WRKY10 Regulates Seed Size through the miR397a-LAC2 Module in Arabidopsis thaliana
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Plasmid Construction
2.3. Dual-Luciferase Assay
2.4. Protein Expression and Purification
2.5. Electrophoretic Mobility Shift Assay
2.6. RNA Isolation and qRT-PCR
2.7. Seed Size Measurement
3. Results
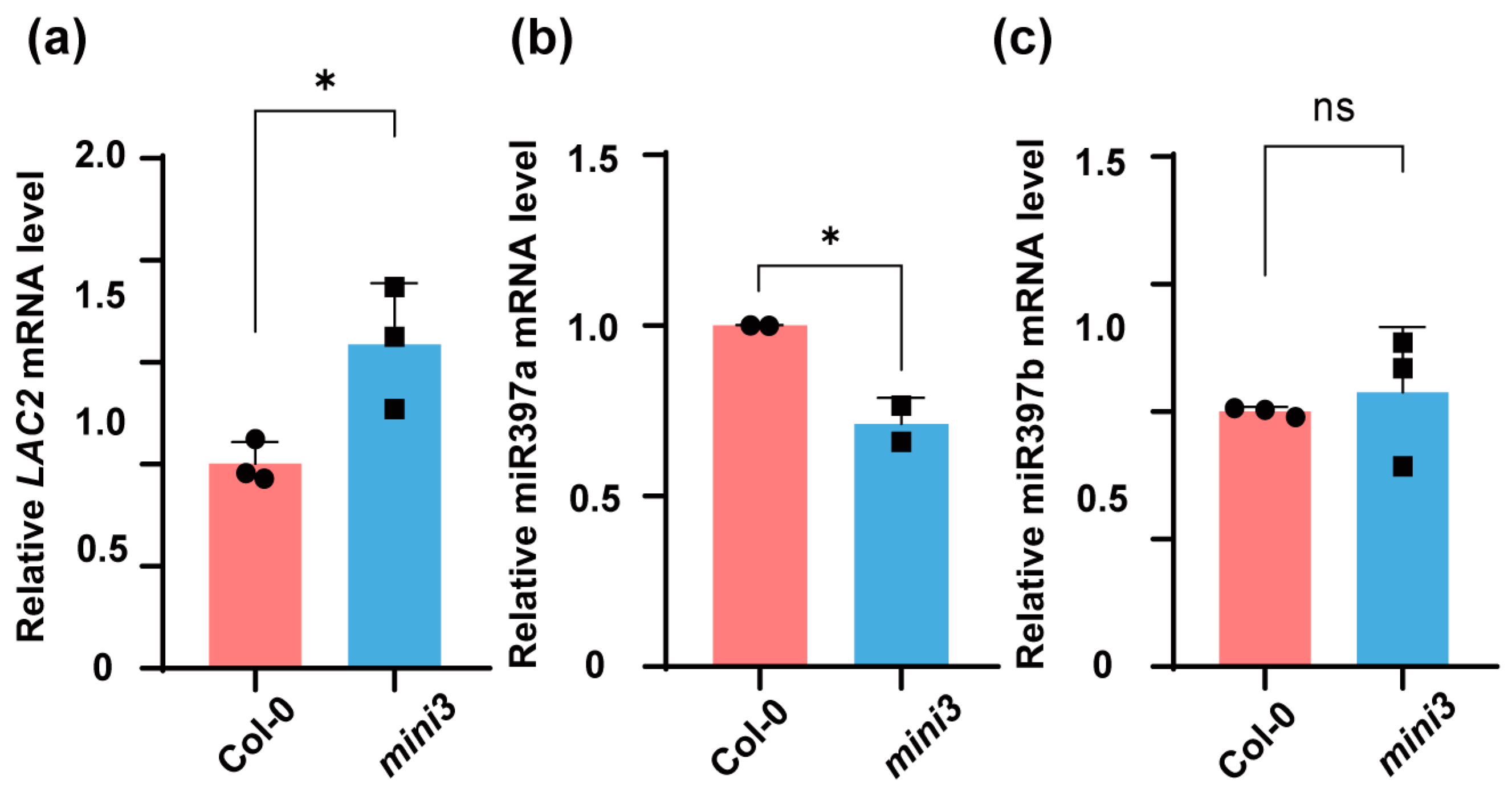
3.1. The LAC2 Is Upregulated in mini3 Seeds
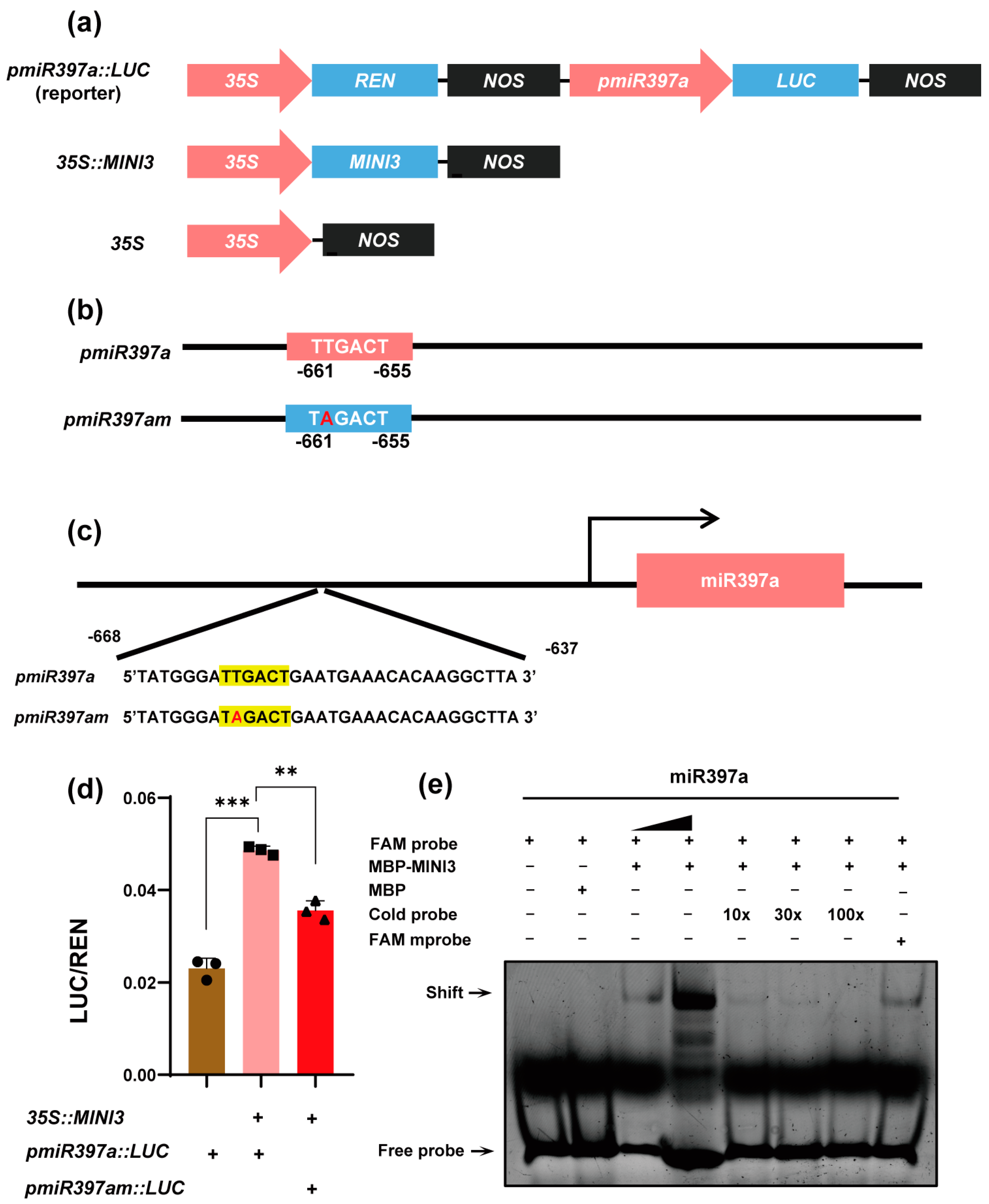
3.2. WRKY10 Directly Regulates the Expression of miR397a
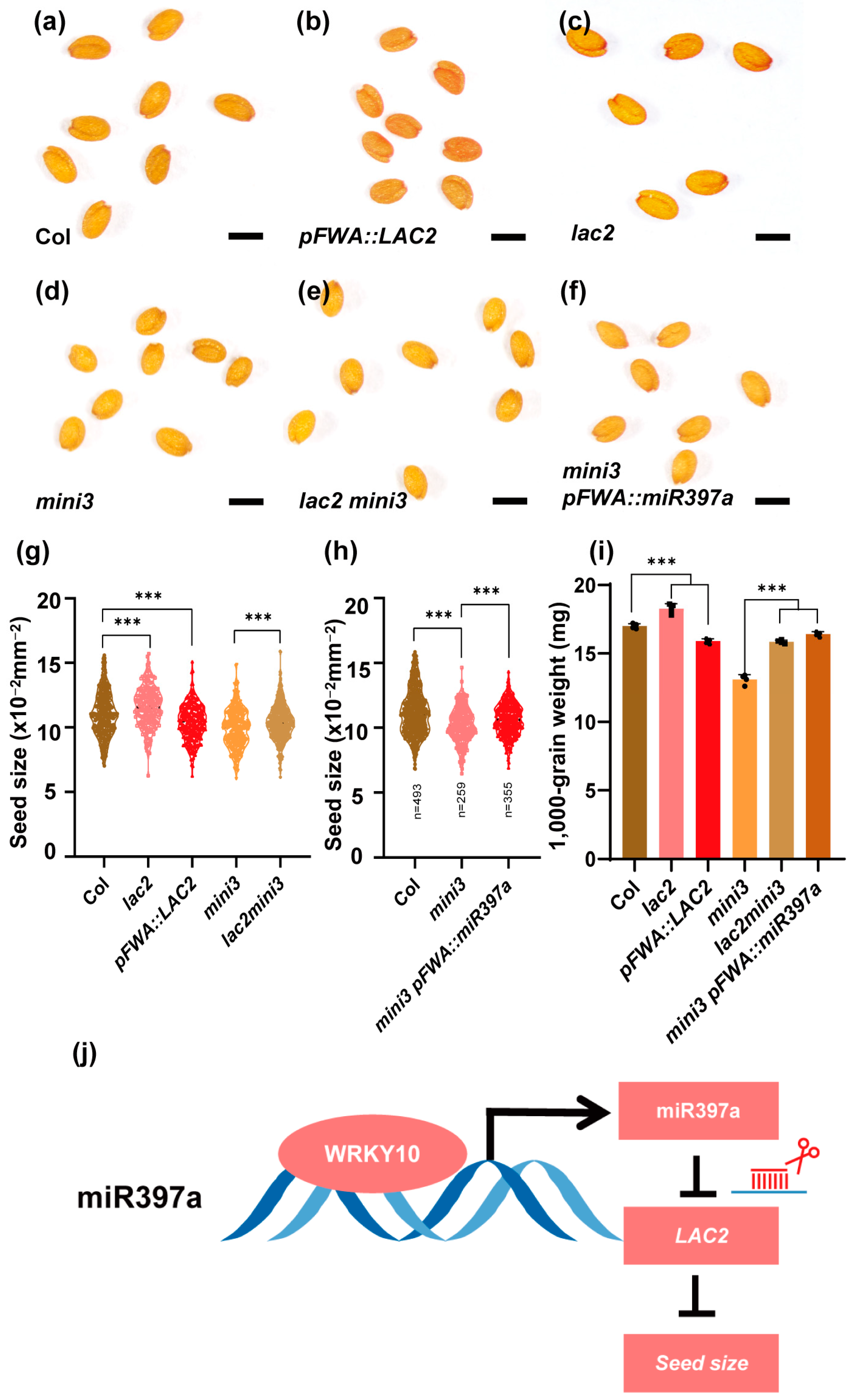
3.3. LAC2 and miR397a Act Downstream of MINI3 and Are Involved in Seed Size Regulation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nowack, M.K.; Grini, P.E.; Jakoby, M.J.; Lafos, M.; Koncz, C.; Schnittger, A. A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 2006, 38, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Saingery, V.; Chambrier, P.; Mayer, U.; Jurgens, G.; Berger, F. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003, 131, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, V. Control of seed size in plants. Proc. Natl. Acad. Sci. USA 2005, 102, 17887–17888. [Google Scholar] [CrossRef]
- Garcia, D.; Fitz Gerald, J.N.; Berger, F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 2005, 17, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Dennis, E.S.; Berger, F.; Peacock, W.J.; Chaudhury, A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 17531–17536. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Garcia, D.; Zhang, H.; Feng, K.; Chaudhury, A.; Berger, F.; Peacock, W.J.; Dennis, E.S.; Luo, M. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010, 63, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nie, X.; Tan, J.L.; Berger, F. Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15479–15484. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Serrani-Yarce, J.C.; Chen, F.; Baxter, D.; Venables, B.J.; Dixon, R.A. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2016, 2, 16050. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cezard, L.; Le Bris, P.; Borrega, N.; Herve, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef]
- Blaschek, L.; Pesquet, E. Phenoloxidases in plants—How structural diversity enables functional specificity. Front. Plant Sci. 2021, 12, 754601. [Google Scholar] [CrossRef] [PubMed]
- Shigeto, J.; Itoh, Y.; Hirao, S.; Ohira, K.; Fujita, K.; Tsutsumi, Y. Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J. Integr. Plant Biol. 2015, 57, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Tobimatsu, Y.; Schuetz, M. Lignin polymerization: How do plants manage the chemistry so well? Curr. Opin. Biotechnol. 2019, 56, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Yu, Y.; Wang, C.Y.; Li, Z.Y.; Liu, Q.; Xu, J.; Liao, J.Y.; Wang, X.J.; Qu, L.H.; Chen, F.; et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhou, J.; Gao, L.; Tang, Y. Plant miR397 and its functions. Funct. Plant Biol. 2021, 48, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Zhang, S.; Yu, Y.; Luo, Y.C.; Liu, Q.; Ju, C.; Zhang, Y.C.; Qu, L.H.; Lucas, W.J.; Wang, X.; et al. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 2014, 12, 1132–1142. [Google Scholar] [CrossRef]
- Gaddam, S.R.; Sharma, A.; Trivedi, P.K. miR397b-LAC2 module regulates cadmium stress response by coordinating root lignification and copper homeostasis in Arabidopsis thaliana. J. Hazard. Mater. 2023, 465, 133100. [Google Scholar] [CrossRef] [PubMed]
- Khandal, H.; Singh, A.P.; Chattopadhyay, D. The MicroRNA397b-LACCASE2 module regulates root lignification under water and phosphate deficiency. Plant Physiol. 2020, 182, 1387–1403. [Google Scholar] [CrossRef]
- Yu, Y. LACCASE2 negatively regulates lignin deposition of Arabidopsis roots. Plant Physiol. 2020, 182, 1190–1191. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Bensmihen, S.; To, A.; Lambert, G.; Kroj, T.; Giraudat, J.; Parcy, F. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 2004, 561, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Christensen, C.A.; King, E.J.; Jordan, J.R.; Drews, G.N. Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 1997, 10, 49–64. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.P.; Yuan, C.; Feng, Y.Z.; Liu, Q.; Wang, C.Y.; Zhou, Y.F.; Huang, Q.J.; Zhu, Q.F.; Zhang, Y.C.; Chen, Y.Q.; et al. MicroRNA397 promotes rice flowering by regulating the photorespiration pathway. Plant Physiol. 2023, 194, 2101–2116. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.Q.; Brown, C.W.; Pegler, J.L.; Eamens, A.L.; Grof, C.P.L. Molecular Manipulation of MicroRNA397 Abundance Influences the Development and Salt Stress Response of Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 7879. [Google Scholar] [CrossRef]
- Ali, S.; Huang, S.; Zhou, J.; Bai, Y.; Liu, Y.; Shi, L.; Liu, S.; Hu, Z.; Tang, Y. miR397-LACs mediated cadmium stress tolerance in Arabidopsis thaliana. Plant Mol. Biol. 2023, 113, 415–430. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Yang, K.; Ye, H.; Yao, J.; Li, J. WRKY10 Regulates Seed Size through the miR397a-LAC2 Module in Arabidopsis thaliana. Genes 2024, 15, 1040. https://doi.org/10.3390/genes15081040
Guo W, Yang K, Ye H, Yao J, Li J. WRKY10 Regulates Seed Size through the miR397a-LAC2 Module in Arabidopsis thaliana. Genes. 2024; 15(8):1040. https://doi.org/10.3390/genes15081040
Chicago/Turabian StyleGuo, Wenbin, Ke Yang, Hang Ye, Jialing Yao, and Jing Li. 2024. "WRKY10 Regulates Seed Size through the miR397a-LAC2 Module in Arabidopsis thaliana" Genes 15, no. 8: 1040. https://doi.org/10.3390/genes15081040
APA StyleGuo, W., Yang, K., Ye, H., Yao, J., & Li, J. (2024). WRKY10 Regulates Seed Size through the miR397a-LAC2 Module in Arabidopsis thaliana. Genes, 15(8), 1040. https://doi.org/10.3390/genes15081040




