Analysis of the Mitochondrial COI Gene and Genetic Diversity of Endangered Goose Breeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Animals
2.2. DNA Extraction
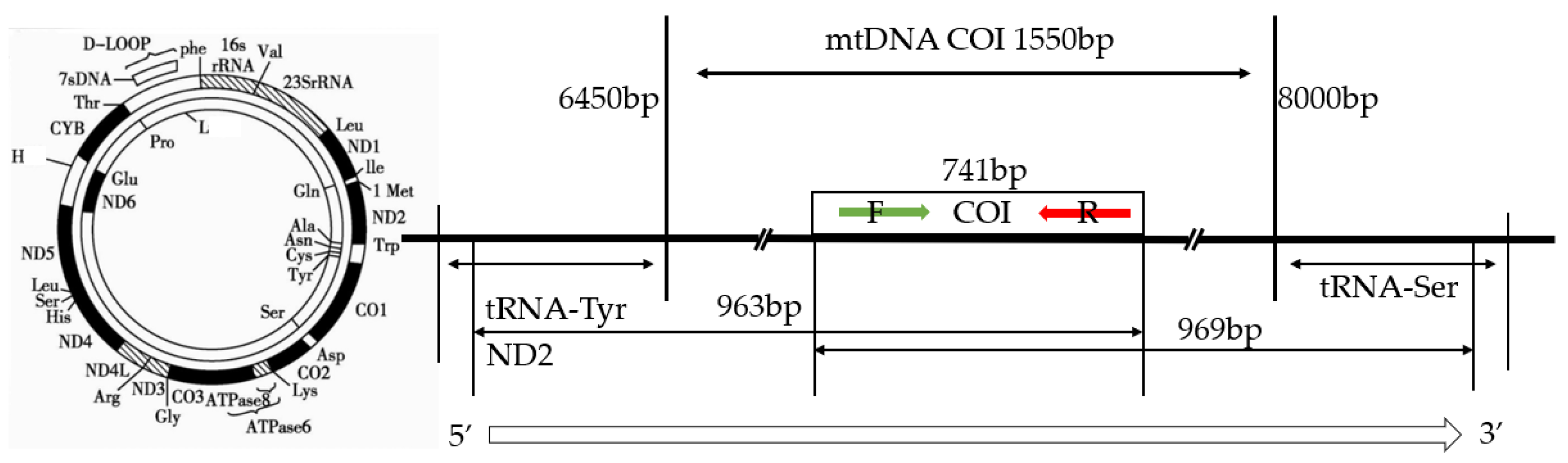
2.3. Primer Design, Gene Amplification, and Sequence Determination
2.4. Statistical Analysis
3. Results
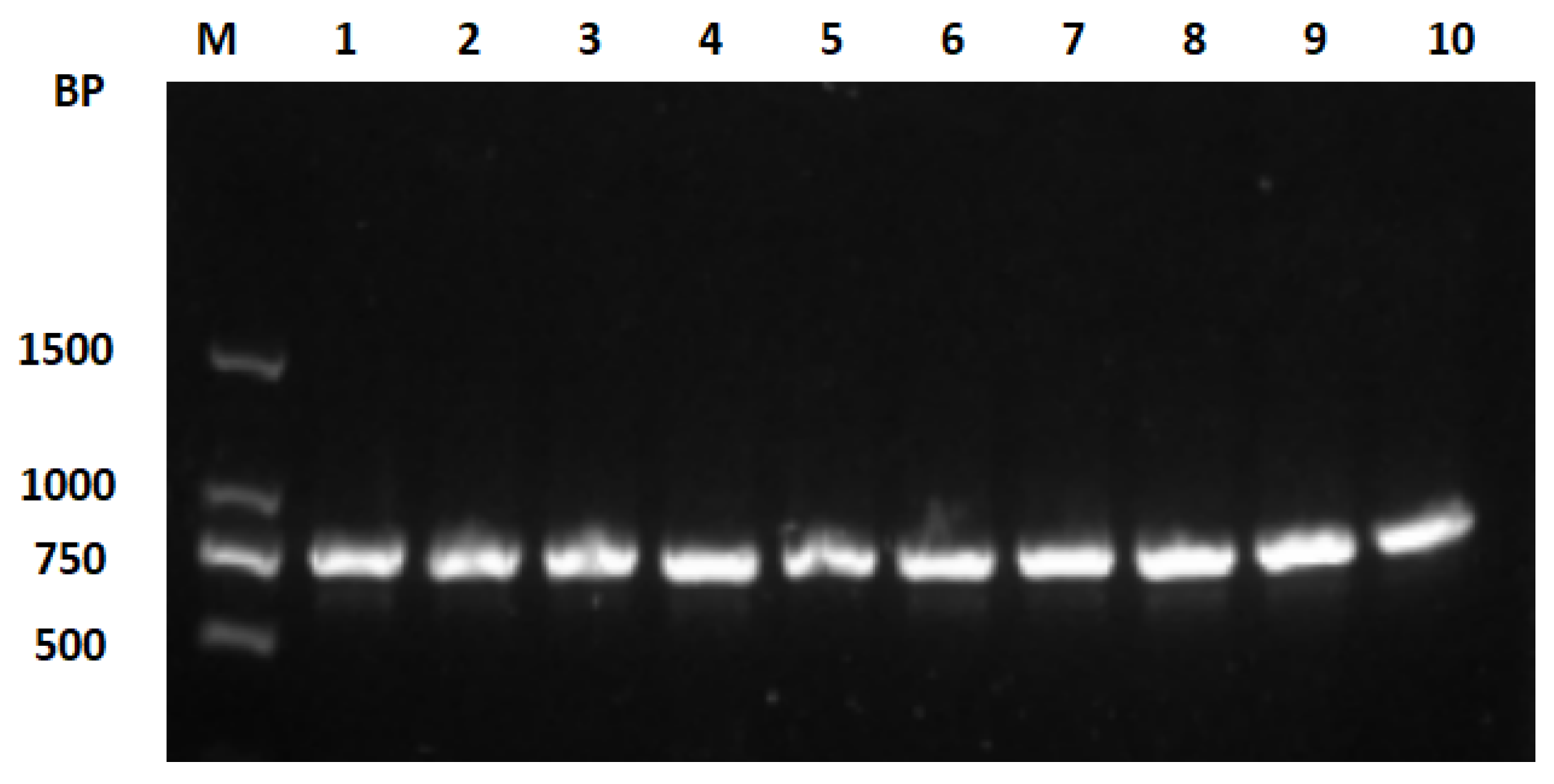
3.1. DNA Extraction, COI Gene Amplification, and Sequencing Results
3.2. Population Genetic Diversity
3.2.1. Locus Information and Nucleic Acid Diversity Analysis
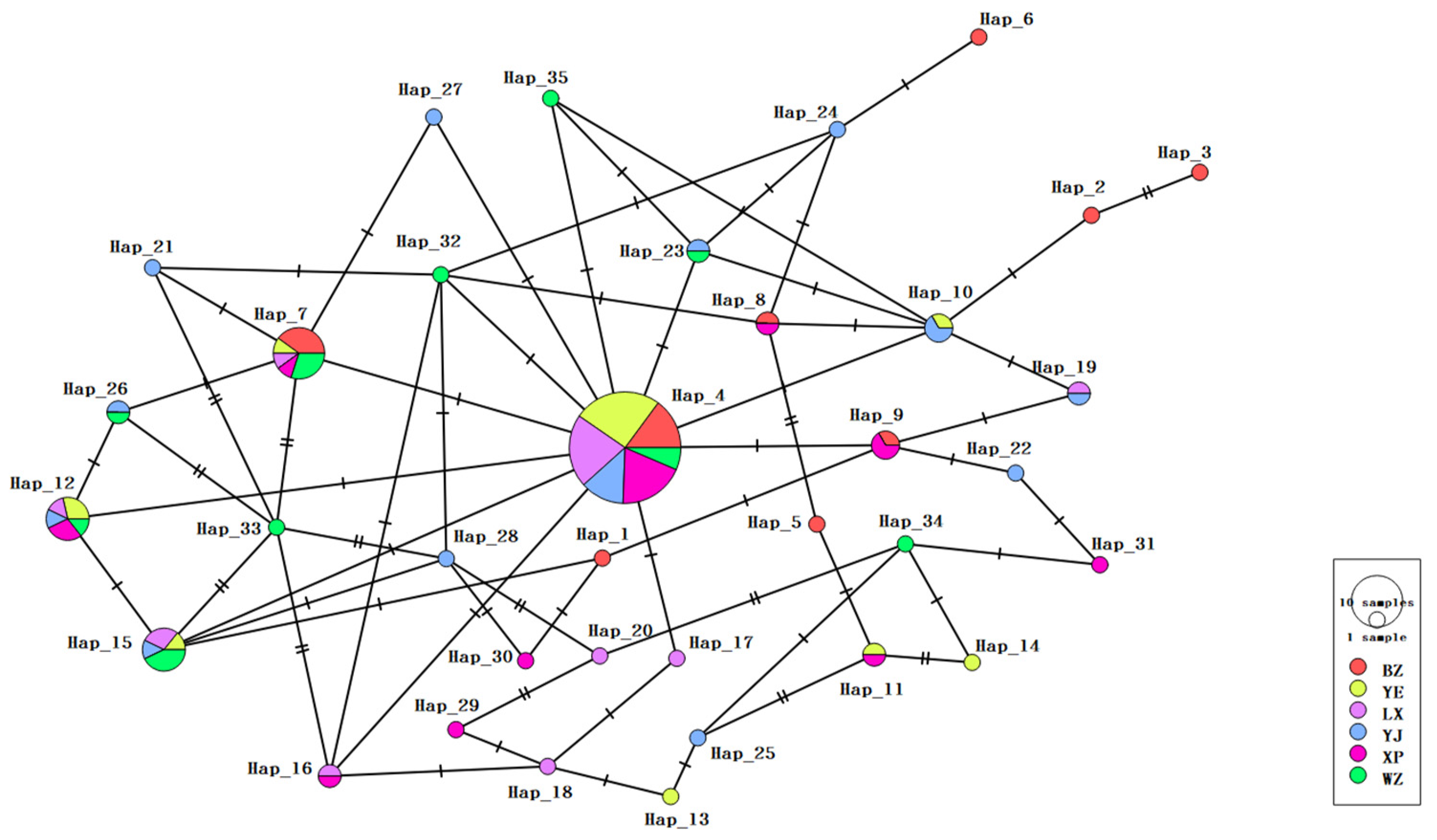
3.2.2. Haplotype Distribution and Frequency Analysis
3.3. Genetic Variation among Populations
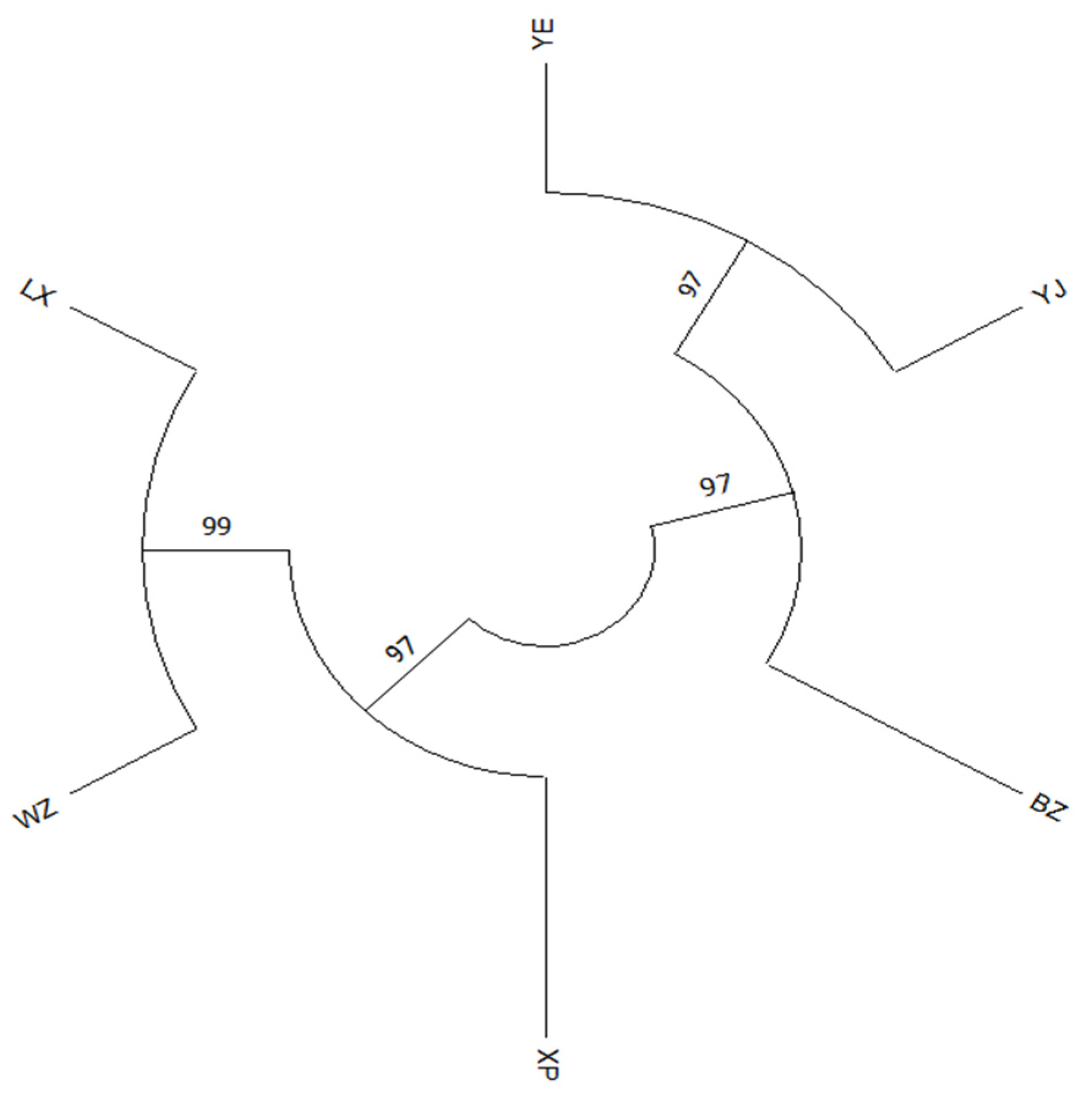
3.4. Clustering Relationship Analysis
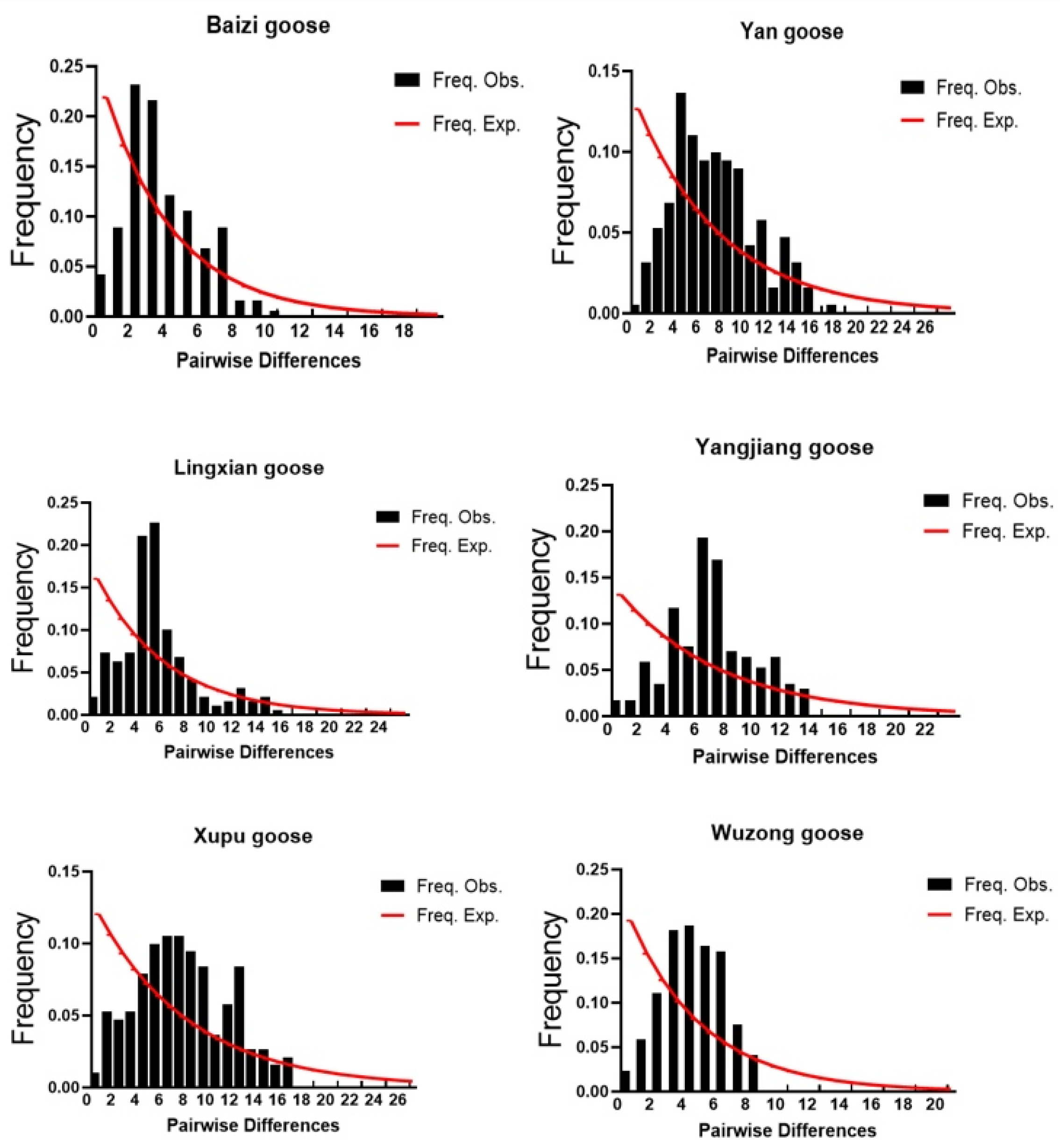
3.5. Analysis of Historical Population Dynamics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, H.F.; Zhu, W.Q.; Chen, K.W.; Xu, W.J.; Song, W. Two maternal origins of Chinese domestic goose. Poult. Sci. 2011, 90, 2705–2710. [Google Scholar] [CrossRef]
- Eda, M.; Itahashi, Y.; Kikuchi, H.; Sun, G.; Hsu, K.-H.; Gakuhari, T.; Yoneda, M.; Jiang, L.; Yang, G.; Nakamura, S. Multiple lines of evidence of early goose domestication in a 7,000-y-old rice cultivation village in the lower Yangtze River, China. Proc. Natl. Acad. Sci. USA 2022, 119, e2117064119. [Google Scholar] [CrossRef]
- Utnik-Banaś, K.; Żmija, J. The geese market in Poland. Roczniki 2018, 2018, 158–163. [Google Scholar] [CrossRef]
- Blackburn, H.D. The national animal germplasm program: Challenges and opportunities for poultry genetic resources. Poult. Sci. 2006, 85, 210–215. [Google Scholar] [CrossRef]
- Qi, S.; Xu, X.; Liu, L.; Wang, G.; Bao, Q.; Zhang, Y.; Zhang, Y.; Zhang, Y.; Xu, Q.; Zhao, W.; et al. The development rule of feathers and application of hair root tissue in sex identification of Yangzhou geese. Poult Sci. 2024, 103, 103529. [Google Scholar] [CrossRef]
- Abbitt, R.J.F.; Scott, J.M. Examining differences between recovered and declining endangered species. Conserv. Biol. 2001, 15, 1274–1284. [Google Scholar] [CrossRef]
- Si, Y.; Wei, J.; Wu, W.; Zhang, W.; Hou, L.; Yu, L.; Wielstra, B. Reducing human pressure on farmland could rescue China’s declining wintering geese. Mov Ecol. 2020, 8, 35. [Google Scholar] [CrossRef]
- Rosenberg, K.V.; Dokter, A.M.; Blancher, P.J.; Sauer, J.R.; Smith, A.C.; Smith, P.A.; Stanton, J.C.; Panjabi, A.; Helft, L.; Parr, M.; et al. Decline of the North American avifauna. Science 2019, 366, 120–124. [Google Scholar] [CrossRef]
- Zhang, Y.; Qi, S.; Liu, L.; Bao, Q.; Wu, T.; Liu, W.; Zhang, Y.; Zhao, W.; Xu, Q.; Chen, G. Genetic Diversity Analysis and Breeding of Geese Based on the Mitochondrial ND6 Gene. Genes 2023, 14, 1605. [Google Scholar] [CrossRef]
- Li, S.; Li, D.; Zhang, L.; Shang, W.; Qin, B.; Jiang, Y. High levels of genetic diversity and an absence of genetic structure among breeding populations of the endangered Rufous-backed Bunting in Inner Mongolia, China: Implications for conservation. Avian Res. 2021, 12, 1–11. [Google Scholar]
- Moniem, H.A.; Fathy, A.; Chen, G. Evaluation of the genetic diversity of some rare geese breeds based on microsatellite markers. Eur. Poult. Sci. Arch. Geflügelkunde 2020, 84, 84. [Google Scholar]
- Dobrzański, J.; Calik, J.; Krawczyk, J.; Szwaczkowski, T. Conservation of goose genetic resources in Poland-past and present status. World’s Poult. Sci. J. 2019, 75, 387–400. [Google Scholar] [CrossRef]
- Zheng, Z.H.; Wu, S.S.; Wei, Y.; Zhong, D.B.; Zheng, X.L. Analysis of the genetic diversity of 15 Aedes albopictus populations in Guangzhou based on the mitochondrial COI gene. Chin. J. Zoonoses 2021, 37, 985–994. [Google Scholar]
- Adams, C.I.; Knapp, M.; Gemmell, N.J.; Jeunen, G.J.; Bunce, M.; Lamare, M.D.; Taylor, H.R. Beyond Biodiversity: Can Environmental DNA (eDNA) Cut It as a Population Genetics Tool? Genes 2019, 10, 192. [Google Scholar] [CrossRef]
- Gonçalves, R.M.; Mastrangelo, T.; Rodrigues, J.C.V.; Paulo, D.F.; Omoto, C.; Corrêa, A.S.; de Azeredo-Espin, A.M.L. Invasion origin, rapid population expansion, and the lack of genetic structure of cotton bollworm (Helicoverpa armigera) in the Americas. Ecol. Evol. 2019, 9, 7378–7401. [Google Scholar] [CrossRef]
- Park, M.H.; Jung, J.H.; Jo, E.; Park, K.M.; Baek, Y.S.; Kim, S.J.; Min, G.S. Utility of mitochondrial CO1 sequences for species discrimination of Spirotrichea ciliates (Protozoa, Ciliophora). Mitochondrial DNA Part A 2019, 30, 148–155. [Google Scholar] [CrossRef]
- Chan, K.O.; Hertwig, S.T.; Neokleous, D.N.; Flury, J.M.; Brown, R.M. Widely used, short 16S rRNA mitochondrial gene fragments yield poor and erratic results in phylogenetic estimation and species delimitation of amphibians. BMC Ecol. Evol. 2022, 22, 37. [Google Scholar] [CrossRef]
- Deef, L.E.M. Phylogenetic Relationships among Puddle Duck Species in Egypt using COI Gene Variations in mtDNA. Russ. J. Genet. 2021, 57, 1189–1197. [Google Scholar] [CrossRef]
- Azimu, W.; Manatbay, B.; Li, Y.; Kaimaerdan, D.; Wang, H.E.; Reheman, A.; Muhatai, G. Genetic diversity and population structure analysis of eight local chicken breeds of Southern Xinjiang. Br. Poult. Sci. 2018, 59, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, Y.; Zhang, W.; Wang, Y.; Liu, X.; Cui, A.; Gong, Y.; Lu, J.; Liu, X.; Huo, X.; et al. Development of Microsatellite Marker System to Determine the Genetic Diversity of Experimental Chicken, Duck, Goose, and Pigeon Populations. Biomed. Res. Int. 2021, 14, 8851888. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Huo, C.; Zhong, Q.; Xu, M.; Yang, Y.; Tian, H.; Zhang, G.; Hu, Y. Isolation and pathogenicity testing of avian reticuloendotheliosis virus from layer chickens in China. J. Vet. Diagn. Investig. 2020, 32, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Fan, Y.; Jia, X.; Ge, Q.; Lu, J.; Tang, M.; Han, W.; Gao, Y. Genetic diversity and origin characteristics of chicken species based on mitochondrial DNA D-loop region. Sci. Agric. Sin. 2021, 24, 5302–5315. [Google Scholar]
- Yang, M.; Ma, Y.; Wu, J. Mitochondrial genetic differentiation across populations of the malaria vector Anopheles lesteri from China (Diptera: Culicidae). Malar. J. 2011, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Van den Biggelaar, R.H.G.A.; Van Eden, W.; Rutten, V.P.M.G.; Jansen, C.A. Macrophage activation assays to evaluate the immunostimulatory capacity of Avibacterium paragallinarum in A multivalent poultry vaccine. Vaccines 2020, 8, 671. [Google Scholar] [CrossRef] [PubMed]
- Beserra, L.A.R.; Barbosa, B.R.P.; Bernardes, N.T.C.G.; Brandão, P.E.; Gregori, F. Occurrence and characterization of rotavirus A in broilers, layers, and broiler breeders from Brazilian poultry farms. Avian Dis. 2014, 58, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wen, J.; Zhang, X.; Zhang, J.; Zhu, T.; Wang, H.; Yang, W.; Cao, G.; Xiong, W.; Liu, Y.; et al. Significant genomic introgression from grey junglefowl (Gallus sonneratii) to domestic chickens (Gallus gallus domesticus). J. Anim. Sci. Biotechnol. 2024, 15, 45. [Google Scholar] [CrossRef]
- Bounas, A.; Catsadorakis, G.; Logotheti, A.; Voslamber, B.; Magoulas, A.; Tsaparis, D. Conservation genetics of a resident population of Greylag Goose (Anser anser) at the southernmost limit of the species’ distribution in Europe. Avian Res. 2018, 9, 1–10. [Google Scholar]
- Honka, J. Evolutionary and Conservation Genetics of European Domestic and Wild Geese; University of Oulu: Honka, Johanna, 2020. [Google Scholar]
- Zhang, Y.; Wang, L.; Bian, Y.; Wang, Z.; Xu, Q.; Chang, G.; Chen, G. Marginal diversity analysis of conservation of Chinese domestic duck breeds. Sci. Rep. 2019, 9, 13141. [Google Scholar] [CrossRef] [PubMed]
- Díez-Del-Molino, D.; von Seth, J.; Gyllenstrand, N.; Widemo, F.; Liljebäck, N.; Svensson, M.; Sjögren-Gulve, P.; Dalén, L. Population genomics reveals lack of greater white-fronted introgression into the Swedish lesser white-fronted goose. Sci. Rep. 2020, 10, 18347. [Google Scholar] [CrossRef]
- Zhuoxian, W.; Jiaqiong, H.; Shihao, Z.; Kaichun, Y.; Fusheng, Z.; Xunhe, H.; Bin, Z. Genetic diversity and population structure of black-bone chickens in China revealed by mitochondrial COI gene sequences. Biodivers. Sci. 2019, 27, 667. [Google Scholar] [CrossRef]
- Yang, J.; Ding, H.; Kan, X. Codon usage patterns and evolution of HSP60 in birds. Int. J. Biol. Macromol. 2021, 183, 1002–1012. [Google Scholar] [CrossRef] [PubMed]
- Teinlek, P.; Siripattarapravat, K.; Tirawattanawanich, C. Genetic diversity analysis of Thai indigenous chickens based on complete sequences of mitochondrial DNA D-loop region. Asian-Australas. J. Anim. Sci. 2018, 31, 804. [Google Scholar] [CrossRef] [PubMed]
- Nisar, A.; Waheed, A.; Khan, S.; Feng, X.; Shah, A.H. Population structure, genetic diversity and phylogenetic analysis of different rural and commercial chickens of Pakistan using complete sequence of mtDNA D-loop. Mitochondrial DNA Part A 2019, 30, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bao, Q.; Cao, Z.; Bian, Y.; Zhang, Y.; Cao, Z.; Chen, G.; Xu, Q. Chinese Domestic Ducks Evolved from Mallard Duck (Anas platyrhynchos) and Spot-Billed Duck (A. zonorhyncha). Animals 2023, 13, 1156. [Google Scholar] [CrossRef] [PubMed]
- Rumanta, M.; Kunda, R.M.; Volkandari, S.D.; Indriawati, I.; Kakisina, P. Genetic characterization and phylogenetic study of Lakor goat from Southwest Maluku Regency based on mitochondrial COI gene. Vet World 2020, 13, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Liang, C.; Dai, M.; Wang, X.; Zhang, X.; Song, Z. DNA barcoding and phylogenetic analysis of bagrid catfish in China based on mitochondrial COI gene. Mitochondrial DNA A DNA Mapp Seq Anal. 2020, 31, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021, 30, 62–82. [Google Scholar] [CrossRef]
- Warzecha, J.; Oczkowicz, M.; Rubis, D.; Fornal, A.; Szmatoła, T.; Bugno-Poniewierska, M. An evaluation of the genetic structure of geese maintained in poland on the basis of microsatellite markers. Animals 2019, 9, 737. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, M.; Di Marco, M.; Watson, J.E.M. Protected areas are now the last strongholds for many imperiled mammal specie. Conserv. Lett. 2020, 13, e12748. [Google Scholar] [CrossRef]
- De, A.K.; Sawhney, S.; Bhattacharya, D.; Sujatha, T.; Sunder, J.; Ponraj, P.; Ravi, S.K.; Mondal, S.; Malakar, D.; Kundu, A. Origin, genetic diversity and evolution of Andaman local duck, a native duck germplasm of an insular region of India. PLoS ONE 2021, 16, e0245138. [Google Scholar] [CrossRef]
- Ran, B.; Zhu, W.; Zhao, X.; Li, L.; Yi, Z.; Li, M.; Wang, T.; Li, D. Studying Genetic Diversity and Relationships between Mountainous Meihua Chickens Using Mitochondrial DNA Control Region. Genes 2023, 14, 998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, S.; Xu, R.; Liu, Y.; Zhang, H.; Sun, M.; Wang, J.; Liu, Z.; Wu, K. Managing genomic diversity in conservation programs of Chinese domestic chickens. Genet. Sel. Evol. 2023, 55, 92. [Google Scholar] [CrossRef] [PubMed]
- Scanes, C.G.; Butler, L.D.; Kidd, M.T. Reproductive management of poultry. In Animal Agriculture; Academic Press: Cambridge, MA, USA, 2020; pp. 349–366. [Google Scholar]
- Sun, Y.; Li, Y.; Zong, Y.; Mehaisen, G.M.K.; Chen, J. Poultry genetic heritage cryopreservation and reconstruction: Advancement and future challenges. J. Anim. Sci. Biotechnol. 2022, 13, 1–18. [Google Scholar]
- Ozbay, G.; Khatiwada, R.; Smith, S.; Chintapenta, L.K. Efficacy of Heavy Use Area Protection (HUAP) Pads in Poultry Farm. Agriculture 2021, 11, 154. [Google Scholar] [CrossRef]
- Bessei, W. Impact of animal welfare on worldwide poultry production. World Poult. Sci. J. 2018, 74, 211–224. [Google Scholar] [CrossRef]


| Breed | Origin | Livestock Purpose | Physical Characteristics |
|---|---|---|---|
| XP | Xupu, Hunan | Meat and liver | Large body; oblong-cylindrical in shape |
| YJ | Yangjiang, Guangdong | Meat (small breed) | Compact body |
| BZ | Jinxiang, Shandong | Eggs | Medium-sized head |
| WZ | Qingyuan, Guangdong | Meat (small breed) | Compact body, ebony feathers |
| YE | Liuan, Anhui | Meat (medium-sized breed) | Large body; rectangular in shape |
| LX | Yanling, Hunan | Meat (small breed) | Small and compact body; cylindrical in shape |
| Breed | MS | NPIS | PIS | GC | K | Pi | Hd |
|---|---|---|---|---|---|---|---|
| XP | 715 | 4 | 16 | 51.01% | 7.305 | 0.0099 ± 0.00130 | 0.989 ± 0.019 |
| YJ | 716 | 4 | 12 | 50.62% | 6.608 | 0.0090 ± 0.00088 | 0.982 ± 0.026 |
| BZ | 709 | 3 | 8 | 50.33% | 3.568 | 0.0049 ± 0.00079 | 0.958 ± 0.033 |
| WZ | 711 | 2 | 7 | 50.54% | 4.199 | 0.0058 ± 0.00041 | 0.977 ± 0.023 |
| YE | 718 | 4 | 15 | 50.71% | 6.905 | 0.0093 ± 0.00110 | 0.995 ± 0.018 |
| LX | 717 | 3 | 14 | 50.93% | 5.237 | 0.0071 ± 0.00110 | 0.979 ± 0.024 |
| Haplotype | Breed and Number of Individuals | Total Number of Individuals with Haplotype |
|---|---|---|
| H1 | [BZ1] | 1 |
| H2 | [BZ1] | 1 |
| H3 | [BZ1] | 1 |
| H4 | [BZ7 YE12 LX10 YJ6 XP9 WZ3] | 47 |
| H5 | [BZ1] | 1 |
| H6 | [BZ1] | 1 |
| H7 | [BZ4 YE1 LX1 XP1 WZ3] | 10 |
| H8 | [BZ1 XP1] | 2 |
| H9 | [BZ1 XP2] | 3 |
| H10 | [YE1 YJ2] | 3 |
| H11 | [YE1 XP1] | 2 |
| H12 | [YE2 LX1 YJ1 XP2 WZ1] | 7 |
| H13 | [YE1] | 1 |
| H14 | [YE1] | 1 |
| H15 | [YE1 LX2 YJ1 WZ3] | 7 |
| H16 | [LX1 XP1] | 2 |
| H17 | [LX1] | 1 |
| H18 | [LX1] | 1 |
| H19 | [LX1 YJ1] | 2 |
| H20 | [LX1] | 1 |
| H21 | [YJ1] | 1 |
| H22 | [YJ1] | 1 |
| H23 | [YJ1 WZ1] | 2 |
| H24 | [YJ1] | 1 |
| H25 | [YJ1] | 1 |
| H26 | [YJ1 WZ1] | 2 |
| H27 | [YJ1] | 1 |
| H28 | [YJ1] | 1 |
| H29 | [XP1] | 1 |
| H30 | [XP1] | 1 |
| H31 | [XP1] | 1 |
| H32 | [WZ1] | 1 |
| H33 | [WZ1] | 1 |
| H34 | [WZ1] | 1 |
| H35 | [WZ1] | 1 |
| Breed | Haplotype Number | Number of Unique Haplotypes | Unique Single/Double Type Frequency |
|---|---|---|---|
| WZ | 15 | 4 (Hap32, Hap33, Hap34, Hap35) | 26.66% |
| XP | 18 | 3 (Hap29, Hap30, Hap31) | 16.66% |
| YJ | 17 | 6 (Hap21, Hap22, Hap24, Hap25, Hap27, Hap28) | 35.29% |
| LX | 17 | 3 (Hap17, Hap18, Hap20) | 17.64% |
| YE | 19 | 2 (Hap13, Hap14) | 10.52% |
| BZ | 15 | 6 (Hap1, Hap2, Hap3, Hap4, Hap5, Hap6) | 40.00% |
| Goose Breed | BZ | YE | LX | YJ | XP | WZ |
|---|---|---|---|---|---|---|
| BZ | −37.34 | 23.25 | −29.21 | 50.14 | Inf | |
| YE | −0.01349 | −14.18 | −28.50 | −15.46 | 24.93 | |
| LX | 0.02105 | −0.03072 | 4.77 | Inf | Inf | |
| YJ | 0.03781 | −0.02864 | −0.00084 | Inf | 67.06 | |
| XP | 0.00087 | −0.03115 | −0.00349 | −0.02390 | 18.40 | |
| WZ | 0.00079 | 0.01966 | 0.00869 | −0.00726 | 0.02645 |
| Source of Variation | Degrees of Freedom | Square Sum | Variance Component | Percentage of Variation |
|---|---|---|---|---|
| Interpopulation | 5 | 3.916 | −0.00684 | −0.76 |
| Intrapopulation | 106 | 96.539 | 0.91075 | 100.76 |
| Total | 111 | 100.455 | 0.90391 | 100.00 |
| Breed | BZ | YE | LX | YJ | XP | WZ |
|---|---|---|---|---|---|---|
| BZ | 0.01002 | 0.00988 | 0.01021 | 0.01013 | 0.01110 | |
| YE | 0.01002 | 0.00918 | 0.00963 | 0.00965 | 0.01054 | |
| LX | 0.00988 | 0.00918 | 0.00920 | 0.00938 | 0.01020 | |
| YJ | 0.01021 | 0.00963 | 0.00920 | 0.00989 | 0.01076 | |
| XP | 0.01013 | 0.00965 | 0.00938 | 0.00989 | 0.01075 | |
| WZ | 0.01110 | 0.01054 | 0.01020 | 0.01076 | 0.01075 |
| Metric | BZ | YE | LX | YJ | XP | WZ |
|---|---|---|---|---|---|---|
| Fu’s Fs | −9.0600 | −12.5530 | −10.0230 | −9.1800 | −9.4990 | −8.4730 |
| p value | p > 0.10 | p > 0.10 | p > 0.10 | p > 0.10 | p > 0.10 | p > 0.10 |
| Tajima’s D | −2.5156 | −0.0777 | −0.5958 | −0.2694 | −0.4158 | −0.1789 |
| p value | p > 0.10 | p > 0.10 | p > 0.10 | p > 0.10 | p > 0.10 | p > 0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Qi, S.; Fan, S.; Li, H.; Zhang, Y.; Zhang, Y.; Xu, Q.; Chen, G. Analysis of the Mitochondrial COI Gene and Genetic Diversity of Endangered Goose Breeds. Genes 2024, 15, 1037. https://doi.org/10.3390/genes15081037
Wu H, Qi S, Fan S, Li H, Zhang Y, Zhang Y, Xu Q, Chen G. Analysis of the Mitochondrial COI Gene and Genetic Diversity of Endangered Goose Breeds. Genes. 2024; 15(8):1037. https://doi.org/10.3390/genes15081037
Chicago/Turabian StyleWu, Hao, Shangzong Qi, Suyu Fan, Haoyu Li, Yu Zhang, Yang Zhang, Qi Xu, and Guohong Chen. 2024. "Analysis of the Mitochondrial COI Gene and Genetic Diversity of Endangered Goose Breeds" Genes 15, no. 8: 1037. https://doi.org/10.3390/genes15081037






