Associations between Radiomics and Genomics in Non-Small Cell Lung Cancer Utilizing Computed Tomography and Next-Generation Sequencing: An Exploratory Study
Abstract
:1. Introduction
2. Patients and Methods
2.1. Study Design and Patients’ Selection Criteria
2.2. Patients Management and Follow-Up
2.3. Genetic Assessment
2.4. Feature Extraction
2.5. Feature/Gene Association Study
2.6. Machine Learning
2.7. Sample Size, Statistical Analysis, and Data Presentation
3. Results
3.1. Clinicopathological Characteristics of the Analyzed Cohort
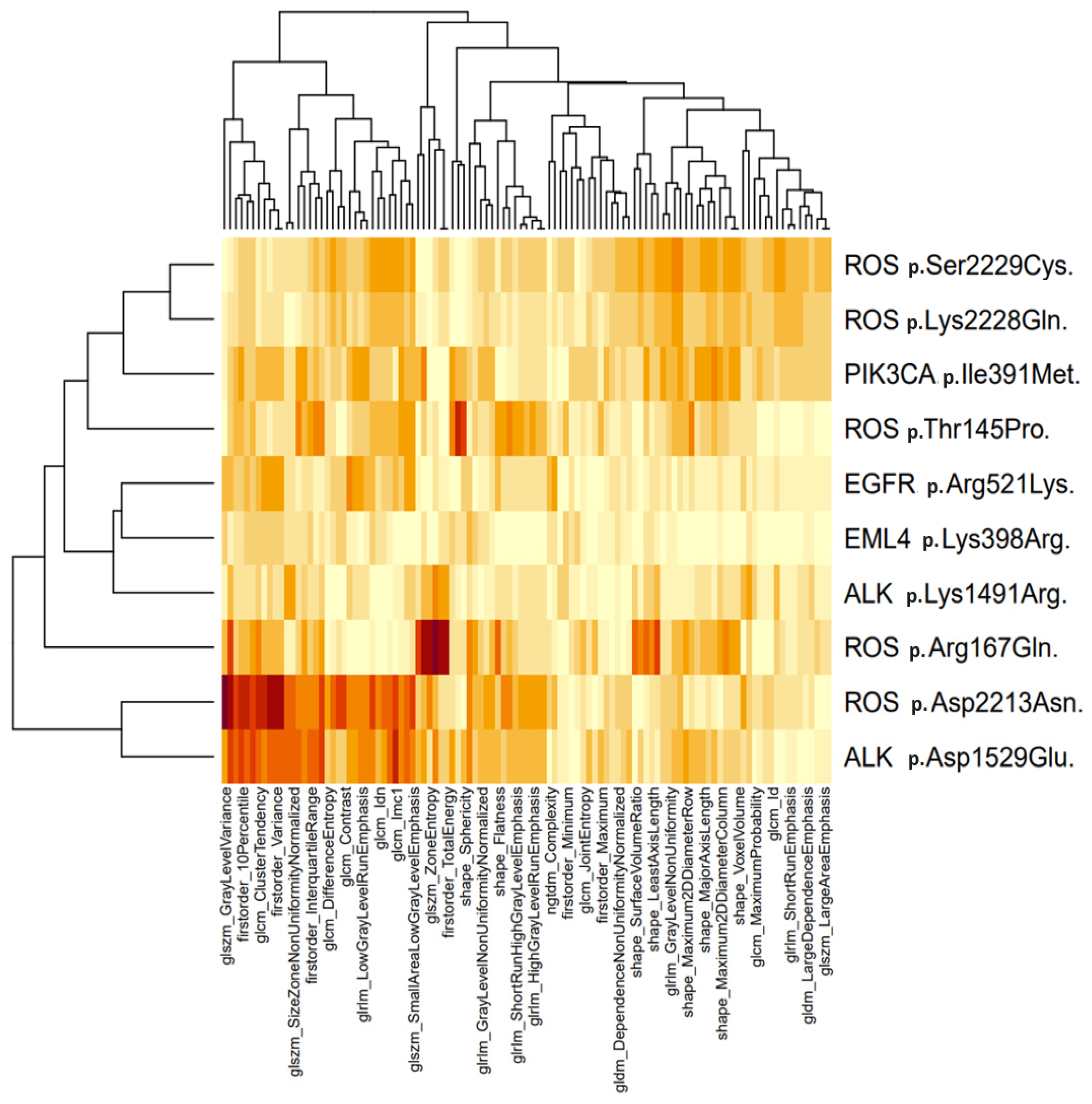
3.2. Molecular and Radiomic Profiling
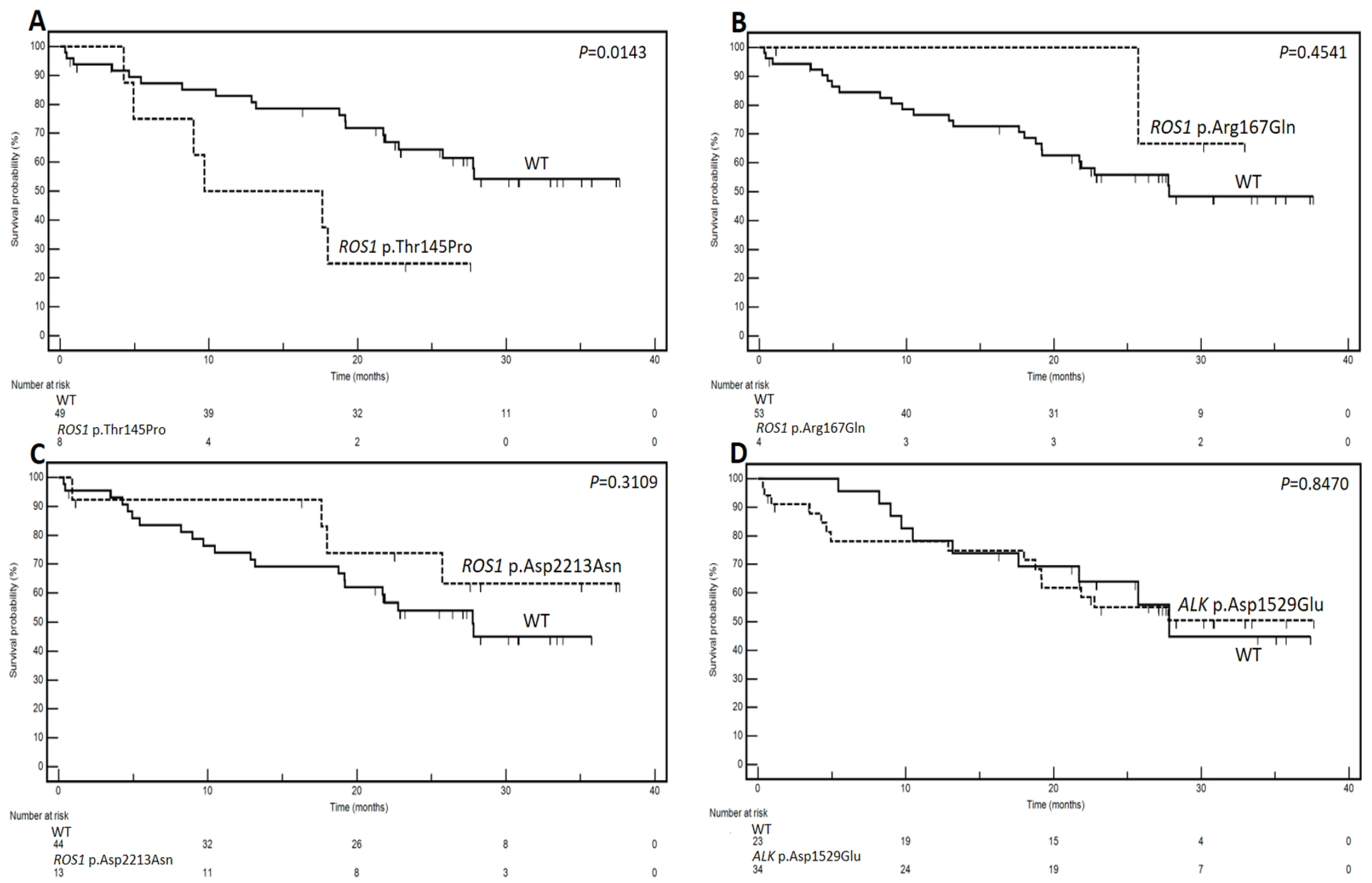
3.3. Genetic Variants and Prognostic Analysis
4. Discussion
5. Study Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef]
- Mithoowani, H.; Febbraro, M. Non-Small-Cell Lung Cancer in 2022: A Review for General Practitioners in Oncology. Curr. Oncol. 2022, 29, 1828–1839. [Google Scholar] [CrossRef]
- Jonna, S.; Subramaniam, D.S. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): An update. Discov. Med. 2019, 27, 167–170. [Google Scholar]
- Pons-Tostivint, E.; Bennouna, J. Treatments for Non-Small-Cell Lung Cancer: The Multiple Options for Precision Medicine. Curr. Oncol. 2022, 29, 7106–7108. [Google Scholar] [CrossRef]
- Nagano, T.; Tachihara, M.; Nishimura, Y. Molecular Mechanisms and Targeted Therapies Including Immunotherapy for Non-Small Cell Lung Cancer. Curr. Cancer Drug Targets 2019, 19, 595–630. [Google Scholar] [CrossRef]
- Qian, L.; Wu, T.; Kong, S.; Lou, X.; Jiang, Y.; Tan, Z.; Wu, L.; Gao, C. Could the underlying biological basis of prognostic radiomics and deep learning signatures be explored in patients with lung cancer? A systematic review. Eur. J. Radiol. 2024, 171, 111314. [Google Scholar] [CrossRef]
- Hatt, M.; Cheze Le Rest, C.; Antonorsi, N.; Tixier, F.; Tankyevych, O.; Jaouen, V.; Lucia, F.; Bourbonne, V.; Schick, U.; Badic, B.; et al. Radiomics in PET/CT: Current Status and Future AI-Based Evolutions. Semin. Nucl. Med. 2021, 51, 126–133. [Google Scholar] [CrossRef]
- Guiot, J.; Vaidyanathan, A.; Deprez, L.; Zerka, F.; Danthine, D.; Frix, A.N.; Lambin, P.; Bottari, F.; Tsoutzidis, N.; Miraglio, B.; et al. A review in radiomics: Making personalized medicine a reality via routine imaging. Med. Res. Rev. 2022, 42, 426–440. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; Sansone, M.; Grassi, R.; Maio, F.; Palaia, R.; Tatangelo, F.; Botti, G.; Grimm, R.; Curley, S.; et al. Magnetic resonance imaging in the assessment of pancreatic cancer with quantitative parameter extraction by means of dynamic contrast-enhanced magnetic resonance imaging, diffusion kurtosis imaging and intravoxel incoherent motion diffusion-weighted imaging. Ther. Adv. Gastroenterol. 2020, 13, 1756284819885052. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; Barretta, M.L.; Picone, C.; Avallone, A.; Belli, A.; Patrone, R.; Ferrante, M.; Cozzi, D.; Grassi, R.; et al. Radiomics in hepatic metastasis by colorectal cancer. Infect. Agents Cancer 2021, 16, 39. [Google Scholar] [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Chen, M.; Copley, S.J.; Viola, P.; Lu, H.; Aboagye, E.O. Radiomics and artificial intelligence for precision medicine in lung cancer treatment. Semin. Cancer Biol. 2023, 93, 97–113. [Google Scholar] [CrossRef]
- Qi, Y.; Zhao, T.; Han, M. The application of radiomics in predicting gene mutations in cancer. Eur. Radiol. 2022, 32, 4014–4024. [Google Scholar] [CrossRef]
- Remon, J.; Soria, J.C.; Peters, S.; ESMO Guidelines Committee. Early and locally advanced non-small-cell lung cancer: An update of the ESMO Clinical Practice Guidelines focusing on diagnosis, staging, systemic and local therapy. Ann. Oncol. 2021, 32, 1637–1642. [Google Scholar] [CrossRef]
- Hendriks, L.E.; Kerr, K.M.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 339–357. [Google Scholar] [CrossRef]
- Hendriks, L.E.; Kerr, K.M.; Menis, J.; Mok, T.S.; Nestle, U.; Passaro, A.; Peters, S.; Planchard, D.; Smit, E.F.; Solomon, B.J.; et al. Non-oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023, 34, 358–376. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Huang, S.; Cai, N.; Pacheco, P.P.; Narrandes, S.; Wang, Y.; Xu, W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genom. Proteom. 2018, 15, 41–51. [Google Scholar] [CrossRef]
- Shur, J.D.; Doran, S.J.; Kumar, S.; Ap Dafydd, D.; Downey, K.; O’Connor, J.P.B.; Papanikolaou, N.; Messiou, C.; Koh, D.M.; Orton, M.R. Radiomics in Oncology: A Practical Guide. Radiographics 2021, 41, 1717–1732. [Google Scholar] [CrossRef]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P.; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. J. Clin. Epidemiol. 2008, 61, 344–349. [Google Scholar] [CrossRef]
- Montella, M.; Ciani, G.; Granata, V.; Fusco, R.; Grassi, F.; Ronchi, A.; Cozzolino, I.; Franco, R.; Zito Marino, F.; Urraro, F.; et al. Preliminary Experience of Liquid Biopsy in Lung Cancer Compared to Conventional Assessment: Light and Shadows. J. Pers. Med. 2022, 12, 1896. [Google Scholar] [CrossRef] [PubMed]
- Padinharayil, H.; Varghese, J.; John, M.C.; Rajanikant, G.K.; Wilson, C.M.; Al-Yozbaki, M.; Renu, K.; Dewanjee, S.; Sanyal, R.; Dey, A.; et al. Non-small cell lung carcinoma (NSCLC): Implications on molecular pathology and advances in early diagnostics and therapeutics. Genes Dis. 2022, 10, 960–989. [Google Scholar] [CrossRef] [PubMed]
- Araghi, M.; Mannani, R.; Heidarnejad Maleki, A.; Hamidi, A.; Rostami, S.; Safa, S.H.; Faramarzi, F.; Khorasani, S.; Alimohammadi, M.; Tahmasebi, S.; et al. Recent advances in non-small cell lung cancer targeted therapy; an update review. Cancer Cell Int. 2023, 23, 162. [Google Scholar] [CrossRef]
- Wang, M.; Herbst, R.S.; Boshoff, C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat. Med. 2021, 27, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Abughanimeh, O.; Kaur, A.; El Osta, B.; Ganti, A.K. Novel targeted therapies for advanced non-small lung cancer. Semin. Oncol. 2022, 49, 326–336. [Google Scholar] [CrossRef]
- Polcaro, G.; Liguori, L.; Manzo, V.; Chianese, A.; Donadio, G.; Caputo, A.; Scognamiglio, G.; Dell’Annunziata, F.; Langella, M.; Corbi, G.; et al. rs822336 binding to C/EBPβ and NFIC modulates induction of PD-L1 expression and predicts anti-PD-1/PD-L1 therapy in advanced NSCLC. Mol. Cancer 2024, 23, 63. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, B. A review of radiomics and genomics applications in cancers: The way towards precision medicine. Radiat Oncol. 2022, 17, 217. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Han, B. Liquid Biopsy Promotes Non-Small Cell Lung Cancer Precision Therapy. Technol. Cancer Res. Treat. 2018, 17, 1533033818801809. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Kirienko, M.; Sollini, M.; Corbetta, M.; Voulaz, E.; Gozzi, N.; Interlenghi, M.; Gallivanone, F.; Castiglioni, I.; Asselta, R.; Duga, S.; et al. Radiomics and gene expression profile to characterise the disease and predict outcome in patients with lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 3643–3655. [Google Scholar] [CrossRef]
- Perez-Johnston, R.; Araujo-Filho, J.A.; Connolly, J.G.; Caso, R.; Whiting, K.; Tan, K.S.; Zhou, J.; Gibbs, P.; Rekhtman, N.; Ginsberg, M.S.; et al. CT-based Radiogenomic Analysis of Clinical Stage I Lung Adenocarcinoma with Histopathologic Features and Oncologic Outcomes. Radiology 2022, 303, 664–672. [Google Scholar] [CrossRef]
- Rinaldi, L.; Guerini Rocco, E.; Spitaleri, G.; Raimondi, S.; Attili, I.; Ranghiero, A.; Cammarata, G.; Minotti, M.; Lo Presti, G.; De Piano, F.; et al. Association between Contrast-Enhanced Computed Tomography Radiomic Features, Genomic Alterations and Prognosis in Advanced Lung Adenocarcinoma Patients. Cancers 2023, 15, 4553. [Google Scholar] [CrossRef] [PubMed]
- Prencipe, B.; Delprete, C.; Garolla, E.; Corallo, F.; Gravina, M.; Natalicchio, M.I.; Buongiorno, D.; Bevilacqua, V.; Altini, N.; Brunetti, A. An Explainable Radiogenomic Framework to Predict Mutational Status of KRAS and EGFR in Lung Adenocarcinoma Patients. Bioengineering 2023, 10, 747. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Ma, J.; Zhang, S.; Li, J.; Dai, H.; Liang, S.; Yu, Y.; Li, W.; Wang, C. Radiogenomic System for Non-Invasive Identification of Multiple Actionable Mutations and PD-L1 Expression in Non-Small Cell Lung Cancer Based on CT Images. Cancers 2022, 14, 4823. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Jenkins, C.; Iyer, S.; Schoenfeld, A.; Keddy, C.; Davare, M.A. ROS1-dependent cancers—Biology, diagnostics and therapeutics. Nat. Rev. Clin. Oncol. 2021, 18, 35–55. [Google Scholar] [CrossRef]
- Saladino, G.; Gervasio, F.L. Modeling the effect of pathogenic mutations on the conformational landscape of protein kinases. Curr. Opin. Struct. Biol. 2016, 37, 108–114. [Google Scholar] [CrossRef]
| Characteristic | No. | % |
|---|---|---|
| Age | ||
| <70 | 31 | 54.4 |
| ≥70 | 26 | 45.6 |
| Gender | ||
| Male | 39 | 68.4 |
| Female | 18 | 31.6 |
| Hystotype | ||
| Adenocarcinoma | 42 | 73.7 |
| Squamous | 15 | 26.3 |
| PS ECOG | ||
| 0 | 32 | 56.1 |
| 1 | 18 | 31.6 |
| 2 | 7 | 12.3 |
| Smoking | ||
| Non smoker | 5 | 8.8 |
| Former smoker | 27 | 47.4 |
| Current smoker | 25 | 43.8 |
| Stage | ||
| I/II | 22 | 38.6 |
| III | 18 | 31.6 |
| IV | 17 | 29.8 |
| T | ||
| 1/2 | 33 | 57.9 |
| 3 | 8 | 14.0 |
| 4 | 11 | 19.3 |
| * Non-definable | 5 | 8.8 |
| N | ||
| 0 | 32 | 56.1 |
| 1 | 4 | 7.0 |
| 2 | 14 | 24.6 |
| 3 | 2 | 3.5 |
| * Non-definable | 5 | 8.8 |
| EGFR mutation status | ||
| Mutated | 4 | 7.1 |
| Wild-type | 53 | 92.9 |
| Type of front-line treatment | ||
| Surgery | 26 | 45.6 |
| Platinum-based CT | 10 | 17.5 |
| Active palliative treatment | 4 | 7.0 |
| CT and/or RT followed by surgery | 4 | 7.0 |
| Target therapy | 4 | 7.0 |
| ICI monotherapy | 3 | 5.3 |
| Non-platinum based CT | 3 | 5.3 |
| Surgery followed by CT and/or RT | 3 | 5.3 |
| Gene Variant | Dicothomization | Median Survivals (Months) | No. of Events/Patients | HR | 95% CI | p at Log-Rank Test |
|---|---|---|---|---|---|---|
| ROS1 p.Thr145Pro | Mutated vs. WT | 9.7 vs. NR | 6/8 vs. 19/49 | 5.35 | 1.39–20.48 | 0.0143 |
| ROS1 p.Arg167Gln | Mutated vs. WT | NR vs. 27.8 | 1/4 vs. 24/53 | 0.57 | 0.13–2.44 | 0.4541 |
| ROS1 p.Asp2213Asn | Mutated vs. WT | NR vs. 27.7 | 4/13 vs. 21/44 | 0.62 | 0.25–1.55 | 0.3109 |
| ALK p.Asp1529Glu | Mutated vs. WT | NR vs. 27.8 | 15/34 vs. 10/23 | 1.08 | 0.48–2.40 | 0.8470 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ottaiano, A.; Grassi, F.; Sirica, R.; Genito, E.; Ciani, G.; Patanè, V.; Monti, R.; Belfiore, M.P.; Urraro, F.; Santorsola, M.; et al. Associations between Radiomics and Genomics in Non-Small Cell Lung Cancer Utilizing Computed Tomography and Next-Generation Sequencing: An Exploratory Study. Genes 2024, 15, 803. https://doi.org/10.3390/genes15060803
Ottaiano A, Grassi F, Sirica R, Genito E, Ciani G, Patanè V, Monti R, Belfiore MP, Urraro F, Santorsola M, et al. Associations between Radiomics and Genomics in Non-Small Cell Lung Cancer Utilizing Computed Tomography and Next-Generation Sequencing: An Exploratory Study. Genes. 2024; 15(6):803. https://doi.org/10.3390/genes15060803
Chicago/Turabian StyleOttaiano, Alessandro, Francesca Grassi, Roberto Sirica, Emanuela Genito, Giovanni Ciani, Vittorio Patanè, Riccardo Monti, Maria Paola Belfiore, Fabrizio Urraro, Mariachiara Santorsola, and et al. 2024. "Associations between Radiomics and Genomics in Non-Small Cell Lung Cancer Utilizing Computed Tomography and Next-Generation Sequencing: An Exploratory Study" Genes 15, no. 6: 803. https://doi.org/10.3390/genes15060803






