CNV Analysis through Exome Sequencing Reveals a Large Duplication Involved in Sex Reversal, Neurodevelopmental Delay, Epilepsy and Optic Atrophy
Abstract
:1. Introduction
2. Material and Methods
2.1. Patient
2.2. Isolation of Genomic DNA
2.3. Exome Sequencing (ES)
2.4. CNV Analysis—ExomeDepth
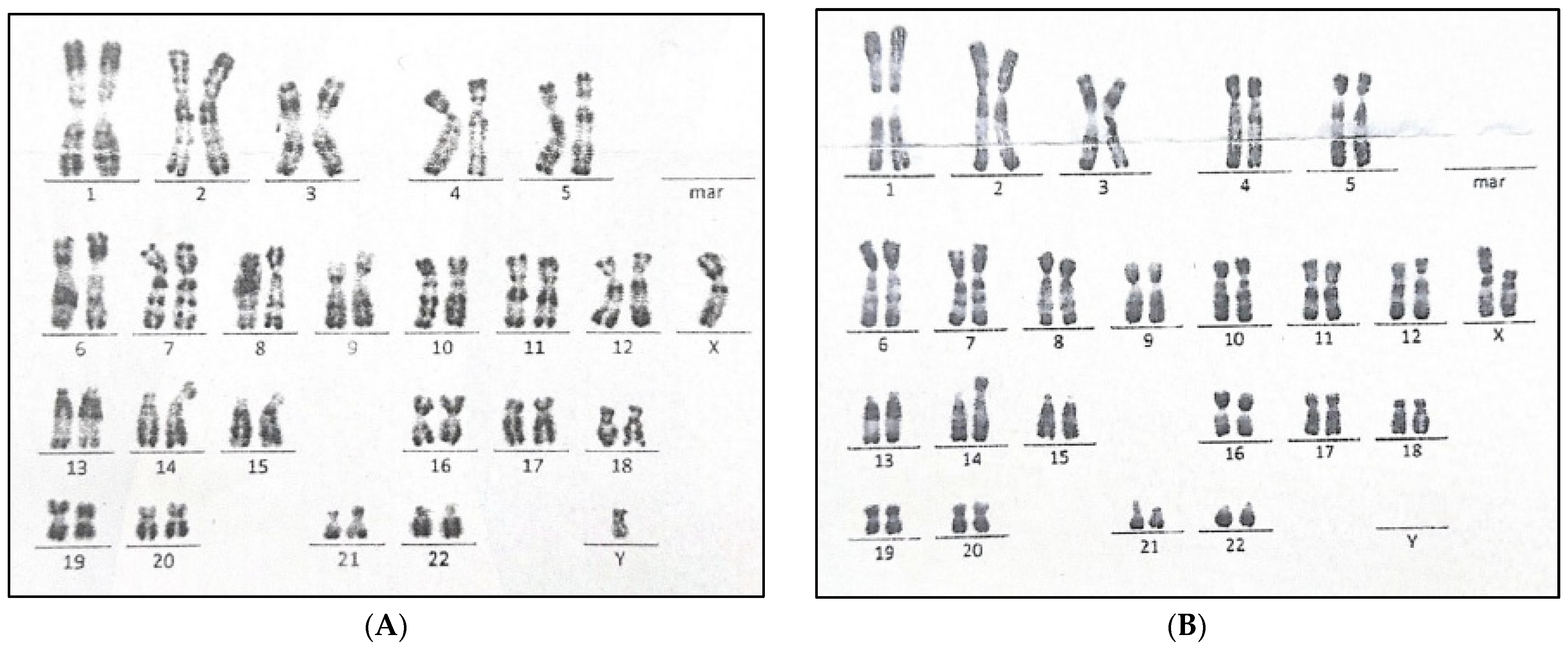
2.5. Blood Karyotyping
3. Results
3.1. Case Presentation
3.2. Genetic Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, T.; Liu, J.H.; Yang, J.; Chen, J.; Ye, Z.Q. 46, XX male sex reversal syndrome: A case report and review of the genetic basis. Andrologia 2009, 41, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Taylor, H.S. Chapter 27—Development of the Genital System. In Principles of Developmental Genetics, 2nd ed.; Moody, S.A., Ed.; Academic Press: Oxford, UK, 2015; pp. 487–504. ISBN 978-0-12-405945-0. [Google Scholar]
- Li, Y.; Zheng, M.; Lau, Y.-F.C. The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Rep. 2014, 8, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Sekido, R.; Lovell-Badge, R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008, 453, 930–934. [Google Scholar] [CrossRef] [PubMed]
- Gonen, N.; Lovell-Badge, R. Chapter Eight—The regulation of Sox9 expression in the gonad. In Current Topics in Developmental Biology; Capel, B., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 134, pp. 223–252. ISBN 0070-2153. [Google Scholar] [CrossRef]
- Barrionuevo, F.; Scherer, G. SOX E genes: SOX9 and SOX8 in mammalian testis development. SOX Transcr. Factors. 2010, 42, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Barrionuevo, F.J.; Hurtado, A.; Kim, G.-J.; Real, F.M.; Bakkali, M.; Kopp, J.L.; Sander, M.; Scherer, G.; Burgos, M.; Jiménez, R. Sox9 and Sox8 protect the adult testis from male-to-female genetic reprogramming and complete degeneration. eLife 2016, 5, e15635. [Google Scholar] [CrossRef]
- Acién, P.; Acién, M. Disorders of Sex Development: Classification, Review, and Impact on Fertility. J. Clin. Med. 2020, 9, 3555. [Google Scholar] [CrossRef] [PubMed]
- Narahara, K.; Kodama, Y.; Kimura, S.; Kimoto, H. Probable inverted tandem duplication of Xp in a 46,Xp+Y boy. Jinrui Idengaku Zasshi. Jpn. J. Hum. Genet. 1979, 24, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, R.; Jenkins, T.; Dawson, B.; Wagner, J.; Dewald, G.; Koo, G.C.; Wachtel, S.S. Female phenotype and multiple abnormalities in sibs with a Y chromosome and partial X chromosome duplication: H–Y antigen and Xg blood group findings. J. Med. Genet. 1980, 17, 291–300. [Google Scholar] [CrossRef]
- Nielsen, K.B.; Langkjaer, F. Inherited partial X chromosome duplication in a mentally retarded male. J. Med. Genet. 1982, 19, 222–224. [Google Scholar] [CrossRef]
- Scherer, G.; Schempp, W.; Baccichetti, C.; Lenzini, E.; Bricarelli, F.D.; Carbone, L.D.; Wolf, U. Duplication of an Xp segment that includes the ZFX locus causes sex inversion in man. Hum. Genet. 1989, 81, 291–294. [Google Scholar] [CrossRef]
- Stern, H.J.; Garrity, A.M.; Saal, H.M.; Wangsa, D.; Disteche, C.M. Duplication of Xp21 and sex reversal: Insight into the mechanism of sex determination. Am. J. Hum. Genet. 1990, 47 (Suppl. A41), 153. [Google Scholar]
- May, K.M.; Grinzaid, K.A.; Blackston, R.D. Sex reversal and multiple abnormalities due to abnormal segregation of t(X; 16)-(pl 1.4;p13.3. Am. J. Hum. Genet. 1991, 49, 19. [Google Scholar]
- Ogata, T.; Hawkins, J.R.; Taylor, A.; Matsuo, N.; Hata, J.; Goodfellow, P.N. Sex reversal in a child with a 46,X,Yp+ karyotype: Support for the existence of a gene(s), located in distal Xp, involved in testis formation. J. Med. Genet. 1992, 29, 226–230. [Google Scholar] [CrossRef]
- Arn, P.; Chen, H.; Tuck-Muller, C.M.; Mankinen, C.; Wachtel, G.; Li, S.; Shen, C.C.; Wachtel, S.S. SRVX, a sex reversing locus in Xp21.2-->p22.11. Hum. Genet. 1994, 93, 389–393. [Google Scholar] [CrossRef]
- Bardoni, B.; Floridia, G.; Guioli, S.; Peverali, G.; Anichini, C.; Cisternino, M.; Casalone, R.; Danesino, C.; Fraccaro, M.; Zuffardi, O. Functional disomy of Xp22-pter in three males carrying a portion of Xp translocated to Yq. Hum. Genet. 1993, 91, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Baumstark, A.; Barbi, G.; Djalali, M.; Geerkens, C.; Mitulla, B.; Mattfeldt, T.; de Almeida, J.C.; Vargas, F.R.; Llerena Júnior, J.C.; Vogel, W.; et al. Xp-duplications with and without sex reversal. Hum. Genet. 1996, 97, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Telvi, L.; Ion, A.; Carel, J.C.; Desguerre, I.; Piraud, M.; Boutin, A.M.; Feingold, J.; Ponsot, G.; Fellous, M.; McElreavey, K. A duplication of distal Xp associated with hypogonadotrophic hypogonadism, hypoplastic external genitalia, mental retardation, and multiple congenital abnormalities. J. Med. Genet. 1996, 33, 767–771. [Google Scholar] [CrossRef]
- Sukumaran, A.; Desmangles, J.C.; Gartner, L.A.; Buchlis, J. Duplication of dosage sensitive sex reversal area in a 46, XY patient with normal sex determining region of Y causing complete sex reversal. J. Pediatr. Endocrinol. 2013, 26, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Bardoni, B.; Zanaria, E.; Guioli, S.; Floridia, G.; Worley, K.C.; Tonini, G.; Ferrante, E.; Chiumello, G.; McCabe, E.R.; Fraccaro, M. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat. Genet. 1994, 7, 497–501. [Google Scholar] [CrossRef]
- Swain, A.; Narvaez, V.; Burgoyne, P.; Camerino, G.; Lovell-Badge, R. Dax1 antagonizes Sry action in mammalian sex determination. Nature 1998, 391, 761–767. [Google Scholar] [CrossRef]
- Ludbrook, L.M.; Bernard, P.; Bagheri-Fam, S.; Ryan, J.; Sekido, R.; Wilhelm, D.; Lovell-Badge, R.; Harley, V.R. Excess DAX1 leads to XY ovotesticular disorder of sex development (DSD) in mice by inhibiting steroidogenic factor-1 (SF1) activation of the testis enhancer of SRY-box-9 (Sox9). Endocrinology 2012, 153, 1948–1958. [Google Scholar] [CrossRef] [PubMed]
- García-Acero, M.; Molina, M.; Moreno, O.; Ramirez, A.; Forero, C.; Céspedes, C.; Prieto, J.C.; Pérez, J.; Suárez-Obando, F.; Rojas, A. Gene dosage of DAX-1, determining in sexual differentiation: Duplication of DAX-1 in two sisters with gonadal dysgenesis. Mol. Biol. Rep. 2019, 46, 2971–2978. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, N.V.; Cotter, P.D. Factors affecting clinical manifestation of chromosomal imbalance in carriers of segmental autosomal mosaicism: Differential impact of gender. J. Appl. Genet. 2022, 63, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, J.P.; Bartoli, M.; Delague, V.; Krahn, M.; Miltgen, M.; Béroud, C.; Salgado, D. VarAFT: A variant annotation and filtration system for human next generation sequencing data. Nucleic Acids Res. 2018, 46, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Miressi, F.; Faye, P.A.; Pyromali, I.; Bourthoumieux, S.; Derouault, P.; Husson, M.; Favreau, F.; Sturtz, F.; Magdelaine, C.; Lia, A.S. A mutation can hide another one: Think Structural Variants! Comput. Struct. Biotechnol. J. 2020, 18, 2095–2099. [Google Scholar] [CrossRef] [PubMed]
- MacColl, C.; Stein, N.; Tarnopolsky, M.; Lu, J.Q. Neurodevelopmental and associated changes in a patient with Xp22.31 duplication. Neurol. Sci. Off. J. Ital.Neurol. Soc. Ita.l Soc. Clin. Neurophysiol. 2020, 41, 713–716. [Google Scholar] [CrossRef] [PubMed]
- Esplin, E.D.; Li, B.; Slavotinek, A.; Novelli, A.; Battaglia, A.; Clark, R.; Curry, C.; Hudgins, L. Nine patients with Xp22.31 microduplication, cognitive deficits, seizures, and talipes anomalies. Am. J. Med. Genet. A 2014, 164, 2097–2103. [Google Scholar] [CrossRef]
- Poeta, L.; Malacarne, M.; Padula, A.; Drongitis, D.; Verrillo, L.; Lioi, M.B.; Chiariello, A.M.; Bianco, S.; Nicodemi, M.; Piccione, M.; et al. Further Delineation of Duplications of ARX Locus Detected in Male Patients with Varying Degrees of Intellectual Disability. Int. J. Mol. Sci. 2022, 23, 3084. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Hayashi, S.; Imoto, I.; Toyama, J.; Okazawa, H.; Nakagawa, E.; Goto, Y.-I.; Inazawa, J. Copy-number variations on the X chromosome in Japanese patients with mental retardation detected by array-based comparative genomic hybridization analysis. J. Hum. Genet. 2010, 55, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Sakka, R.; Abdelhedi, F.; Sellami, H.; Pichon, B.; Lajmi, Y.; Mnif, M.; Kebaili, S.; Derbel, R.; Kamoun, H.; Gdoura, R.; et al. An unusual familial Xp22.12 microduplication including EIF1AX: A novel candidate dosage-sensitive gene for premature ovarian insufficiency. Eur. J. Med. Genet. 2022, 65, 104613. [Google Scholar] [CrossRef] [PubMed]
- Tzschach, A.; Chen, W.; Erdogan, F.; Hoeller, A.; Ropers, H.-H.; Castellan, C.; Ullmann, R.; Schinzel, A. Characterization of interstitial Xp duplications in two families by tiling path array CGH. Am. J. Med. Genet. 2008, 146, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, D.; de Brouwer, A.P.M.; Kleefstra, T.; Oudakker, A.R.; Frints, S.G.M.; Schrander-Stumpel, C.T.R.M.; Fryns, J.P.; Jensen, L.R.; Chelly, J.; Moraine, C.; et al. Chromosomal copy number changes in patients with non-syndromic X linked mental retardation detected by array CGH. J. Med. Genet. 2006, 43, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Popovici, C.; Busa, T.; Boute, O.; Thuresson, A.-C.; Perret, O.; Sigaudy, S.; Södergren, T.; Andrieux, J.; Moncla, A.; Philip, N. Whole ARX gene duplication is compatible with normal intellectual development. Am. J. Med. Genet. A 2014, 164, 2324–2327. [Google Scholar] [CrossRef] [PubMed]
- Sismani, C.; Anastasiadou, V.; Kousoulidou, L.; Parkel, S.; Koumbaris, G.; Zilina, O.; Bashiardes, S.; Spanou, E.; Kurg, A.; Patsalis, P.C. 9 Mb familial duplication in chromosome band Xp22.2-22.13 associated with mental retardation, hypotonia and developmental delay, scoliosis, cardiovascular problems and mild dysmorphic facial features. Eur. J. Med. Genet. 2011, 54, e510–e515. [Google Scholar] [CrossRef] [PubMed]
- Chatron, N.; Thibault, L.; Lespinasse, J.; Labalme, A.; Schluth-Bolard, C.; Till, M.; Edery, P.; Touraine, R.; des Portes, V.; Lesca, G.; et al. Genetic Counselling Pitfall: Co-Occurrence of an 11.8-Mb Xp22 Duplication and an Xp21.2 Duplication Disrupting IL1RAPL1. Mol. Syndromol. 2017, 8, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Achermann, J.C.; Vilain, E.J. NR0B1-Related Adrenal Hypoplasia Congenita; Adam, M.P., Ed.; University of Seattle: Seattle, WA, USA, 2001. [Google Scholar]
- Massimino, C.R.; Smilari, P.; Greco, F.; Marino, S.; Vecchio, D.; Bartuli, A.; Parisi, P.; Cho, S.Y.; Pavone, P. Poland Syndrome with Atypical Malformations Associated to a de novo 1.5 Mb Xp22.31 Duplication. Neuropediatrics 2020, 51, 359–363. [Google Scholar] [CrossRef]
| Cytogenetic Location | Gene | Gene Name | Gene MIM# | Function | Dosage Sensitivity | Phenotype | Phenotype MIM# |
|---|---|---|---|---|---|---|---|
| Xp22.31 | ANOS1 | anosmin 1 | 300836 | migration of GNRH neurons to the hypothalamus | Triplosensitivity | Hypogonadotropic hypogonadism 1 with or without anosmia (Kallmann syndrome 1) | 308700 |
| Xp21.3 | ARX | aristaless related homeobox | 300382 | cerebral development and patterning | Triplosensitivity | Developmental and epileptic encephalopathy 1 | 308350 |
| Xp22.13 | CDKL5 | cyclin dependent kinase like 5 | 300203 | Involved in neural maturation and synaptogenesis | Triplosensitivity | Developmental and epileptic encephalopathy 2 | 300672 |
| Xp22.12 | CNKSR2 | connector enhancer of kinase suppressor of Ras 2 | 300724 | Plays a role CNS neuronal postsynaptic density (PSD) | Triplosensitivity | Intellectual developmental disorder, X-linked syndromic, Houge type | 301008 |
| Xp22.2 | HCCS | holocytochrome c synthase | 300056 | Plays a role in mitochondrial respiratory chain (heme attachment to cytochrome C) | Triplosensitivity | Linear skin defects with multiple congenital anomalies 1 | 309801 |
| Xp22.2 | MID1 | midline 1 | 300552 | Plays a role in linking cytoskeleton-associated mRNA transport and translation control factors with mTOR gene | Triplosensitivity | Opitz GBBB syndrome | 300000 |
| Xp21.2 | NR0B1 | nuclear receptor subfamily 0 group B member 1 | 300473 | Special type of the nuclear receptor (NR) superfamily by acting as a coregulatory protein that inhibits the transcriptional activity of other NRs | Triplosensitivity | 46XY sex reversal 2, dosage-sensitive | 300018 |
| Xp22.2 | OFD1 | OFD1 centriole and centriolar satellite protein | 300170 | Plays a role in regulation of microtubule dynamics | Triplosensitivity | Retinitis pigmentosa 23 | 300424 |
| Xp22.11 | PTCHD1 | patched domain containing 1 | 300828 | Plays a role in hedgehog signaling pathway | Triplosensitivity | {Autism, susceptibility to, X-linked 4} | 300830 |
| Xp22.12 | RPS6KA3 | ribosomal protein S6 kinase A3 | 300075 | Plays a role in cell cycle progression, differentiation, and cell survival | Triplosensitivity | Coffin-Lowry syndrome | 303600 |
| Xp22.13 | RS1 | retinoschisin 1 | 300839 | Retina cell-cell adhesion and interactions | Triplosensitivity | Retinoschisis | 312700 |
| Xp22.11 | SMS | spermine synthase | 300105 | Involved in the synthesis of polyamines from arginine and methionine | Triplosensitivity | Intellectual developmental disorder, X-linked syndromic, Snyder-Robinson type | 309583 |
| Xp22.2 | AP1S2 | adaptor related protein complex 1 subunit sigma 2 | 300629 | Recruitment of clathrin and sorting signals recognition | Haploinsufficiency | Pettigrew syndrome | 304340 |
| Xp22.2 | CLCN4 | chloride voltage-gated channel 4 | 302910 | encodes for voltage-gated chloride channel | Haploinsufficiency | Raynaud-Claes syndrome | 300114 |
| Xp22.2 | FANCB | FA complementation group B | 300515 | Part of Fanconi anemia core complex | Haploinsufficiency | Fanconi anemia, complementation group B | 300514 |
| Xp21.2 | GK | glycerol kinase | 300474 | Catalyzes the phosphorylation of glycerol to glycerol-3-phosphate | Haploinsufficiency | Glycerol kinase deficiency | 307030 |
| Xp21.3-p21.2 | IL1RAPL1 | interleukin 1 receptor accessory protein like 1 | 300206 | Plays a role in synaptic regulation and regulation | Haploinsufficiency | Intellectual developmental disorder, X-linked 21 | 300143 |
| Xp22.2 | MSL3 | MSL complex subunit 3 | 300609 | Plays a major role in acetylation of histone H4 | Haploinsufficiency | Basilicata-Akhtar syndrome | 301032 |
| Xp22.2-p22.13 | NHS | NHS actin remodeling regulator | 300457 | Plays a role in actin remodeling and cell morphology | Haploinsufficiency | Cataract 40, X-linked | 302200 |
| Xp22.12 | PDHA1 | pyruvate dehydrogenase E1 subunit α 1 | 300502 | Catalyzing the irreversible conversion of pyruvate into acetyl-CoA | Haploinsufficiency | Pyruvate dehydrogenase E1-α deficiency | 312170 |
| Xp22.11 | PHEX | phosphate regulating endopeptidase X-linked | 300550 | Encodes for an integral membrane zinc-dependent endopeptidase protein | Haploinsufficiency | Hypophosphatemic rickets, X-linked dominant | 307800 |
| Xp22.2 | PIGA | phosphatidylinositol glycan anchor biosynthesis class A | 311770 | Plays a role in GPI (Glycosylphosphatidylinositol) anchoring biosynthesis | Haploinsufficiency | Multiple congenital anomalies-hypotonia-seizures syndrome 2 | 300868 |
| Xp22.31 | STS | steroid sulfatase | 300747 | Encodes for steroid sulfatase protein that plays a role in estrogen, androgen, and cholesterol synthesis | Haploinsufficiency | Ichthyosis, X-linked | 308100 |
| Xp22.2 | TRAPPC2 | trafficking protein particle complex subunit 2 | 300202 | Member of TRAPP complex that plays a role in intracellular vesicle trafficking | Haploinsufficiency | Spondyloepiphyseal dysplasia tarda | 313400 |
| Year | Citation | Patient | Location | Size | Genes | Clinical Manifestations |
|---|---|---|---|---|---|---|
| 2006 | 31 | 1 | ChrX:9,700,000–16,400,000 | 7 MB | MID1, ARHGAP6, MSL3L1 | Mental Retardation |
| Facial dysmorphism | ||||||
| Hearing loss/Presbycusis | ||||||
| Pectus excavatum | ||||||
| Arachnodactyly | ||||||
| Atrophy of interdigital muscles | ||||||
| 2011 | 33 | 2 | ChrX:9,750,000–18,710,000 | 9 MB | 59 genes of which seven are involved in syndromic X-linked intellectual deficiency, namely MID1, HCCS, OFD1, FANCB, AP1S2, CDKL5 and NHS. | Mental retardation |
| Developmental delay | ||||||
| Dysmorphic features | ||||||
| Facial dysmorphism (hypertelorism, broad nasal bridge, widow’s peak) | ||||||
| Genitourinary abnormalities | ||||||
| Heart defects | ||||||
| Short stature | ||||||
| Scoliosis | ||||||
| Hypertelorism | ||||||
| Neurodevelopmental disorders | ||||||
| Diaphragmatic hernia | ||||||
| 2022 | 29 | 3 | ChrX:19,563,240–20,597,641 | 1 MB | SH3KBP1, EIF1AX and RPS6KA3 | Delayed speech |
| Language development delay | ||||||
| Seizure | ||||||
| Joint laxity | ||||||
| 4 | ChrX: 19,825,290–20,930,431 | 1.1 MB | SH3KBP1, EIF1AX and RPS6KA3 | Specific learning disability | ||
| 5 | ChrX: 19,651,193–20,700,691 | 1.05 MB | SH3KBP1, EIF1AX and RPS6KA3 | Intellectual disability | ||
| 2008 | 30 | 6 | ChrX: 1,5000,000–23,500,000 | 8.5 MB | CDKL5, RPS6KA3 | Facial dysmorphism |
| Bilateral inguinal hernia | ||||||
| Downslanted palpebral fissures | ||||||
| Upslanted palpebral fissures Bilateral inguinal hernia | ||||||
| Epilepsy | ||||||
| Brain tumor at age 3 yrs | ||||||
| 2022 | 27 | 7 | ChrX: 24,513,979–27,864,451 | 3.35 MB | PDK3, PCYT1B, POLA1, SCARNA23, ARX, MAGEB18, MAGEB6B, MAGEB6, MAGEB5, PPP4R3C, DCAF8L2 and MAGEB10 | Autism |
| 8 | ChrX: 24,810,754–27,125,219 | 2.3 MB | POLA1, ARX, MAGEB18, MAGEB6B, MAGEB6 and MAGEB5 | Intellectual disabilities | ||
| Short stature | ||||||
| Current case | 4 | 9 | ChrX: 7,137,718–30,739,112 | 23.6 MB | Around 143 genes, of which 11 are triplosensitive: ANOS1, ARX, CDKL5, CNKSR2, HCCS, MID1, NR0B1, OFD1, PTCHD1, RPS6KA3, RS1, SMS, AP1S2, CLCN4, FANCB, GK, IL1RAPL1, MSL3, NHS, PDHA1, PHEX, PIGA, STS, TRAPPC2. | Neurodevelopmental delay |
| Intellectual disability | ||||||
| Psychomoter delay | ||||||
| Hypotonia | ||||||
| Failure to thrive | ||||||
| CNS malformation (abnormal T2 signals in the basal ganglia, thalami, and brainstem, atrophy of the hippocampus | ||||||
| Seizures | ||||||
| Strabismus | ||||||
| Facial dysmorphism | ||||||
| Short stature | ||||||
| Scoliosis | ||||||
| Joint hypermobility/laxity, | ||||||
| Optic nerve hypoplasia | ||||||
| Feeding difficulty | ||||||
| GE reflux |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehawej, C.; Maalouf, J.E.; Abdelkhalik, M.; Mahfouz, P.; Chouery, E.; Megarbane, A. CNV Analysis through Exome Sequencing Reveals a Large Duplication Involved in Sex Reversal, Neurodevelopmental Delay, Epilepsy and Optic Atrophy. Genes 2024, 15, 901. https://doi.org/10.3390/genes15070901
Mehawej C, Maalouf JE, Abdelkhalik M, Mahfouz P, Chouery E, Megarbane A. CNV Analysis through Exome Sequencing Reveals a Large Duplication Involved in Sex Reversal, Neurodevelopmental Delay, Epilepsy and Optic Atrophy. Genes. 2024; 15(7):901. https://doi.org/10.3390/genes15070901
Chicago/Turabian StyleMehawej, Cybel, Joy El Maalouf, Mohamad Abdelkhalik, Peter Mahfouz, Eliane Chouery, and Andre Megarbane. 2024. "CNV Analysis through Exome Sequencing Reveals a Large Duplication Involved in Sex Reversal, Neurodevelopmental Delay, Epilepsy and Optic Atrophy" Genes 15, no. 7: 901. https://doi.org/10.3390/genes15070901
APA StyleMehawej, C., Maalouf, J. E., Abdelkhalik, M., Mahfouz, P., Chouery, E., & Megarbane, A. (2024). CNV Analysis through Exome Sequencing Reveals a Large Duplication Involved in Sex Reversal, Neurodevelopmental Delay, Epilepsy and Optic Atrophy. Genes, 15(7), 901. https://doi.org/10.3390/genes15070901








