Molecular Mechanisms of Resistance against PSII-Inhibiting Herbicides in Amaranthus retroflexus from the Czech Republic
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Dose–Response Experiment
2.2. Statistical Analysis
2.3. psbA Gene Partial Sequencing
2.4. 3D Modeling and Molecular Docking Studies
2.5. Chlorophyll Fluorescence Measurement
3. Results
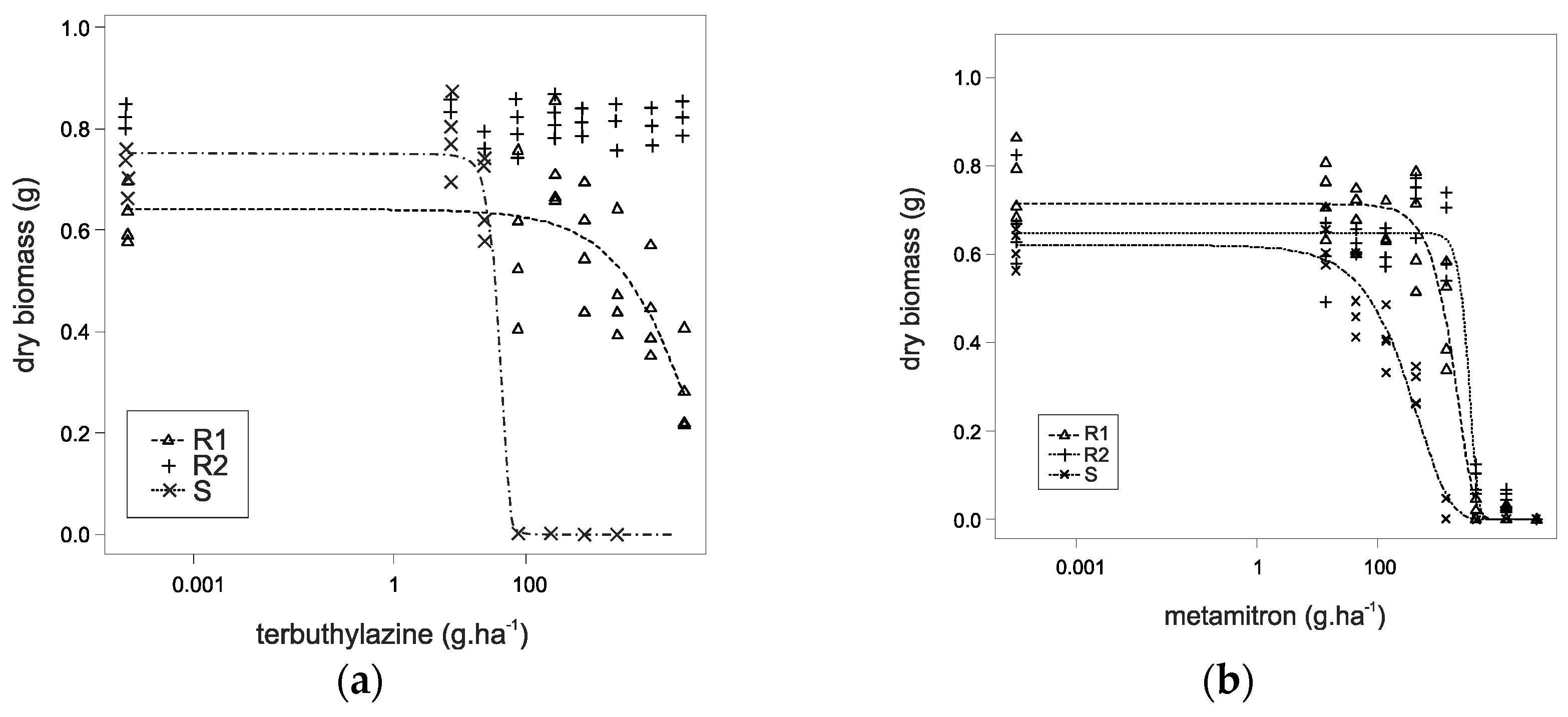
3.1. Dose–Response Test
3.2. Sequencing of psbA Gene from A. retroflexus
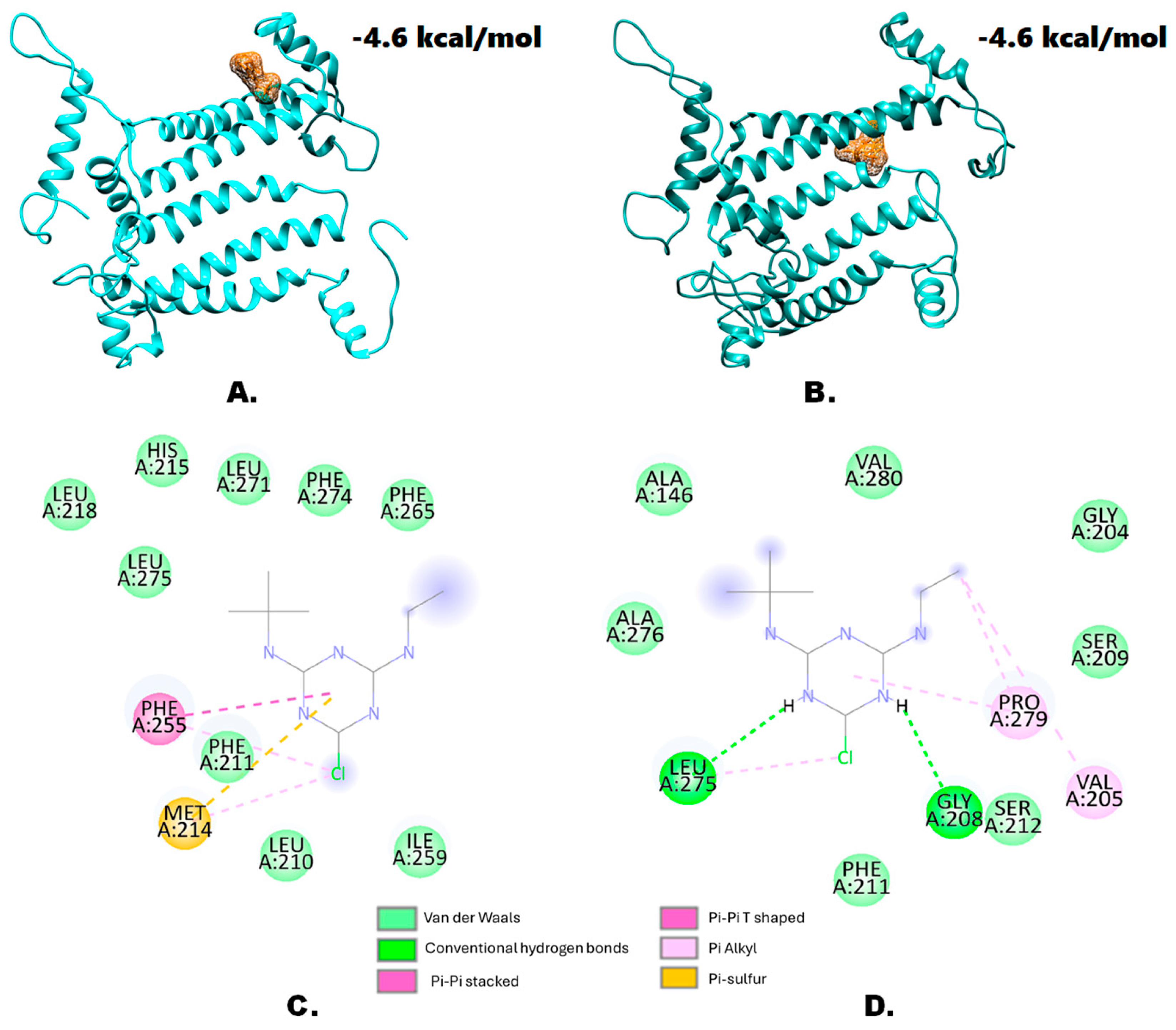
3.3. Three-Dimensional Structural Modeling of the Wild-Type and the Mutant-Type D1 Protein variants and Molecular Docking Experiments
3.4. Effects of the S264G Mutation on Protein Structural Features
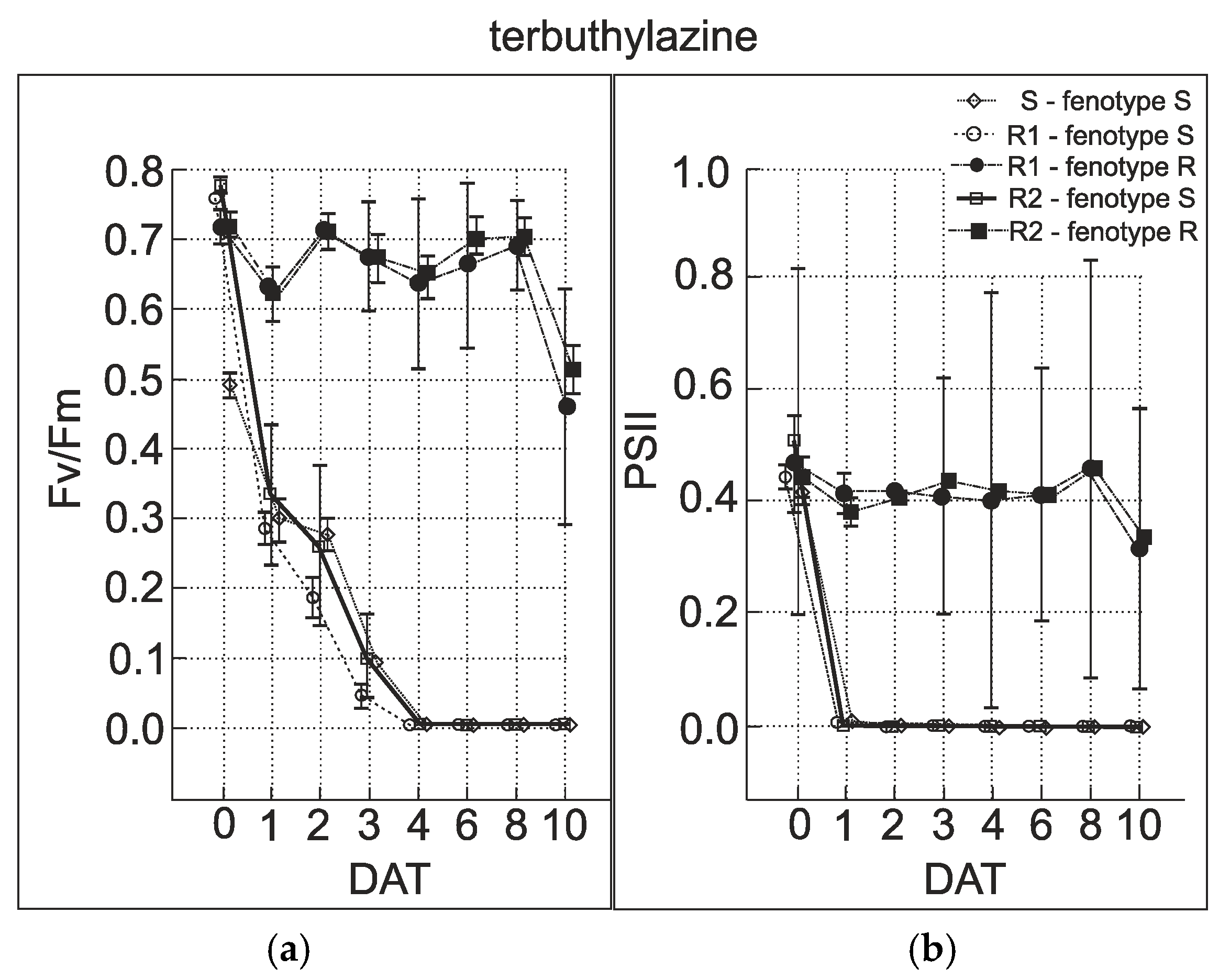
3.5. Chlorophyll Fluorescence Measurement
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chauhan, B. Biology and Management of Problematic Crop Weed Species; Elsevier: Amsterdam, The Netherlands, 2021; ISBN 978-0-12-822935-4. [Google Scholar]
- CABI. Amaranthus retroflexus (Redroot Pigweed). CABI Compend. 2021, 11, 4652. [Google Scholar] [CrossRef]
- Weaver, S.E.; McWilliams, E.L. The Biology of Canadian Weeds.: 44. Amaranthus retroflexus L., A. powellii S. Wats. and A. hybridus L. Can. J. Plant Sci. 1980, 60, 1215–1234. [Google Scholar] [CrossRef]
- Vencill, W.; Grey, T.; Culpepper, A.S.; Gaines, C.; Westra, P. Herbicide-Resistance in the Amaranthaceae. J. Plant Dis. Protection 2008, 9 (Suppl. S21), 41–44. [Google Scholar]
- Tranel, P.J. Herbicide Resistance in Amaranthus tuberculatus†. Pest Manag. Sci. 2021, 77, 43–54. [Google Scholar] [CrossRef]
- De Prado, R.; Lopez-Martinez, N.; Gimenez-Espinosa, R. Herbicide-Resistant Weeds in Europe: Agricultural, Physiological and Biochemical Aspects. In Weed and Crop Resistance to Herbicides; De Prado, R., Jorrín, J., García-Torres, L., Eds.; Springer: Dordrecht, The Netherlands, 1997; pp. 17–27. ISBN 978-94-011-5538-0. [Google Scholar]
- Cao, Y.; Wei, S.; Huang, H.; Li, W.; Zhang, C.; Huang, Z. Target-Site Mutation and Enhanced Metabolism Confer Resistance to Thifensulfuron-Methyl in a Multiple-Resistant Redroot Pigweed (Amaranthus retroflexus) Population. Weed Sci. 2021, 69, 161–166. [Google Scholar] [CrossRef]
- Jones, E.A.L.; Andres, R.J.; Dunne, J.C.; Leon, R.G.; Everman, W.J. Confirmation and Detection of Novel Acetolactate Synthase- and Protoporphyrinogen Oxidase–Inhibiting Herbicide-Resistant Redroot Pigweed (Amaranthus retroflexus) Populations in North Carolina. Weed Sci. 2023, 71, 84–94. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, J.; Zhang, C.; Huang, H.; Wei, S.; Zhou, X.; Chen, J.; Wang, X. Target-Site Basis for Resistance to Imazethapyr in Redroot Amaranth (Amaranthus retroflexus L.). Pestic. Biochem. Physiol. 2016, 128, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Thiel, H.; Varrelmann, M. Identification of a New PSII Target Site psbA Mutation Leading to D1 Amino Acid Leu218Val Exchange in the Chenopodium Album D1 Protein and Comparison to Cross-Resistance Profiles of Known Modifications at Positions 251 and 264. Pest Manag. Sci. 2014, 70, 278–285. [Google Scholar] [CrossRef]
- Mengistu, L.W.; Mueller-Warrant, G.W.; Liston, A.; Barker, R.E. psbA Mutation (Valine219 to Isoleucine) in Poa annua Resistant to Metribuzin and Diuron. Pest Manag. Sci. 2000, 56, 209–217. [Google Scholar] [CrossRef]
- Mechant, E.; Tania, D.; Olivier, H.; Robert, O.; Robert, B. Target Site Resistance to Metamitron in Chenopodium album L. J. Plant Dis. Prot. 2008, XXI, 37–40. [Google Scholar]
- Perez-Jones, A.; Intanon, S.; Mallory-Smith, C. Molecular Analysis of Hexazinone-Resistant Shepherd’s-Purse (Capsella bursa-pastoris) Reveals a Novel psbA Mutation. Weed Sci. 2009, 57, 574–578. [Google Scholar] [CrossRef]
- Blauth, S.L.; Steffens, J.C.; Churchill, G.A.; Mutschler, M.A. Identification of QTLs Controlling Acylsugar Fatty Acid Composition in an Intraspecific Population of Lycopersicon pennellii (Corr.) D’Arcy. Theor. Appl. Genet. 1999, 99, 373–381. [Google Scholar] [CrossRef]
- Park, K.W.; Mallory-Smith, C.A. psbA Mutation (Asn266 to Thr) in Senecio Vulgaris L. Confers Resistance to Several PS II-Inhibiting Herbicides. Pest Manag. Sci. 2006, 62, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Yu, Q.; Han, H.; Owen, M.J.; Powles, S.B. A Novel psbA Mutation (Phe274–Val) Confers Resistance to PSII Herbicides in Wild Radish (Raphanus raphanistrum). Pest Manag. Sci. 2019, 75, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Schwenger-Erger, C.; Thiemann, J.; Barz, W.; Johanningmeier, U.; Naber, D. Metribuzin Resistance in Photoautotrophic Chenopodium Rubrum Cell Cultures. Characterization of Double and Triple Mutations in the psbA Gene. FEBS Lett. 1993, 329, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Sen, M.K.; Hamouzová, K.; Mikulka, J.; Bharati, R.; Košnarová, P.; Hamouz, P.; Roy, A.; Soukup, J. Enhanced Metabolism and Target Gene Overexpression Confer Resistance against Acetolactate Synthase-Inhibiting Herbicides in Bromus sterilis. Pest Manag. Sci. 2021, 77, 2122–2128. [Google Scholar] [CrossRef] [PubMed]
- Košnarová, P.; Hamouz, P.; Hamouzová, K.; Linn, A.; Sen, M.K.; Mikulka, J.; Šuk, J.; Soukup, J. Apera spica-venti in the Czech Republic Develops Resistance to Three Herbicide Modes of Action. Weed Res. 2021, 61, 420–429. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput.-Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Battaglino, B.; Grinzato, A.; Pagliano, C. Binding Properties of Photosynthetic Herbicides with the QB Site of the D1 Protein in Plant Photosystem II: A Combined Functional and Molecular Docking Study. Plants 2021, 10, 1501. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Cao, H.-F.; Wang, C.-F.; Gao, S.; Wang, J.-Y.; Zhao, L.-X.; Ye, F.; Fu, Y. In Silico Approach of Novel HPPD/PDS Dual Target Inhibitors by Pharmacophore, AILDE and Molecular Docking. J. Taiwan Inst. Chem. Eng. 2023, 143, 104711. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, T.; Yang, X.; Wang, L.; Long, Y.; Hasi, A.; Pei, X. A LuALS Mutation with High Sulfonylurea Herbicide Resistance in Linum usitatissimum L. Int. J. Mol. Sci. 2023, 24, 2820. [Google Scholar] [CrossRef]
- Statsoft.com. 2016. STATISTICA|New Features in STATISTICA 12. Available online: http://www.statsoft.com/Products/STATISTICA-Features/Version-12 (accessed on 1 February 2024).
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Chemical Biology: Methods and Protocols; Hempel, J.E., Williams, C.H., Hong, C.C., Eds.; Springer: New York, NY, USA, 2015; pp. 243–250. ISBN 978-1-4939-2269-7. [Google Scholar]
- Adamczewski, K.; Kaczmarek, S.; Kierzek, R.; Matysiak, K. Significant Increase of Weed Resistance to Herbicides in Poland. J. Plant Prot. Res. 2019, 59, 129293. [Google Scholar] [CrossRef]
- Scarabel, L.; Varotto, S.; Sattin, M. A European Biotype of Amaranthus retroflexus Cross-Resistant to ALS Inhibitors and Response to Alternative Herbicides. Weed Res. 2007, 47, 527–533. [Google Scholar] [CrossRef]
- Li, W.; Cao, Y.; Liu, Z.; Wei, S.; Huang, H.; Lan, Y.; Sun, Y.; Huang, Z. Investigation of Resistance Mechanisms to Bentazone in Multiple Resistant Amaranthus retroflexus Populations. Pestic. Biochem. Physiol. 2022, 186, 105164. [Google Scholar] [CrossRef]
- Prado, R.; de Jorrín, J.; Garcia-Torres, L. Weed and Crop Resistance to Herbicides; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1997; ISBN 978-0-7923-4581-7. [Google Scholar]
- Kocurek, V.; Smutný, V.; Filová, J. Chlorophyll Fluorescence as an Instrument for the Assessment of Herbicide Efficacy. Cereal Res. Commun. 2009, 37, 289–292. [Google Scholar]
- Linn, A.I.; Košnarová, P.; Soukup, J.; Gerhards, R. Detecting Herbicide-Resistant Apera spica-venti with a Chlorophyll Fluorescence Agar Test. Plant Soil Environ. 2018, 64, 386–392. [Google Scholar] [CrossRef]
- Hassannejad, S.; Lotfi, R.; Ghafarbi, S.P.; Oukarroum, A.; Abbasi, A.; Kalaji, H.M.; Rastogi, A. Early Identification of Herbicide Modes of Action by the Use of Chlorophyll Fluorescence Measurements. Plants 2020, 9, 529. [Google Scholar] [CrossRef] [PubMed]
- Van Oorschot, J.L.P.; Van Leeuwen, P.H. Inhibition of Photosynthesis in Intact Plants of Biotypes Resistant or Susceptible to Atrazine and Cross-Resistance to Other Herbicides. Weed Res. 1988, 28, 223–230. [Google Scholar] [CrossRef]
- Norsworthy, J.K.; Talbert, R.E.; Hoagland, R.E. Chlorophyll Fluorescence for Rapid Detection of Propanil-Resistant Barnyardgrass (Echinochloa crus-galli). Weed Sci. 1998, 46, 163–169. [Google Scholar] [CrossRef]
- Linn, A.I.; Mink, R.; Peteinatos, G.G.; Gerhards, R. In-Field Classification of Herbicide-Resistant Papaver rhoeas and Stellaria media Using an Imaging Sensor of the Maximum Quantum Efficiency of Photosystem II. Weed Res. 2019, 59, 357–366. [Google Scholar] [CrossRef]
- Wang, P.; Peteinatos, G.; Li, H.; Brändle, F.; Pfündel, E.; Drobny, H.G.; Gerhards, R. Rapid Monitoring of Herbicide-Resistant Alopecurus Myosuroides Huds. Using Chlorophyll Fluorescence Imaging Technology. J. Plant Dis. Prot. 2018, 125, 187–195. [Google Scholar] [CrossRef]
- Oettmeier, W. Herbicide Resistance and Supersensitivity in Photosystem II. Cell. Mol. Life Sci. CMLS 1999, 55, 1255–1277. [Google Scholar] [CrossRef] [PubMed]
- Johanningmeier, U.; Sopp, G.; Brauner, M.; Altenfeld, U.; Orawski, G.; Oettmeier, W. Herbicide Resistance and Supersensitivity in Ala250 or Ala251 Mutants of the D1 Protein in Chlamydomonas reinhardtii. Pestic. Biochem. Physiol. 2000, 66, 9–19. [Google Scholar] [CrossRef]
- Aiach, A.; Ohmann, E.; Bodner, U.; Johanningmeier, U. A Herbicide Resistant Euglena Mutant Carrying a Ser to Thr Substitution at Position 265 in the D1 Protein of Photosystem II. Z. Für Naturforschung C 1992, 47, 245–248. [Google Scholar] [CrossRef]
- Bowie, J.U. Membrane Protein Folding: How Important Are Hydrogen Bonds? Curr. Opin. Struct. Biol. 2011, 21, 42–49. [Google Scholar] [CrossRef]
- Hubbard, R.E.; Kamran Haider, M. Hydrogen Bonds in Proteins: Role and Strength. In Encyclopedia of Life Sciences; Wiley: Hoboken, NJ, USA, 2010; ISBN 978-0-470-01617-6. [Google Scholar]
- Rost, B. Review: Protein Secondary Structure Prediction Continues to Rise. J. Struct. Biol. 2001, 134, 204–218. [Google Scholar] [CrossRef]




| Active Ingredient (a.i) | 0.01 N | 0.0316 N | 0.1 N | 0.316 N | 1 N | 3.16 N | 10 N | 31.6 N |
|---|---|---|---|---|---|---|---|---|
| terbuthylazine | 7.5 | 23.7 | 75 | 237 | 750 | 2370 | 7500 | 23,700 |
| metamitron | 14 | 44.24 | 140 | 442.4 | 1440 | 4424 | 14,000 | 44,240 |
| Herbicide | Biotype | GR50 (g a.i. ha−1) 1 | CI 2 | RF 3 |
|---|---|---|---|---|
| terbuthylazine | R1 | 5512.5 | 4081.5–6943.5 | 120.49 |
| R2 | >23,700 | - | >518.032 | |
| S | 45.75 | 33.75–56.25 | - | |
| metamitron | R1 | 3617.6 1 | 3362.8–3871 | 3.275 |
| R2 | 4825.8 | 4142.6–5511.8 | 4.369 | |
| S | 1104.6 | 1104.6–1020.6 | - |
| Protein Name | Template | Sequence Identity | Description | GMQE | QMEANDisCoGlobal | Ramachandran Favored |
|---|---|---|---|---|---|---|
| Wild type | 4yuu.1.A | 89.24% | Crystal structure of oxygen-evolving photosystem II from a red alga | 0.84 | 0.75 ± 0.05 | 96.49% |
| S264G | 4yuu.1.A | 88.95% | Crystal structure of oxygen-evolving photosystem II from a red alga | 0.84 | 0.74 ± 0.05 | 96.49% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikulka, J.; Sen, M.K.; Košnarová, P.; Hamouz, P.; Hamouzová, K.; Sur, V.P.; Šuk, J.; Bhattacharya, S.; Soukup, J. Molecular Mechanisms of Resistance against PSII-Inhibiting Herbicides in Amaranthus retroflexus from the Czech Republic. Genes 2024, 15, 904. https://doi.org/10.3390/genes15070904
Mikulka J, Sen MK, Košnarová P, Hamouz P, Hamouzová K, Sur VP, Šuk J, Bhattacharya S, Soukup J. Molecular Mechanisms of Resistance against PSII-Inhibiting Herbicides in Amaranthus retroflexus from the Czech Republic. Genes. 2024; 15(7):904. https://doi.org/10.3390/genes15070904
Chicago/Turabian StyleMikulka, Jakub, Madhab Kumar Sen, Pavlína Košnarová, Pavel Hamouz, Kateřina Hamouzová, Vishma Pratap Sur, Jaromír Šuk, Soham Bhattacharya, and Josef Soukup. 2024. "Molecular Mechanisms of Resistance against PSII-Inhibiting Herbicides in Amaranthus retroflexus from the Czech Republic" Genes 15, no. 7: 904. https://doi.org/10.3390/genes15070904
APA StyleMikulka, J., Sen, M. K., Košnarová, P., Hamouz, P., Hamouzová, K., Sur, V. P., Šuk, J., Bhattacharya, S., & Soukup, J. (2024). Molecular Mechanisms of Resistance against PSII-Inhibiting Herbicides in Amaranthus retroflexus from the Czech Republic. Genes, 15(7), 904. https://doi.org/10.3390/genes15070904








