A-to-I Editing Is Subtype-Specific in Non-Hodgkin Lymphomas
Abstract
:1. Introduction
2. Results
2.1. Workflow
2.2. A Number of Sites Display Differential Editing among Different Subtypes in NHL
2.3. More Sites Are Differentially Edited between NHL and Normal
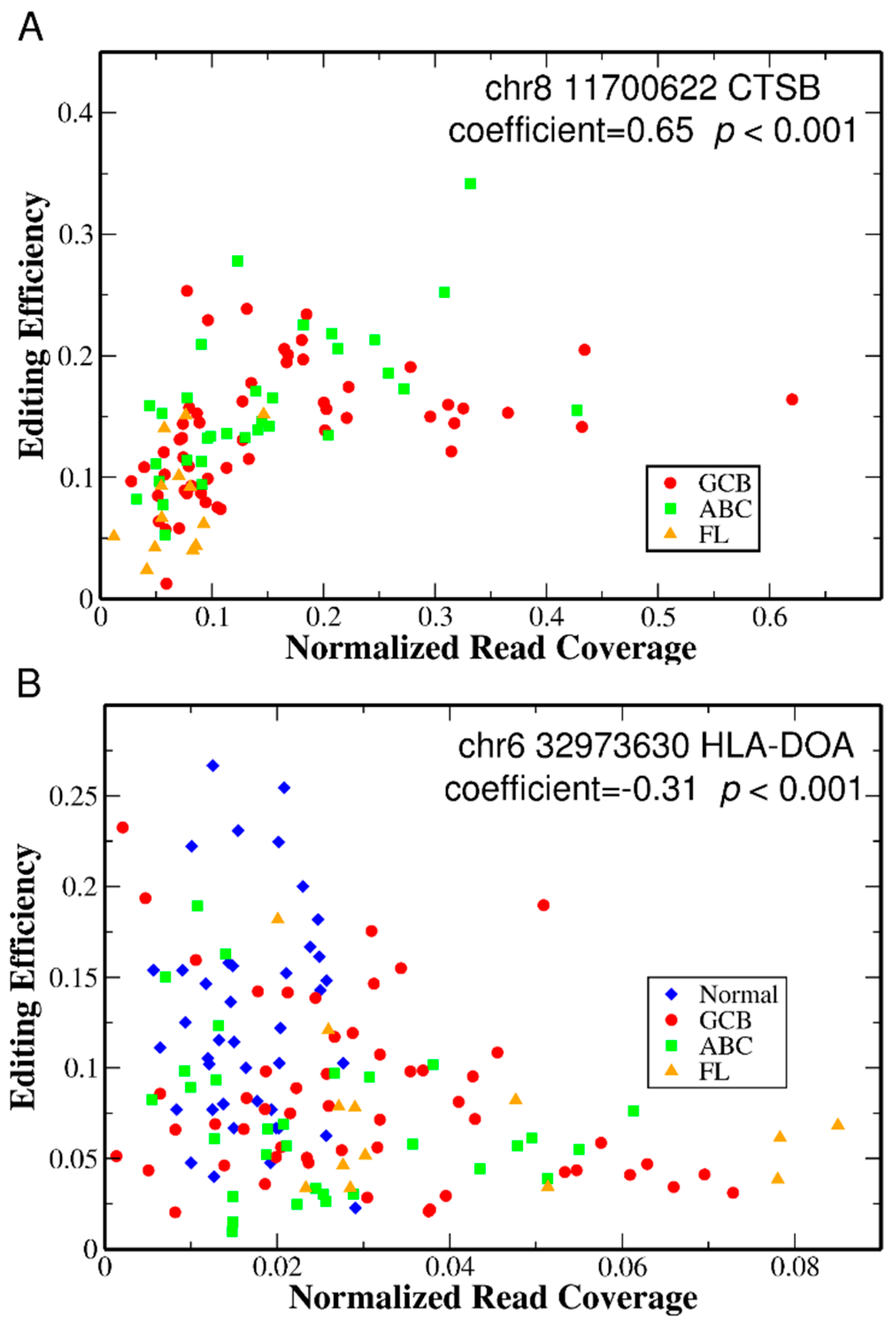
2.4. Gene Expression Is Highly Correlated with Editing Efficiency of Differentially Edited Sites in UTRs
2.5. The Clinical Status of Samples Is Predicted with High Accuracy Based on RNA Editing Profiles Alone
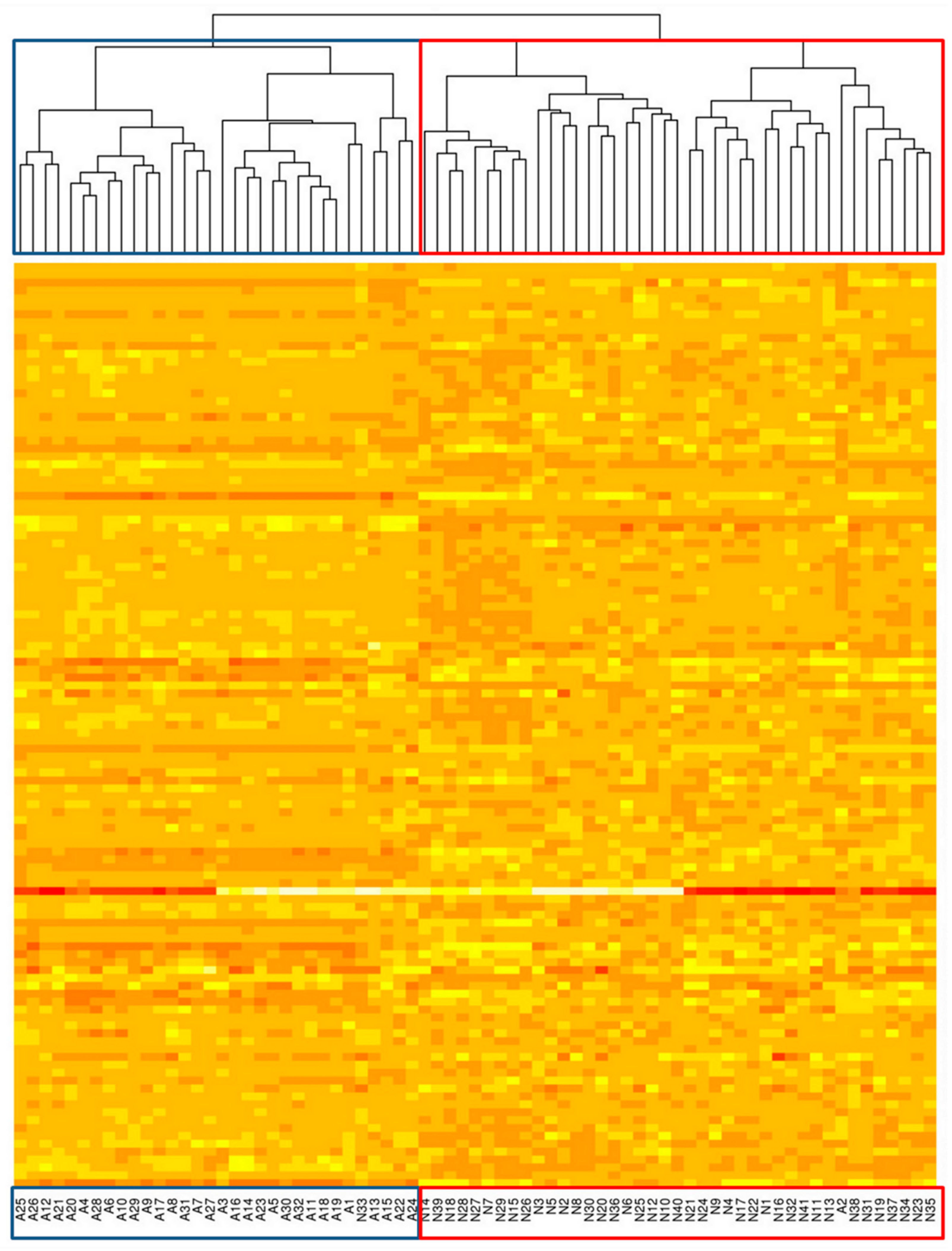
2.6. Unsupervised Clustering Can Differentiate NHL and Normal Samples but Not NHL Subtypes
3. Discussion and Conclusions
4. Materials and Methods
4.1. Mapping RNA-Seq Reads to the Reference
4.2. Filtering, Editing Site Selection, and Editing Efficiency
- We removed duplicate reads (defined as reads having the exact same sequence with their mate and mapping to the same position in the reference), and kept the read with the highest base quality;
- To ensure the mapping uniqueness of a read, we only counted reads with a mapping quality score of at least 10;
- We discarded a read if the editing position was within 2 bp of the 5′ or 3′ end;
- We only counted a read if the editing site of the read had a base quality score of at least 20.
4.3. Grouping Samples
4.4. Further Filtering of Editing Sites for Statistical Comparison
- (mAA + mAG + mGG)/M > 40% (a significant number of samples showed editing efficiencies between 0 and 0.1, 0.4 and 0.6, or 0.9 and 1, consistent with homozygotic or heterozygotic SNPs);
- At least two of the three conditions mAA/M > 5%, mAG/M > 5%, and mGG/M > 5% were satisfied (to ensure that there was variation between the configurations of an SNP in the sample population).
4.5. Statistical Comparison of Groups
4.6. Correlating Editing Efficiency with Gene Expression
4.7. Leave-One-Out cross Validation
- If read coverage was less than 10, we scored the site as “0”;
- If read coverage was at least 10 and editing efficiency was closer to the mean of Group I than to the mean of Group II, we scored the site as “1”;
- If read coverage was at least 10 and editing efficiency was closer to the mean of Group II than to the mean of Group I, we scored the site as “−1”;
4.8. Clustering
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gott, J.M.; Emeson, R.B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000, 34, 499–531. [Google Scholar] [CrossRef] [PubMed]
- Knoop, V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci. CMLS 2011, 68, 567–586. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002, 71, 817–846. [Google Scholar] [CrossRef] [PubMed]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef] [PubMed]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004, 2, e391. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.D.; Kim, T.T.; Walsh, T.; Kobayashi, Y.; Matise, T.C.; Buyske, S.; Gabriel, A. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004, 14, 1719–1725. [Google Scholar] [CrossRef] [PubMed]
- Blow, M.; Futreal, P.A.; Wooster, R.; Stratton, M.R. A survey of RNA editing in human brain. Genome Res. 2004, 14, 2379–2387. [Google Scholar] [CrossRef]
- Li, J.B.; Levanon, E.Y.; Yoon, J.K.; Aach, J.; Xie, B.; Leproust, E.; Zhang, K.; Gau, Y.; Church, G.M. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 2009, 324, 1210–1213. [Google Scholar] [CrossRef]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014, 24, 365–376. [Google Scholar] [CrossRef]
- Ramaswami, G.; Zhang, R.; Piskol, R.; Keegan, L.P.; Deng, P.; O’Connell, M.A.; Li, J.B. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods 2013, 10, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA editing in human tissues: Towards the inosinome atlas. Sci. Rep. 2015, 5, 14941. [Google Scholar] [CrossRef] [PubMed]
- Greenberger, S.; Levanon, E.Y.; Paz-Yaacov, N.; Barzilai, A.; Safran, M.; Osenberg, S.; Amariglio, N.; Rechavi, G.; Eisenberg, E. Consistent levels of A-to-I RNA editing across individuals in coding sequences and non-conserved Alu repeats. BMC Genom. 2010, 11, 608. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Locatelli, F. ADARs: Allies or enemies? The importance of A-to-I RNA editing in human disease: From cancer to HIV-1. Biol. Rev. Camb. Philos. Soc. 2012, 87, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.; Kawahara, Y.; Tamburro, K.M.; Nishikura, K. A-to-I RNA editing and human disease. RNA Biol. 2006, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Paz, N.; Levanon, E.Y.; Amariglio, N.; Heimberger, A.B.; Ram, Z.; Constantini, S.; Barbash, Z.S.; Adamsky, K.; Safran, M.; Hirschberg, A.; et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007, 17, 1586–1595. [Google Scholar] [CrossRef]
- Cenci, C.; Barzotti, R.; Galeano, F.; Corbelli, S.; Rota, R.; Massimi, L.; Di Rocco, C.; O’Connell, M.A.; Gallo, A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 2008, 283, 7251–7260. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Y.; Lin, C.H.; Chan, T.H.; Chow, R.K.; Song, Y.; Liu, M.; Yuan, Y.F.; Kong, K.L.; Qi, L.; et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013, 19, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, J.; Shi, X.; Feng, F.; Kau, K.W.; Chen, Y.; Chen, Y.; Jiang, L.; Cui, F.; Zhang, Y.; et al. RNA editing of AZIN1 induces the malignant progression of non-small-cell lung cancers. Tumour Biol. 2017, 39, 1010428317700001. [Google Scholar] [CrossRef]
- Qin, Y.R.; Qiao, J.J.; Chan, T.H.M.; Zhu, Y.H.; Li, F.F.; Liu, H.; Fei, J.; Li, Y.; Guan, X.Y.; Chen, L. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014, 74, 840–851. [Google Scholar] [CrossRef]
- Shigeyasu, K.; Okugawa, Y.; Toden, S.; Miyoshi, J.; Toiyama, Y.; Nagasaka, T.; Takahashi, N.; Kusunoki, M.; Takayama, T.; Yamada, Y.; et al. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight 2018, 3, e99976. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Shigeyasu, K.; Okugawa, Y.; Yoshida, K.; Mori, Y.; Yano, S.; Noma, K.; Umeda, Y.; Kondo, Y.; Kishimoto, H.; et al. Activation of AZIN1 RNA editing is a novel mechanism that promotes invasive potential of cancer-associated fibroblasts in colorectal cancer. Cancer Lett. 2019, 444, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, H.; Feng, Q.; Wang, S.; Shao, Y.; Wu, J.; Jin, G.; Lin, W.; Peng, X.; Xu, X. A novel mechanism for A-to-I RNA-edited AZIN1 in promoting tumor angiogenesis in colorectal cancer. Cell Death Dis. 2022, 13, 294. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, T.; Rahman, M.F.; Tostar, U.; Sonkoly, E.; Stahle, M.; Pivarcsi, A.; Palaniswamy, R.; Zaphiropoulos, P.G. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol. 2013, 10, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, M.; Gilead, L.; Markel, G.; Nemlich, Y.; Feiler, Y.; Enk, C.D.; Denichenko, P.; Kami, R.; Ingber, A. Insulin-like growth factor-binding protein-7 (IGFBP7) transcript: A-to-I editing events in normal and cancerous human keratinocytes. Arch. Dermatol. Res. 2013, 305, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Gumireddy, K.; Li, A.; Kossenkov, A.V.; Sakurai, M.; Yan, J.; Li, Y.; Xu, H.; Wang, J.; Zhang, P.J.; Zhang, L.; et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat. Commun. 2016, 7, 10715. [Google Scholar] [CrossRef] [PubMed]
- Han, S.W.; Kim, H.P.; Shin, J.Y.; Jeong, E.G.; Lee, W.C.; Kim, K.Y.; Park, S.Y.; Lee, D.W.; Won, J.K.; Jeong, S.Y.; et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J. Exp. Med. 2014, 211, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Bahn, J.H.; Lee, J.H.; Li, G.; Greer, C.; Peng, G.; Xiao, X. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012, 22, 142–150. [Google Scholar] [CrossRef]
- Li, M.; Wang, I.X.; Li, Y.; Bruzel, A.; Richards, A.L.; Toung, J.M.; Cheung, V.G. Widespread RNA and DNA sequence differences in the human transcriptome. Science 2011, 333, 53–58. [Google Scholar] [CrossRef]
- Peng, Z.; Cheng, Y.; Tan, B.C.; Kang, L.; Tian, Z.; Zhu, Y.; Zhang, W.; Liang, Y.; Hu, X.; Tan, X.; et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012, 30, 253–260. [Google Scholar] [CrossRef]
- Ramaswami, G.; Lin, W.; Piskol, R.; Tan, M.H.; Davis, C.; Li, J.B. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods 2012, 9, 579–581. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, S.; Wei, J.; Song, G.; Zhao, Y. A-to-I RNA editing in cancer: From evaluating the editing level to exploring the editing effects. Front. Oncol. 2021, 10, 632187. [Google Scholar] [CrossRef] [PubMed]
- Kiran, A.; Baranov, P.V. DARNED: A DAtabase of RNA EDiting in humans. Bioinformatics 2010, 26, 1772–1776. [Google Scholar] [CrossRef] [PubMed]
- Kiran, A.M.; O’Mahony, J.J.; Sanjeev, K.; Baranov, P.V. Darned in 2013, inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res. 2013, 41, D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Ramaswami, G.; Li, J.B. RADAR: A rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014, 42, D109–D113. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; D’Erchia, A.M.; Giudice, C.L.; Pesole, G. REDIportal: A comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017, 45, D750–D757. [Google Scholar] [CrossRef] [PubMed]
- Paz-Yaacov, N.; Bazak, L.; Buchumenski, L.; Porath, H.T.; Danan-Gotthold, M.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 2015, 13, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, D.; Gacquer, D.; Rothe, F.; Lefort, A.; Libert, F.; Brown, D.; Kheddoumi, N.; Shlien, A.; Konopka, T.; Salgado, R.; et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell Rep. 2015, 13, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Diao, L.X.; Yu, S.X.; Xu, X.Y.; Li, J.; Zhang, R.; Yang, Y.; Werner, H.M.J.; Eterovic, A.K.; Yoan, Y.; et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 2015, 28, 515–528. [Google Scholar] [CrossRef]
- Frezza, V.; Chellini, L.; Del Verme, A.; Paronetto, M.P. RNA Editing in Cancer Progression. Cancers 2023, 15, 5277. [Google Scholar] [CrossRef]
- Pecori, R.; Ren, W.; Pirmoradian, M.; Wang, X.; Liu, D.; Berglund, M.; Li, W.; Tasakis, R.N.; Di Giorgio, S.; Ye, X.; et al. ADAR1-mediated RNA editing promotes B cell lymphomagenesis. iScience 2023, 26, 106864. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, E.; Field, M.; et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Toung, J.M.; Morley, M.; Li, M.; Cheung, V.G. RNA-sequence analysis of human B-cells. Genome Res. 2011, 21, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Pesole, G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, X. Genome Sequence-Independent Identification of RNA Editing Sites. Nat. Methods 2015, 12, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Piechotta, M.; Naarmann-de Vries, I.; Wang, Q.; Altmüller, J.; Dieterich, C. RNA modification mapping with JACUSA2. Genome Biol. 2022, 23, 115. [Google Scholar] [CrossRef] [PubMed]
- John, D.; Weirick, T.; Dimmeler, S.; Uchida, S. RNAEditor: Easy detection of RNA editing events and the introduction of editing islands. Brief. Bioinform. 2017, 18, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Kofman, E.; Yee, B.; Medina-Munoz, H.C.; Yeo, G.W. FLARE: A fast and flexible workflow for identifying RNA editing foci. BMC Bioinform. 2023, 24, 370. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bararia, D.; Hildebrand, J.A.; Stolz, S.; Haebe, S.; Alig, S.; Trevisani, C.P.; Osorio-Barrios, F.; Bartoschek, M.D.; Mentz, M.; Pastore, A.; et al. Cathepsin S alterations induce a tumor-promoting immune microenvironment in follicular lymphoma. Cell. Rep. 2020, 31, 107522. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.; Li, J.; Yin, T.; Wang, Y.; Cheng, L. PRKCSH serves as a potential immunological and prognostic biomarker in pan-cancer. Sci. Rep. 2024, 14, 1778. [Google Scholar] [CrossRef]
- Stellos, K.; Gatsiou, A.; Stamatelopoulos, K.; Matic, L.P.; John, D.; Lunella, F.F.; Jae, N.; Rossbach, O.; Amrhein, C.; Sigala, F.; et al. Adenosine-to-insosine RNA editing controls cathepsin S expression in atherosclerosis by anabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016, 22, 1140–1150. [Google Scholar] [CrossRef]
- Chan, T.H.; Lin, C.H.; Qi, L.; Fei, J.; Li, Y.; Yong, K.J.; Liu, M.; Song, Y.; Chow, R.K.K.; Ng, V.H.E.; et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut 2013, 63, 832–843. [Google Scholar] [CrossRef]
- Schrider, D.R.; Gout, J.F.; Hahn, M.W. Very few RNA and DNA sequence differences in the human transcriptome. PLoS ONE 2011, 6, e25842. [Google Scholar] [CrossRef]
- Kleinman, C.L.; Majewski, J. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 2012, 335, 1302. [Google Scholar] [CrossRef]
- Lin, W.; Piskol, R.; Tan, M.H.; Li, J.B. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 2012, 335, 1302. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Gilad, Y.; Pritchard, J.K. Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science 2012, 335, 1302. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Eisenberg, E.; Adamsky, K.; Cohen, L.; Amariglio, N.; Hirshberg, A.; Rechavi, G.; Levanon, E.Y. Identification of RNA editing sites in the SNP database. Nucleic Acids Res. 2005, 33, 4612–4617. [Google Scholar] [CrossRef]
- Gommans, W.M.; Tatalias, N.E.; Sie, C.P.; Dupuis, D.; Vendetti, N.; Smith, L.; Kaushal, R.; Maas, S. Screening of human SNP database identifies recoding sites of A-to-I RNA editing. RNA 2008, 14, 2074–2085. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-Seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Tran, S.S.; Zhou, Q.; Xiao, X. Statistical inference of differential RNA-editing sites from RNA-sequencing data by hierarchical modeling. Bioinformatics 2020, 36, 2798–2804. [Google Scholar] [CrossRef] [PubMed]


| Comparison of Groups | Number of Sites Tested | Number of Sites with Significant Differences (FDR p-Value < 0.05) | Known Cancer-Related Genes | Number of Sites in Each Category | |||
|---|---|---|---|---|---|---|---|
| UTRs | Introns | Intergenic Regions | Repetitive Elements | ||||
| ABC vs. FL | 543 | 28 (16/12) 2 | CTSS, CTSB, STK4, SAMHD1 | 21 | 4 | 3 | 25 |
| GCB vs. FL | 546 | 16 (14/2) 2 | CTSS, CTSB, PRKCSH | 14 | 2 | 0 | 15 |
| GCB vs. ABC | 502 | 68 (62/6) 2 | NOP14, SAMHD1, VHL | 30 | 21 | 15 | 67 |
| Comparison of Groups | Number of Sites Tested | Number of Sites with Significant Differences (FDR p-Value < 0.05) | Known Cancer Related Genes | Number of Sites in Each Category | ||||
|---|---|---|---|---|---|---|---|---|
| Coding Regions | UTRs | Introns | Intergenic Regions | Repetitive Elements | ||||
| NHL vs. Normal | 398 | 59 (18/41) 2 | STK4, AZIN1, CTSS, NOP14, PRKCSH | 2 | 22 | 28 | 7 | 56 |
| GCB vs. Normal | 464 | 74 (19/55) 2 | STK4, AZIN1, CTSS, NOP14, PRKCSH, VHL, TP53 | 2 | 27 | 35 | 12 | 71 |
| ABC vs. Normal | 484 | 69 (20/49) 2 | STK4, AZIN1, CTSS, PRKCSH, VHL | 2 | 25 | 33 | 9 | 66 |
| FL vs. Normal | 496 | 84 (32/52) 2 | AZIN1, CTSS, PRKCSH, VHL, TP53 | 2 | 35 | 35 | 12 | 79 |
| Number of Sites Tested | Significant Correlation (FDR p-Value < 0.05) | Positive Correlation | Negative Correlation |
|---|---|---|---|
| 88 | 39 | 32 | 7 |
| Groups | Total Number of Samples | Number of Samples Correctly Predicted | p-Value (Fisher’s Exact Test) |
|---|---|---|---|
| ABC vs. FL | 45 | 38 (84%) | 2.50 × 10−5 |
| GCB vs. FL | 67 | 57 (85%) | 2.41 × 10−6 |
| GCB vs. ABC | 86 | 65 (76%) | 4.54 × 10−5 |
| NHL vs. Normal | 140 | 130 (93%) | 4.35 × 10−25 |
| GCB vs. Normal | 95 | 86 (91%) | 1.56 × 10−17 |
| ABC vs. Normal | 73 | 70 (96%) | 5.26 × 10−17 |
| FL vs. Normal | 54 | 53 (98%) | 1.26 × 10−11 |
| Clinical Status | Number of Samples |
|---|---|
| GCB | 54 |
| ABC | 32 |
| FL | 13 |
| Normal | 41 |
| NHL (GCB + ABC + FL) | 99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Bundschuh, R. A-to-I Editing Is Subtype-Specific in Non-Hodgkin Lymphomas. Genes 2024, 15, 864. https://doi.org/10.3390/genes15070864
Chen C, Bundschuh R. A-to-I Editing Is Subtype-Specific in Non-Hodgkin Lymphomas. Genes. 2024; 15(7):864. https://doi.org/10.3390/genes15070864
Chicago/Turabian StyleChen, Cai, and Ralf Bundschuh. 2024. "A-to-I Editing Is Subtype-Specific in Non-Hodgkin Lymphomas" Genes 15, no. 7: 864. https://doi.org/10.3390/genes15070864
APA StyleChen, C., & Bundschuh, R. (2024). A-to-I Editing Is Subtype-Specific in Non-Hodgkin Lymphomas. Genes, 15(7), 864. https://doi.org/10.3390/genes15070864





