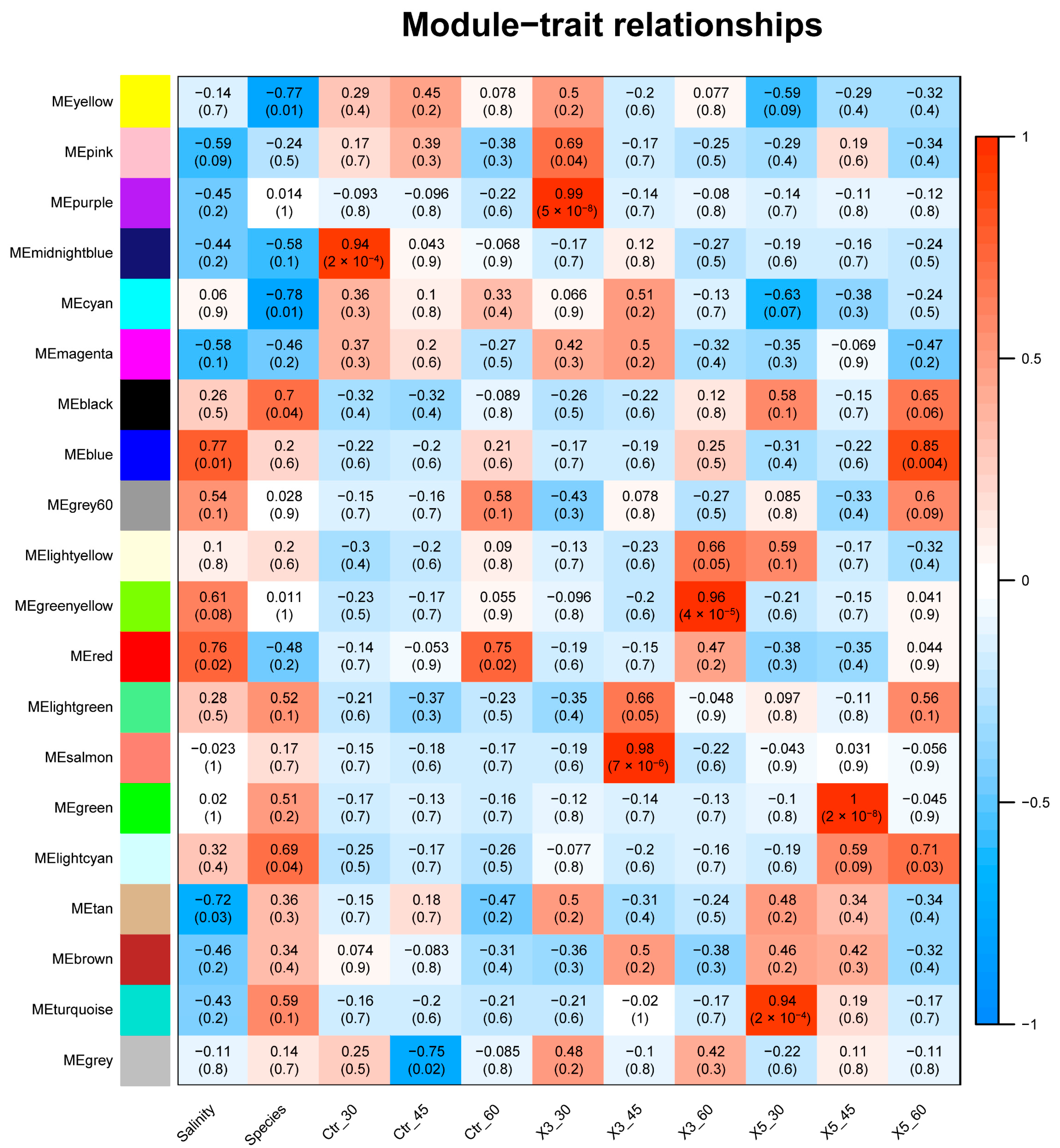
3.2. Correlation Analysis between Samples, Traits, and Modules
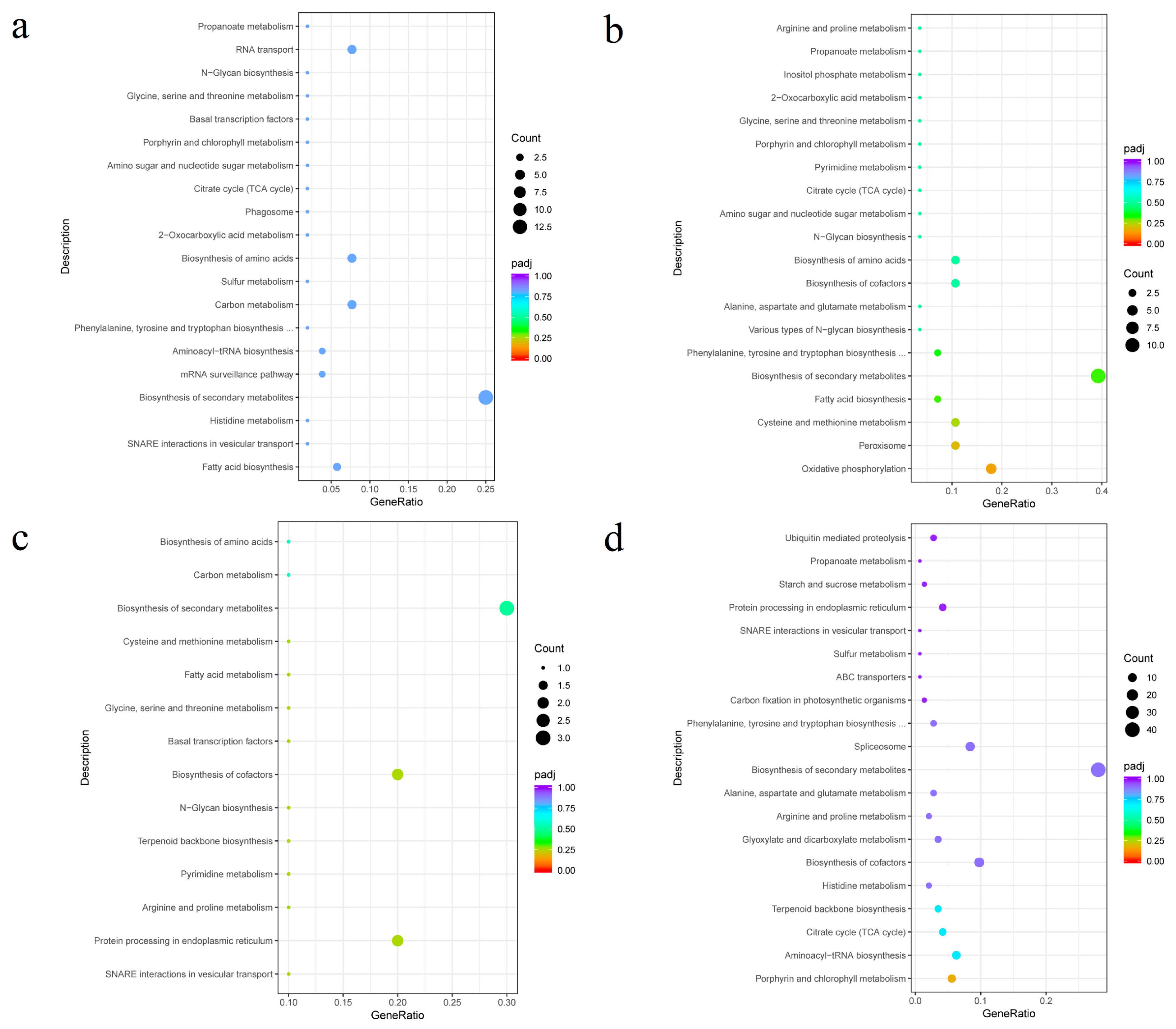
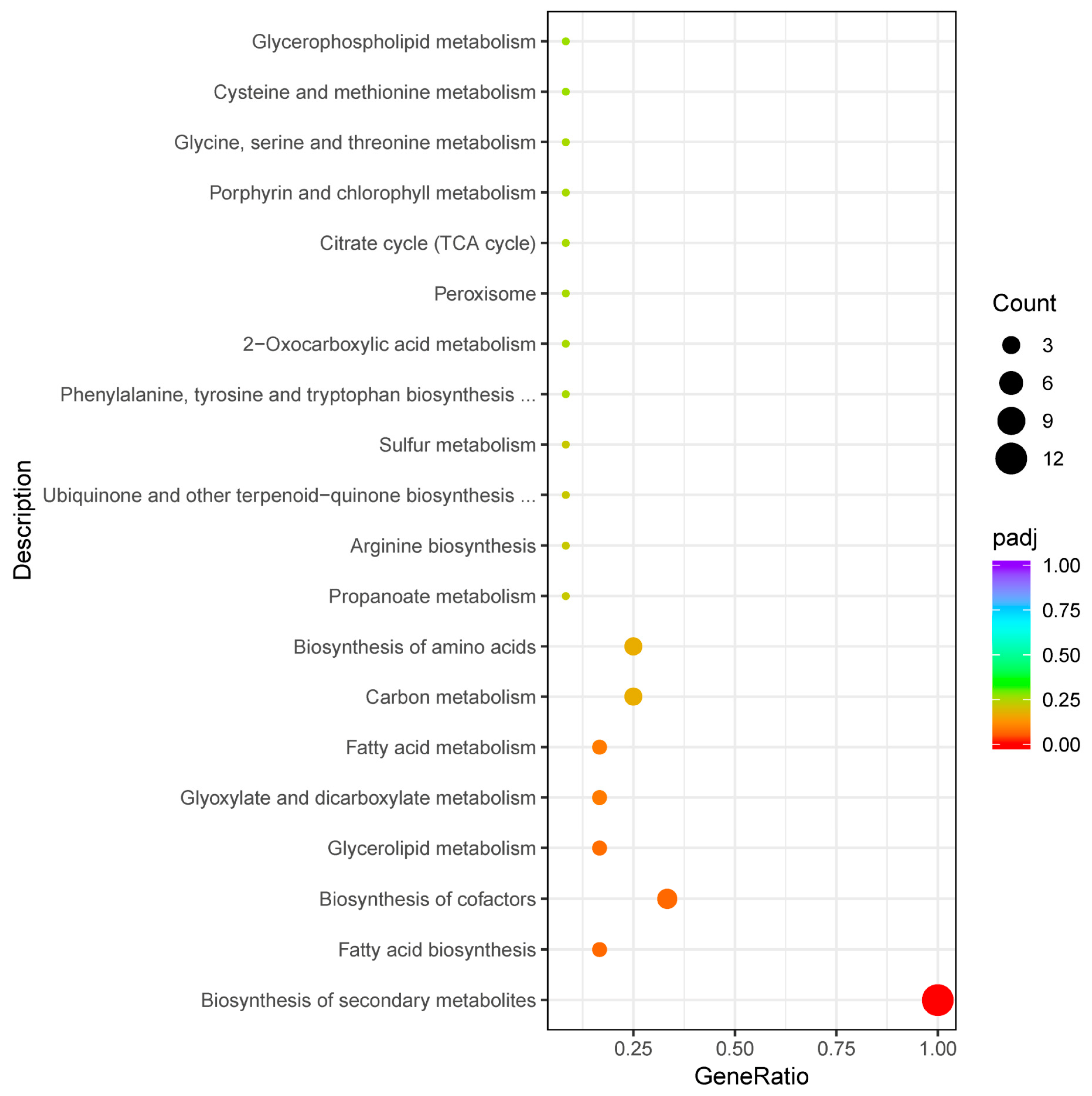
3.3. Module Gene Expression Pattern Analysis and Enrichment Pathway Analysis
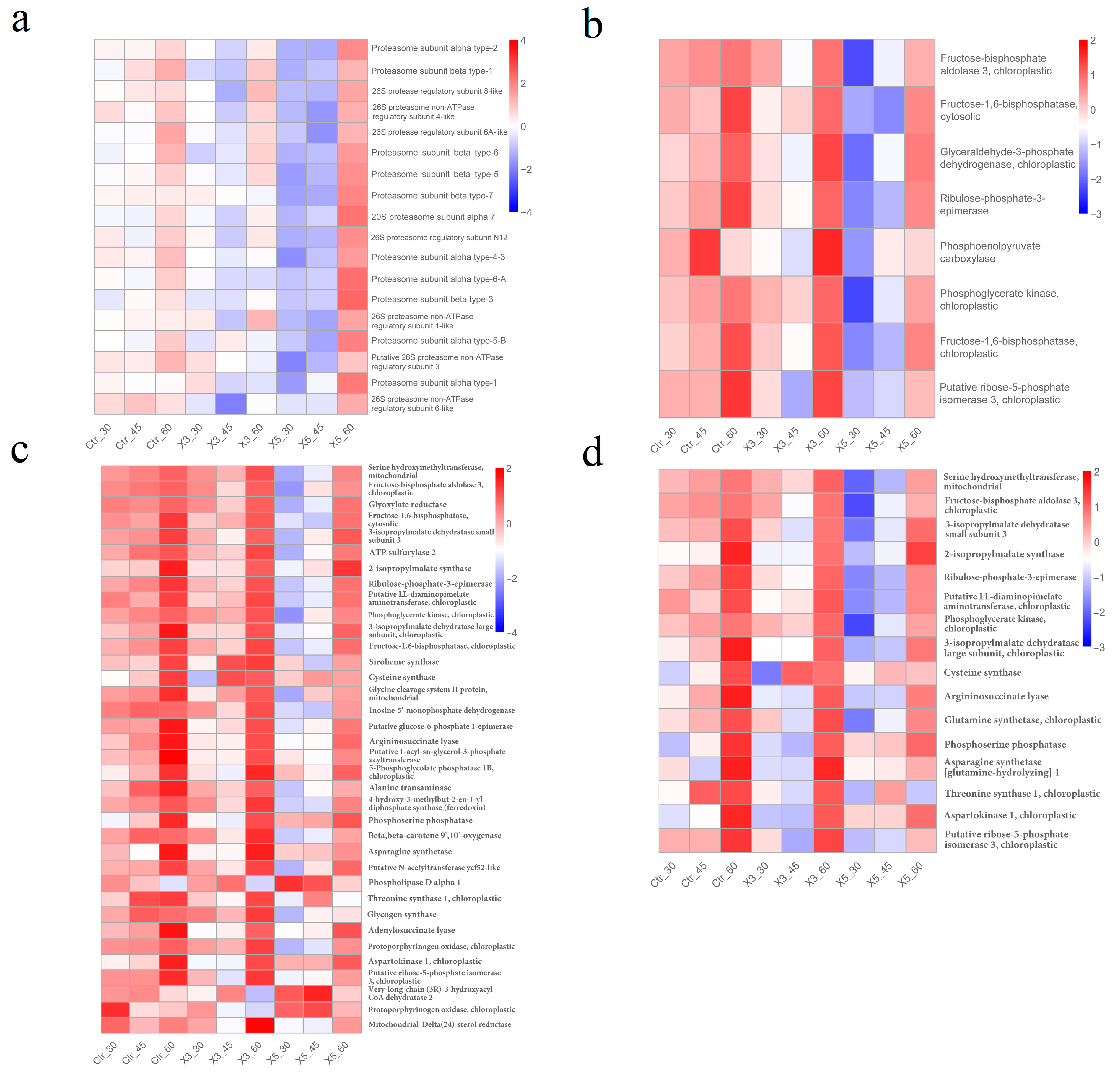
3.4. Expression Analysis of Genes in the Most Significant Enrichment Pathway Correlated with Salinity and Strain
3.5. Association Analysis of Genes and Traits
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, R.; Wang, Q.; Shen, H.; Yang, Y.; Liu, P.; Dong, Y. Environmental benefits of macroalgae products: A case study of agar based on life cycle assessment. Algal Res. 2024, 78, 103384. [Google Scholar] [CrossRef]
- Pang, T.; Lu, L.; Xue, J.; Xin, X.; Liu, J. A new mode of cultivating Gracilariopsis lemaneiformis for saving environmental and economic costs: Keeping a full stand of vegetative frond cuttings in northern China during the winter. Aquaculture 2023, 571, 739459. [Google Scholar] [CrossRef]
- Kang, Y.; Li, H.; Wu, J.; Xu, X.T.; Sun, X.; Zhao, X.D.; Xu, N.J. Transcriptome profiling reveals the antitumor mechanism of polysaccharide from marine algae Gracilariopsis lemaneiformis. PLoS ONE 2016, 11, e0158279. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Liao, X.; Zou, D.; Huang, B.; Liu, Z. The regulations of varied carbon-nitrogen supplies to physiology and amino acid contents in Gracilariopsis lemaneiformis (Gracilariales, Rhodophyta). Algal Res. 2020, 47, 101818. [Google Scholar] [CrossRef]
- Xiao, B.H.; Hu, Y.Y.; Feng, X.Q.; Sui, Z.H. Breeding of new strains of Gracilariopsis lemaneiformis with high agar content by ARTP mutagenesis and high osmotic pressure screening. Mar. Biotechnol. 2023, 25, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.A.; Sarkhosh, A.; Khan, N.; Balal, R.M.; Ali, S.; Rossi, L.; Gómez, C.; Mattson, N.; Nasim, W.; Garcia-Sanchez, F. Insights into the physiological and biochemical impacts of salt stress on plant growth and development. Agronomy 2020, 10, 938. [Google Scholar] [CrossRef]
- Ahluwalia, O.; Singh, P.C.; Bhatia, R. A review on drought stress in plants: Implications, mitigation and the role of plant growth promoting rhizobacteria. Resources. Environ. Sustain. 2021, 5, 100032. [Google Scholar] [CrossRef]
- Wani, K.I.; Naeem, M.; Castroverde, C.D.M.; Kalaji, H.M.; Albaqami, M.; Aftab, T. Molecular mechanisms of nitric oxide (NO) signaling and reactive oxygen species (ROS) homeostasis during abiotic stresses in plants. Int. J. Mol. Sci. 2021, 22, 9656. [Google Scholar] [CrossRef]
- Wang, L.; Du, M.; Wang, B.; Duan, H.; Zhang, B.; Wang, D.; Li, Y.; Wang, J. Transcriptome analysis of halophyte Nitraria tangutorum reveals multiple mechanisms to enhance salt resistance. Sci. Rep. 2022, 12, 14031. [Google Scholar] [CrossRef]
- Kaur, M.; Saini, K.C.; Ojah, H.; Sahoo, R.; Gupta, K.; Kumar, A.; Bast, F. Abiotic stress in algae: Response, signaling and transgenic approaches. J. Appl. Phycol. 2022, 34, 1843–1869. [Google Scholar] [CrossRef]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Khan, M.A.R. Understanding the roles of osmolytes for acclimatizing plants to changing environment: A review of potential mechanism. Plant Signal. Behav. 2021, 16, 1913306. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.A.; Thangavel, P.; Rasheed, F.; Masood, A.; Pirasteh-Anosheh, H.; Khan, N.A. Osmolytes: Efficient Oxidative Stress-Busters in Plants. In Global Climate Change and Plant Stress Management; Wiley: New York, NY, USA, 2023; pp. 399–409. [Google Scholar] [CrossRef]
- Khan, S.; Siraj, S.; Shahid, M.; Haque, M.M.; Islam, A. Osmolytes: Wonder molecules to combat protein misfolding against stress conditions. Int. J. Biol. Macromol. 2023, 234, 123662. [Google Scholar] [CrossRef]
- Ren, L.; Wang, M.R.; Wang, Q.C. ROS-induced oxidative stress in plant cryopreservation: Occurrence and alleviation. Planta 2021, 254, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Ali, A.; Kour, N.; Bornhorst, J.; AlHarbi, K.; Rinklebe, J.; Moneim, D.A.E.; Ahmad, P.; Chung, Y.S. Heavy metal induced oxidative stress mitigation and ROS scavenging in plants. Plants 2023, 12, 3003. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, N.N.; Vetoshkina, D.V.; Marenkova, T.V.; Borisova-Mubarakshina, M.M. Antioxidants of Non-Enzymatic Nature: Their Function in Higher Plant Cells and the Ways of Boosting Their Biosynthesis. Antioxidants 2023, 12, 2014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gao, Y.; Zhang, L.; Zhou, Y. The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [CrossRef] [PubMed]
- Ali, O.; Cheddadi, I.; Landrein, B.; Long, Y. Revisiting the relationship between turgor pressure and plant cell growth. New Phytol. 2023, 238, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lu, S.; Wang, J.; Chen, Z.; Zhang, Y.; Duan, J.; Liu, P.; Wang, X.Y.; Guo, J. Responses of physiological, morphological and anatomical traits to abiotic stress in woody plants. Forests 2023, 14, 1784. [Google Scholar] [CrossRef]
- Provasoli, L. Media and prospects for the cultivation of marine algae. In Cultures and Collections of Algae, Proceedings of the US Japan Conference, Hakonee, Japan, 12–15 September 1966; Japanese Society of Plant Physiology: Kyoto, Japan, 1968; pp. 63–75. [Google Scholar]
- Berges, J.A.; Franklin, D.J.; Harrison, P.J. Evolution of an artificial seawater medium: Improvements in enriched seawater, artificial water over the last two decades. J. Phycol. 2001, 37, 1138–1145. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Sun, G.; Shi, Z.; Ni, H.; Jiang, S. Identification and validation of key modules and hub genes associated with the pathological stage of oral squamous cell carcinoma by weighted gene co-expression network analysis. PeerJ 2020, 8, e8505. [Google Scholar] [CrossRef] [PubMed]
- Sahu, I.; Glickman, M.H. Proteasome in action: Substrate degradation by the 26S proteasome. Biochem. Soc. Trans. 2021, 49, 629–644. [Google Scholar] [CrossRef] [PubMed]
- De Porcellinis, A.J.; Nørgaard, H.; Brey, L.M.F.; Erstad, S.M.; Jones, P.R.; Heazlewood, J.L.; Sakuragi, Y. Overexpression of bifunctional fructose-1, 6-bisphosphatase/sedoheptulose-1, 7-bisphosphatase leads to enhanced photosynthesis and global reprogramming of carbon metabolism in Synechococcus sp. PCC 7002. Metab. Eng. 2018, 47, 170–183. [Google Scholar] [CrossRef]
- Rojas-Pirela, M.; Andrade-Alviárez, D.; Rojas, V.; Kemmerling, U.; Cáceres, A.J.; Michels, P.A.; Concepción, J.L.; Quiñones, W. Phosphoglycerate kinase: Structural aspects and functions, with special emphasis on the enzyme from Kinetoplastea. Open Biol. 2020, 10, 200302. [Google Scholar] [CrossRef] [PubMed]
- Zhan, N.; Huang, L.; Wang, Z.; Xie, Y.; Shang, X.; Liu, G.; Wu, Z. Comparative transcriptomics and bioinformatics analysis of genes related to photosynthesis in Eucalyptus camaldulensis. PeerJ 2022, 10, e14351. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wu, P.; Guo, A.; Liu, L. Myristic acid regulates triglyceride production in bovine mammary epithelial cells through the ubiquitination pathway. Agriculture 2023, 13, 1870. [Google Scholar] [CrossRef]
- Adams, N.B.; Bisson, C.; Brindley, A.A.; Farmer, D.A.; Davison, P.A.; Reid, J.D.; Hunter, C.N. The active site of magnesium chelatase. Nat. Plants 2020, 6, 1491–1502. [Google Scholar] [CrossRef]
- Herrmann, J.M.; Riemer, J. Apoptosis inducing factor and mitochondrial NADH dehydrogenases: Redox-controlled gear boxes to switch between mitochondrial biogenesis and cell death. Biol. Chem. 2021, 402, 289–297. [Google Scholar] [CrossRef]
- Mnatsakanyan, N.; Jonas, E.A. ATP synthase c-subunit ring as the channel of mitochondrial permeability transition: Regulator of metabolism in development and degeneration. J. Mol. Cell. Cardiol. 2020, 144, 109–118. [Google Scholar] [CrossRef]
- Prasad, M.S.; Bhole, R.P.; Khedekar, P.B.; Chikhale, R.V. Mycobacterium enoyl acyl carrier protein reductase (InhA): A key target for antitubercular drug discovery. Bioorg. Chem. 2021, 115, 105242. [Google Scholar] [CrossRef] [PubMed]
- Saini, A.; Kumar, A.; Singh, G.; Giri, S.K. Survival strategies and stress adaptations in halophilic Archaebacteria. In Microbial Stress Response: Mechanisms and Data Science; American Chemical Society: Washington, DC, USA, 2023; pp. 1–21. [Google Scholar] [CrossRef]
- Jiang, X.; Yang, L.; Wang, Y.; Jiang, F.; Lai, J.; Pan, K. Proteomics Provide Insight into the Interaction between Selenite and the Microalgae Dunaliella salina. Processes 2023, 11, 563. [Google Scholar] [CrossRef]
- Mergner, J.; Kuster, B. Plant proteome dynamics. Annu. Rev. Plant Biol. 2022, 73, 67–92. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant abiotic stress proteomics: The major factors determining alterations in cellular proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Terzi, H.; Yıldız, M. Alterations in the root proteomes of Brassica napus cultivars under salt stress. Bot. Serbica 2021, 45, 87–96. [Google Scholar] [CrossRef]
- Huang, L.Y.; Li, Z.Z.; Liu, Q.; Pu, G.B.; Zhang, Y.Q.; Li, J. Research on the adaptive mechanism of photosynthetic apparatus under salt stress: New directions to increase crop yield in saline soils. Ann. Appl. Biol. 2019, 175, 1–17. [Google Scholar] [CrossRef]
- Shetty, P.; Gitau, M.M.; Maróti, G. Salinity stress responses and adaptation mechanisms in eukaryotic green microalgae. Cells 2019, 8, 1657. [Google Scholar] [CrossRef] [PubMed]
- Zahra, N.; Al Hinai, M.S.; Hafeez, M.B.; Rehman, A.; Wahid, A.; Siddique, K.H.; Farooq, M. Regulation of photosynthesis under salt stress and associated tolerance mechanisms. Plant Physiol. Biochem. 2022, 178, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Ahmed, M.Z.; Hussain, T.; Aziz, I.; Ahmad, N.; Gul, B.; Nielsen, B.L. Effects of salinity stress on chloroplast structure and function. Cells 2021, 10, 2023. [Google Scholar] [CrossRef]
- Gambichler, V.; Zuccarello, G.C.; Karsten, U.; Gambichler, V.; Zuccarello, G.C.; Karsten, U. Physiological responses to salt stress by native and introduced red algae in New Zealand. Algae 2021, 36, 137–146. [Google Scholar] [CrossRef]
- Yi, Z.; Su, Y.; Brynjolfsson, S.; Olafsdóttir, K.; Fu, W. Bioactive polysaccharides and their derivatives from microalgae: Biosynthesis, applications, and challenges. Stud. Nat. Prod. Chem. 2021, 71, 67–85. [Google Scholar] [CrossRef]
- Zhang, Q.; Tian, S.; Chen, G.; Tang, Q.; Zhang, Y.; Fleming, A.J.; Zhu, X.G.; Wang, P. A regulatory circuit involving the NADH dehydrogenase-like complex balances C4 photosynthetic carbon flow and cellular redox in maize. bioRxiv 2023. [Google Scholar] [CrossRef]
- Xia, L.; Kong, X.; Song, H.; Han, Q.; Zhang, S. Advances in proteome-wide analysis of plant lysine acetylation. Plant Commun. 2022, 3, 100266. [Google Scholar] [CrossRef]
- Ali, Q.; Malik, A. Genetic response of growth phases for abiotic environmental stress tolerance in cereal crop plants. Genetika 2021, 53, 419–456. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Zhang, D.; Zou, D.; Wang, J.; Zheng, H.; Jia, Y.; Qu, Z.J.; Sun, B.; Zhao, H. Photosynthetic carbon fixation and sucrose metabolism supplemented by weighted gene co-expression network analysis in response to water stress in rice with overlapping growth stages. Front. Plant Sci. 2022, 13, 864605. [Google Scholar] [CrossRef] [PubMed]
- Haeder, D.P. Photosynthesis in Plants and Algae. Anticancer Res. 2022, 42, 5035–5041. [Google Scholar] [CrossRef]
- Obi, C.D.; Bhuiyan, T.; Dailey, H.A.; Medlock, A.E. Ferrochelatase: Mapping the intersection of iron and porphyrin metabolism in the mitochondria. Front. Cell Dev. Biol. 2022, 10, 961. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Duan, N.; Chen, X.; Li, X.; Zhao, N.; Cao, W.; Li, H.Q.; Liu, B.; Tan, F.S.; Zhao, X.L.; et al. Transcriptome Profiling and Chlorophyll Metabolic Pathway Analysis Reveal the Response of Nitraria tangutorum to Increased Nitrogen. Plants 2023, 12, 895. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.S.; Wang, Y.L.; Wang, X.; Liu, L. Hexameric structure of the ATPase motor subunit of magnesium chelatase in chlorophyll biosynthesis. Protein Sci. 2020, 29, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Shi, J.C.; Wang, D.J.; Liang, X.; Wei, F.; Gong, C.M.; Qiu, L.J.; Zhou, H.C.; Folta, K.M.; Wen, Y.Q.; et al. A point mutation in the gene encoding magnesium chelatase I subunit influences strawberry leaf color and metabolism. Plant Physiol. 2023, 192, 2737–2755. [Google Scholar] [CrossRef]
- Wilson, D.F.; Matschinsky, F.M. Metabolic homeostasis: Oxidative phosphorylation and the metabolic requirements of higher plants and animals. J. Appl. Physiol. 2018, 125, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Wang, R.S.; Handy, D.E.; Loscalzo, J. NAD (H) and NADP (H) redox couples and cellular energy metabolism. Antioxid. Redox Signal. 2018, 28, 251–272. [Google Scholar] [CrossRef] [PubMed]
- Koike, S.; Jahn, R. SNARE proteins: Zip codes in vesicle targeting? Biochem. J. 2022, 479, 273–288. [Google Scholar] [CrossRef] [PubMed]



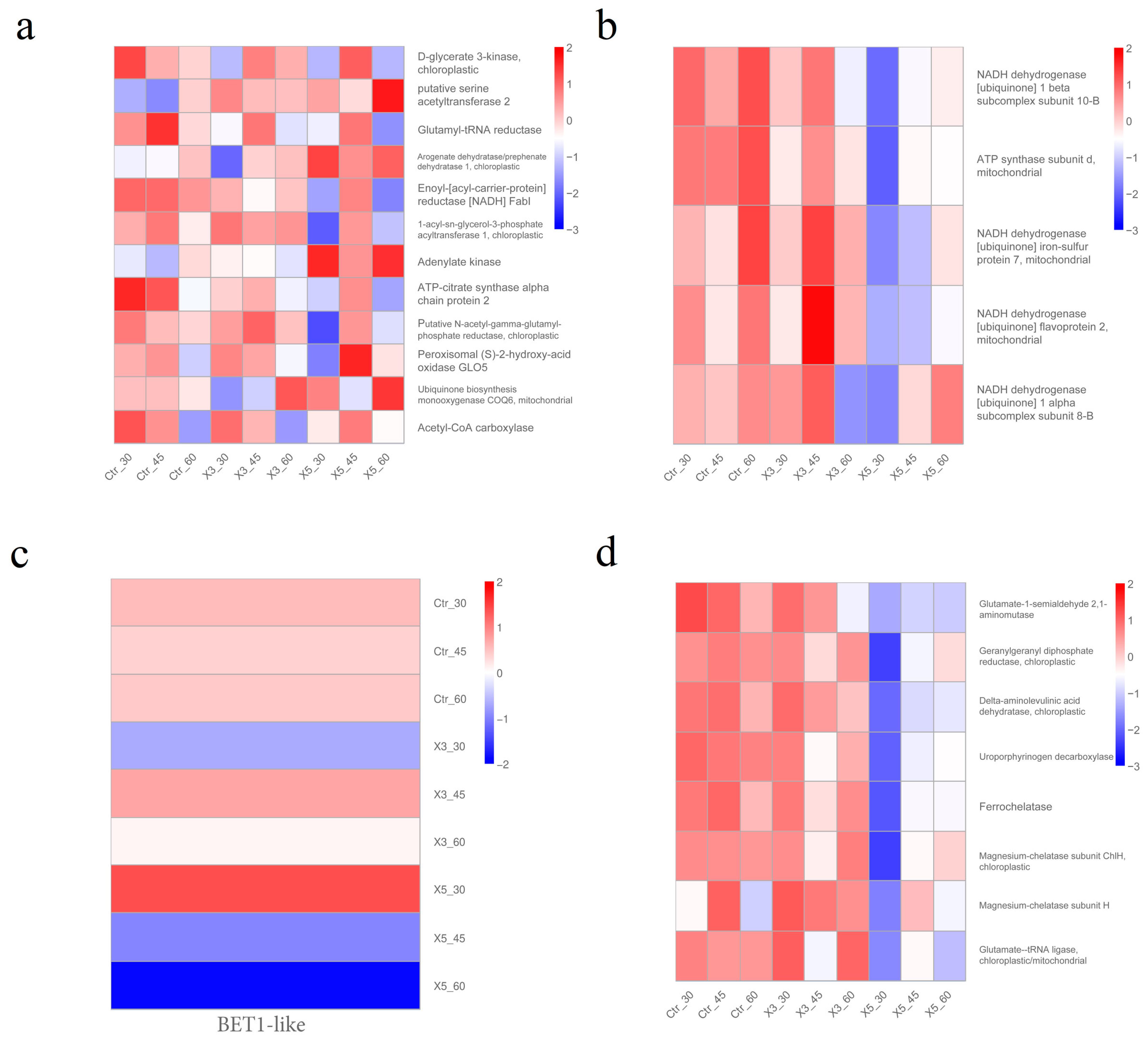

| Gene_Id | Characteristic | Correlation | p-Value | Metabolic Pathway | Function |
|---|---|---|---|---|---|
| LXC000032 | Salinity | 0.7282 | 0.0261 | Carbon fixation in photosynthetic organisms | Fructose-1,6-bisphosphatase, cytosolic [Gracilariopsis chorda] |
| LXC001520 | Salinity | 0.8079 | 0.0084 | Proteasome | Proteasome subunit β type-1 [G. chorda] |
| LXC007344 | Salinity | 0.6991 | 0.0361 | Proteasome | Proteasome subunit β type-5 [G. chorda] |
| LXC003712 | Salinity | 0.7958 | 0.0103 | Carbon fixation in photosynthetic organisms | Glyceraldehyde-3-phosphate dehydrogenase, chloroplastic [G. chorda] |
| LXC002690 | Salinity | 0.7717 | 0.0148 | Carbon fixation in photosynthetic organisms | hypothetical Ribulose-phosphate 3-epimerase [G. chorda] |
| LXC003646 | Salinity | 0.7152 | 0.0303 | Carbon fixation in photosynthetic organisms | Phosphoglycerate kinase, chloroplastic [G. chorda] |
| LXC004308 | Salinity | 0.7890 | 0.0114 | Carbon fixation in photosynthetic organisms | Fructose-1,6-bisphosphatase, chloroplastic [G. chorda] |
| Gene_Id | Characteristic | Correlation | p-Value | Metabolic Pathway | Function |
|---|---|---|---|---|---|
| LXC007989 | Strains | −0.79982 | 0.009656 | Oxidative phosphorylation | NADH dehydrogenase [ubiquinone] 1 β subcomplex subunit 10-B [Gracilariopsis chorda] |
| LXC000548 | Strains | −0.87707 | 0.001898 | Oxidative phosphorylation | ATP synthase subunit d, mitochondrial [G. chorda] |
| LXC002968 | Strains | −0.79724 | 0.010072 | Porphyrin and chlorophyll metabolism | Glutamate-1-semialdehyde 2,1-aminomutase [G. chorda] |
| LXC000696 | Strains | −0.85331 | 0.003438 | Porphyrin and chlorophyll metabolism | Geranylgeranyl diphosphate reductase, chloroplastic [G. chorda] |
| LXC005913 | Strains | −0.80719 | 0.008533 | Porphyrin and chlorophyll metabolism | Delta-aminolevulinic acid dehydratase, chloroplastic [G. chorda] |
| LXC003053 | Strains | −0.88065 | 0.001717 | Porphyrin and chlorophyll metabolism | Uroporphyrinogen decarboxylase [G. chorda] |
| LXC005216 | Strains | −0.77977 | 0.013206 | Porphyrin and chlorophyll metabolism | Ferrochelatase [G. chorda] |
| LXC001791 | Strains | −0.72984 | 0.025602 | Porphyrin and chlorophyll metabolism | Magnesium–chelatase subunit ChlH, chloroplastic [G. chorda] |
| LXC005122 | Strains | −0.71535 | 0.030264 | Fatty acid biosynthesis | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI [G. chorda] |
| LXC004426 | Strains | −0.68286 | 0.042645 | Porphyrin and chlorophyll metabolism | Glutamate–tRNA ligase, chloroplastic/mitochondrial [G. chorda] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, B.; Feng, X.; Li, P.; Sui, Z. Analysis of Hyperosmotic Tolerance Mechanisms in Gracilariopsis lemaneiformis Based on Weighted Co-Expression Network Analysis. Genes 2024, 15, 781. https://doi.org/10.3390/genes15060781
Xiao B, Feng X, Li P, Sui Z. Analysis of Hyperosmotic Tolerance Mechanisms in Gracilariopsis lemaneiformis Based on Weighted Co-Expression Network Analysis. Genes. 2024; 15(6):781. https://doi.org/10.3390/genes15060781
Chicago/Turabian StyleXiao, Baoheng, Xiaoqing Feng, Pingping Li, and Zhenghong Sui. 2024. "Analysis of Hyperosmotic Tolerance Mechanisms in Gracilariopsis lemaneiformis Based on Weighted Co-Expression Network Analysis" Genes 15, no. 6: 781. https://doi.org/10.3390/genes15060781
APA StyleXiao, B., Feng, X., Li, P., & Sui, Z. (2024). Analysis of Hyperosmotic Tolerance Mechanisms in Gracilariopsis lemaneiformis Based on Weighted Co-Expression Network Analysis. Genes, 15(6), 781. https://doi.org/10.3390/genes15060781






