Identification of Nonsynonymous SNPs in Immune-Related Genes Associated with Pneumonia Severity in Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pig Populations and Measurement of Trait Data
2.2. Targeted Resequencing
2.3. Genotyping
2.4. Data Analysis
3. Results
3.1. Characteristics of Pig Populations and Traits
3.2. Extraction and Narrowing Down of Target Genes and Polymorphisms
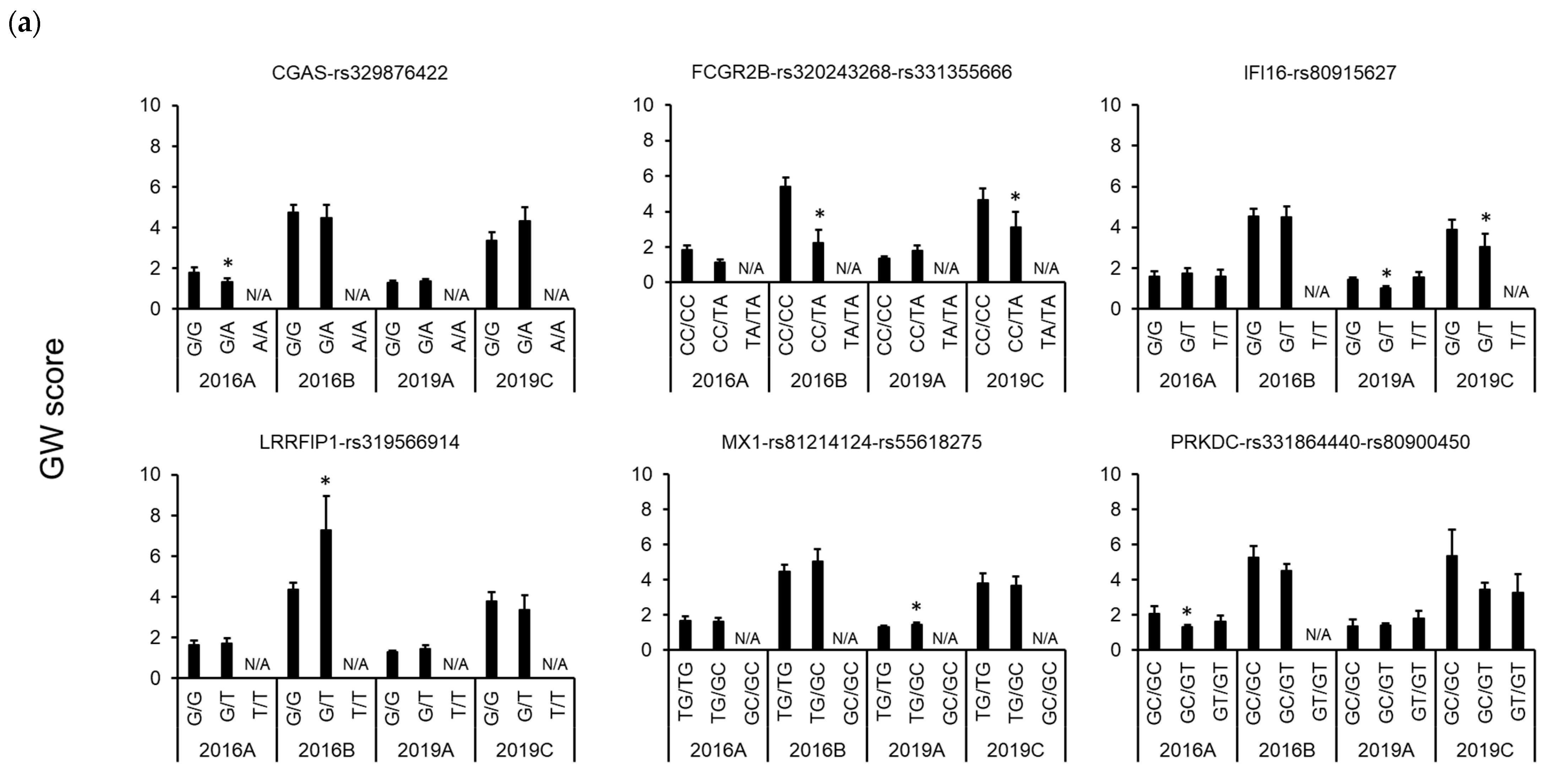
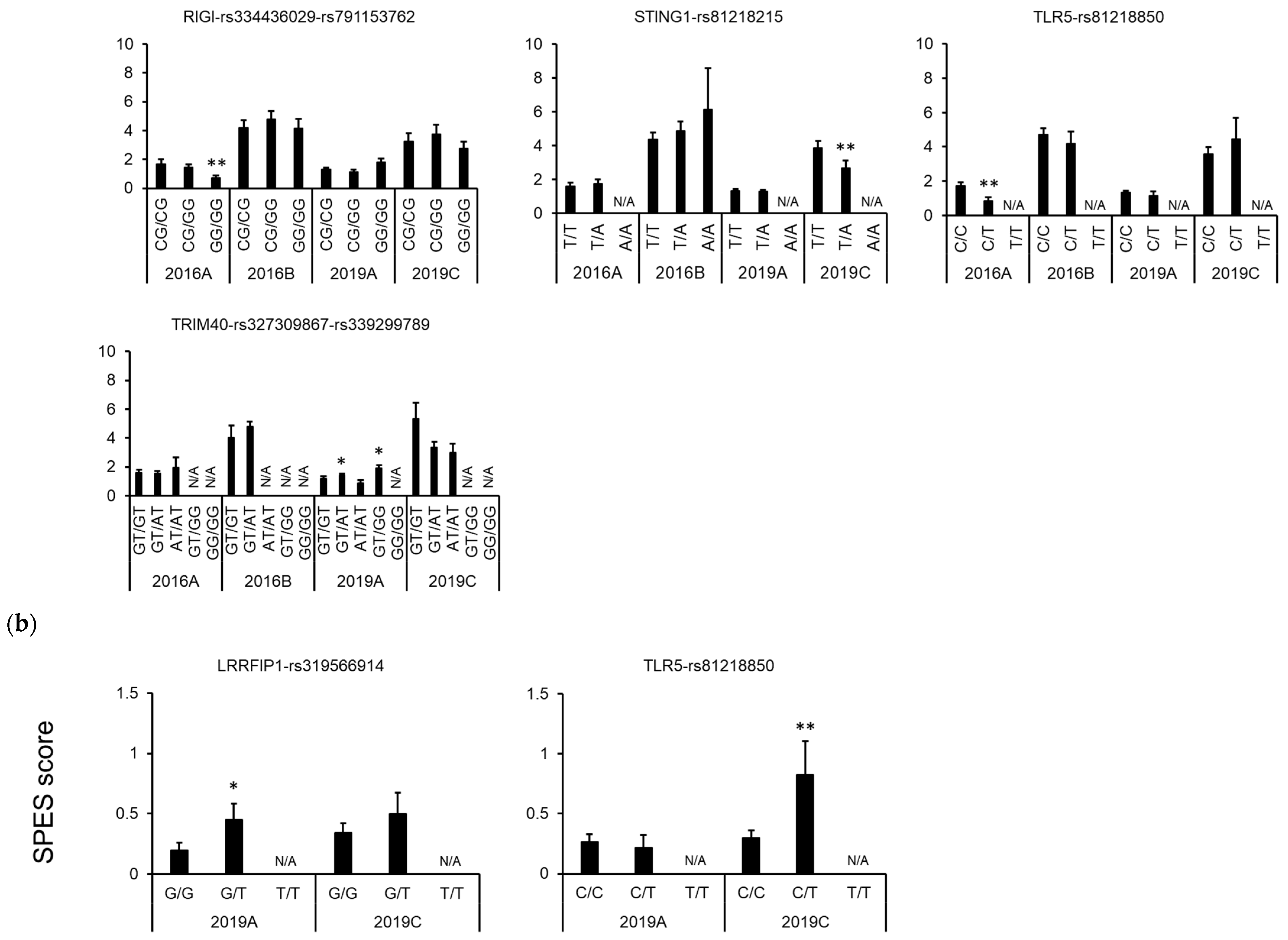
3.3. Association between Polymorphisms in Immune-Related Genes and Traits
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, F.; Li, P.; Yin, Y.; Du, X.; Cao, G.; Wu, S.; Zhao, Y. Molecular breeding of livestock for disease resistance. Virology 2023, 587, 109862. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K.; Kaisho, T. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2001, 2, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A., Jr.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef]
- Uenishi, H.; Shinkai, H.; Morozumi, T.; Muneta, Y. Genomic survey of polymorphisms in pattern recognition receptors and their possible relationship to infections in pigs. Vet. Immunol. Immunopathol. 2012, 148, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Shinkai, H.; Yoshioka, G.; Matsumoto, T.; Takenouchi, T.; Tanaka, J.; Shimizu, M.; Kitazawa, H.; Uenishi, H. Polymorphisms in pattern recognition receptor genes are associated with respiratory disease severity in pig farms. Animals 2022, 12, 3163. [Google Scholar] [CrossRef]
- Shinkai, H.; Suzuki, R.; Akiba, M.; Okumura, N.; Uenishi, H. Porcine Toll-like receptors: Recognition of Salmonella enterica serovar Choleraesuis and influence of polymorphisms. Mol. Immunol. 2011, 48, 1114–1120. [Google Scholar] [CrossRef]
- Suzuki, K.; Shinkai, H.; Yoshioka, G.; Matsumoto, T.; Tanaka, J.; Hayashi, N.; Kitazawa, H.; Uenishi, H. NOD2 genotypes affect the symptoms and mortality in the porcine circovirus 2-spreading pig population. Genes 2021, 12, 1424. [Google Scholar] [CrossRef]
- Ainslie-Garcia, M.H.; Farzan, A.; Jafarikia, M.; Lillie, B.N. Single nucleotide variants in innate immune genes associated with Salmonella shedding and colonization in swine on commercial farms. Vet. Microbiol. 2018, 219, 171–177. [Google Scholar] [CrossRef]
- Pandey, S.; Kawai, T.; Akira, S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb. Perspect. Biol. 2015, 7, a016246. [Google Scholar] [CrossRef]
- Takaoka, A.; Yamada, T. Regulation of signaling mediated by nucleic acid sensors for innate interferon-mediated responses during viral infection. Int. Immunol. 2019, 31, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef]
- Lukacsi, S.; Macsik-Valent, B.; Nagy-Balo, Z.; Kovacs, K.G.; Kliment, K.; Bajtay, Z.; Erdei, A. Utilization of complement receptors in immune cell-microbe interaction. FEBS Lett. 2020, 594, 2695–2713. [Google Scholar] [CrossRef]
- Brencicova, E.; Diebold, S.S. Nucleic acids and endosomal pattern recognition: How to tell friend from foe? Front. Cell. Infect. Microbiol. 2013, 3, 37. [Google Scholar] [CrossRef]
- van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM proteins and their roles in antiviral host defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Gu, Z.; Zhang, H.; Hu, H. To TRIM the immunity: From innate to adaptive immunity. Front. Immunol. 2020, 11, 02157. [Google Scholar] [CrossRef]
- Schmit, K.; Michiels, C. TMEM Proteins in Cancer: A Review. Front. Pharmacol. 2018, 9, 1345. [Google Scholar] [CrossRef]
- Keel, B.N.; Nonneman, D.J.; Lindholm-Perry, A.K.; Oliver, W.T.; Rohrer, G.A. Porcine single nucleotide polymorphisms and their functional effect: An update. BMC Res. Notes 2018, 11, 860. [Google Scholar] [CrossRef]
- Goodwin, R.F.; Whittlestone, P. Enzootic pneumonia of pigs: Immunization attempts inoculating Mycoplasma suipneumoniae antigen by various routes and with different adjuvants. Br. Vet. J. 1973, 129, 456–464. [Google Scholar] [CrossRef]
- Djordjevic, S.P.; Eamens, G.J.; Romalis, L.F.; Nicholls, P.J.; Taylor, V.; Chin, J. Serum and mucosal antibody responses and protection in pigs vaccinated against Mycoplasma hyopneumoniae with vaccines containing a denatured membrane antigen pool and adjuvant. Aust. Vet. J. 1997, 75, 504–511. [Google Scholar] [CrossRef]
- Okamura, T.; Onodera, W.; Tayama, T.; Kadowaki, H.; Kojima-Shibata, C.; Suzuki, E.; Uemoto, Y.; Mikawa, S.; Hayashi, T.; Awata, T.; et al. A genome-wide scan for quantitative trait loci affecting respiratory disease and immune capacity in Landrace pigs. Anim. Genet. 2012, 43, 721–729. [Google Scholar] [CrossRef]
- Fraile, L.; Alegre, A.; Lopez-Jimenez, R.; Nofrarias, M.; Segales, J. Risk factors associated with pleuritis and cranio-ventral pulmonary consolidation in slaughter-aged pigs. Vet. J. 2010, 184, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Merialdi, G.; Dottori, M.; Bonilauri, P.; Luppi, A.; Gozio, S.; Pozzi, P.; Spaggiari, B.; Martelli, P. Survey of pleuritis and pulmonary lesions in pigs at abattoir with a focus on the extent of the condition and herd risk factors. Vet. J. 2012, 193, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Sibila, M.; Aragon, V.; Fraile, L.; Segales, J. Comparison of four lung scoring systems for the assessment of the pathological outcomes derived from Actinobacillus pleuropneumoniae experimental infections. BMC Vet. Res. 2014, 10, 165. [Google Scholar] [CrossRef]
- Groenen, M.A.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.J.; et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 15, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Nagelkerke, S.Q.; Schmidt, D.E.; de Haas, M.; Kuijpers, T.W. Genetic Variation in Low-To-Medium-Affinity Fcgamma Receptors: Functional Consequences, Disease Associations, and Opportunities for Personalized Medicine. Front. Immunol. 2019, 10, 2237. [Google Scholar] [CrossRef]
- Zhang, L.; Xia, Y.; Li, W.; Sun, Y.; Kong, L.; Xu, P.; Xia, P.; Yue, J. Activation of Fc gamma receptor IIb up-regulates the production of interferon-alpha and interferon-gamma in porcine alveolar macrophages during PRRSV infection. Dev. Comp. Immunol. 2020, 109, 103696. [Google Scholar] [CrossRef]
- Burdette, D.L.; Vance, R.E. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2013, 14, 19–26. [Google Scholar] [CrossRef]
- Unterholzner, L.; Keating, S.E.; Baran, M.; Horan, K.A.; Jensen, S.B.; Sharma, S.; Sirois, C.M.; Jin, T.; Latz, E.; Xiao, T.S.; et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010, 11, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Y.; Sun, S.; Luo, J.; Jiang, S.; Zhang, J.; Liu, X.; Shao, Q.; Cao, Q.; Zheng, W.; et al. The Innate Immune DNA Sensing cGAS-STING Signaling Pathway Mediates Anti-PRRSV Function. Viruses 2021, 13, 1829. [Google Scholar] [CrossRef]
- Huang, B.; Zhang, L.; Lu, M.; Li, J.; Lv, Y. PCV2 infection activates the cGAS/STING signaling pathway to promote IFN-β production and viral replication in PK-15 cells. Vet. Microbiol. 2018, 227, 34–40. [Google Scholar] [CrossRef]
- Chang, X.; Shi, X.; Zhang, X.; Wang, L.; Li, X.; Wang, A.; Deng, R.; Zhou, E.; Zhang, G. IFI16 Inhibits Porcine Reproductive and Respiratory Syndrome Virus 2 Replication in a MAVS-Dependent Manner in MARC-145 Cells. Viruses 2019, 11, 1160. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, G.; Yin, J.; Li, L.; Huang, K.; Du, Q.; Tong, D.; Huang, Y. Pseudorabies virus infection activates the NLRP3 and IFI16 inflammasomes to trigger pyroptosis. Vet. Microbiol. 2023, 284, 109826. [Google Scholar] [CrossRef]
- Zheng, W.; Zhou, R.; Li, S.; He, S.; Luo, J.; Zhu, M.; Chen, N.; Chen, H.; Meurens, F.; Zhu, J. Porcine IFI16 Negatively Regulates cGAS Signaling Through the Restriction of DNA Binding and Stimulation. Front. Immunol. 2020, 11, 1669. [Google Scholar] [CrossRef]
- Yang, P.; An, H.; Liu, X.; Wen, M.; Zheng, Y.; Rui, Y.; Cao, X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a β-catenin-dependent pathway. Nat. Immunol. 2010, 11, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Plourde, M.; Vohl, M.C.; Bellis, C.; Carless, M.; Dyer, T.; Dolley, G.; Marette, A.; Despres, J.P.; Bouchard, C.; Blangero, J.; et al. A variant in the LRRFIP1 gene is associated with adiposity and inflammation. Obesity 2013, 21, 185–192. [Google Scholar] [CrossRef]
- Prata, D.P.; Costa-Neves, B.; Cosme, G.; Vassos, E. Unravelling the genetic basis of schizophrenia and bipolar disorder with GWAS: A systematic review. J. Psychiatr. Res. 2019, 114, 178–207. [Google Scholar] [CrossRef]
- Stambuk, C.R.; Staiger, E.A.; Heins, B.J.; Huson, H.J. Exploring physiological and genetic variation of digital cushion thickness in Holstein and Jersey cows and bulls. J. Dairy Sci. 2020, 103, 9177–9194. [Google Scholar] [CrossRef]
- Nakajima, E.; Morozumi, T.; Tsukamoto, K.; Watanabe, T.; Plastow, G.; Mitsuhashi, T. A naturally occurring variant of porcine Mx1 associated with increased susceptibility to influenza virus in vitro. Biochem. Genet. 2007, 45, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R.; Ma, Y.; Pannicke, U.; Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003, 4, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Mansur, D.S.; Peters, N.E.; Ren, H.; Smith, G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife 2012, 1, e00047. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Jia, M.; Song, H.; Yu, Z.; Wang, W.; Li, Q.; Zhang, L.; Zhao, W.; Cao, X. The E3 Ubiquitin Ligase TRIM40 Attenuates Antiviral Immune Responses by Targeting MDA5 and RIG-I. Cell Rep. 2017, 21, 1613–1623. [Google Scholar] [CrossRef]

| Year | Farm | Sex | Sampling Season | Total | |||
|---|---|---|---|---|---|---|---|
| Summer | Fall | Winter | Spring | ||||
| 2016 | A | Female | 48 | 27 | 15 | 24 | 114 |
| Male | 61 | 30 | 48 | 26 | 165 | ||
| Unknown | 0 | 0 | 1 | 0 | 1 | ||
| B | Female | 57 | 25 | 25 | 21 | 128 | |
| Male | 56 | 30 | 36 | 29 | 151 | ||
| Unknown | 0 | 0 | 0 | 0 | 0 | ||
| 2019 | A | Female | 0 | 0 | 66 | 12 | 78 |
| Male | 0 | 0 | 76 | 23 | 99 | ||
| Unknown | 0 | 0 | 1 | 0 | 1 | ||
| C | Female | 45 | 23 | 0 | 0 | 68 | |
| Male | 41 | 16 | 0 | 0 | 57 | ||
| Unknown | 0 | 0 | 0 | 0 | 0 | ||
| Gene | Chromosome: Position | dbSNPID | Resistance | Susceptibility | He | Ne | Function | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Amino Acid | Frequency | Allele | Amino Acid | Frequency | |||||||
| CGAS | 1:92483865 | rs329876422 | A | Met | 0.156 | G | Val | 0.844 | 0.263 | 1.357 | PRR for cytosolic DNA | |
| FCGR2B | 4:88887587 | rs320243268 | T | His | 0.260 | C | Arg | 0.740 | 0.385 | 1.625 | Receptor for antibody heavy chain | |
| 4:88892821 | rs331355666 | A | Ser | 0.054 | C | Ala | 0.946 | 0.102 | 1.114 | |||
| IFI16 | 4:91393489 | rs80915627 | T | Asp | 0.180 | G | Ala | 0.820 | 0.295 | 1.419 | PRR for cytosolic DNA | |
| LRRFIP1 | 15:137424903 | rs319566914 | G | Ala | 0.885 | T | Ser | 0.115 | 0.204 | 1.256 | PRR for cytosolic DNA | |
| MX1 | 13:204856712 | rs81214124 | T | Glu | 0.816 | G | Ala | 0.184 | 0.300 | 1.429 | Involvement in antiviral activity | |
| 13:204856656 | rs55618275 | G | Asp | 0.816 | C | Glu | 0.184 | 0.300 | 1.429 | |||
| PRKDC | 4:79757139 | rs80900450 | T | Met | 0.420 | C | Thr | 0.580 | 0.487 | 1.950 | PRR for cytosolic DNA; involvement in V(D)J recombination in T and B cells | |
| RIGI | 10:33900015 | rs334436029 | G | Cys | 0.335 | C | Ser | 0.665 | 0.446 | 1.804 | PRR for viral RNA | |
| STING1 | 2:141360482 | rs81218215 | A | Asp | 0.170 | T | Val | 0.830 | 0.282 | 1.393 | PRR for cytosolic DNA | |
| TRIM40 | 7:22713980 | rs327309867 | G | Ser | 0.548 | A | Leu | 0.452 | 0.495 | 1.982 | Involvement in intracellular protein degradation | |
| 7:22713916 | rs339299789 | T | Ser | 0.994 | G | Arg | 0.006 | 0.012 | 1.012 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shinkai, H.; Suzuki, K.; Itoh, T.; Yoshioka, G.; Takenouchi, T.; Kitazawa, H.; Uenishi, H. Identification of Nonsynonymous SNPs in Immune-Related Genes Associated with Pneumonia Severity in Pigs. Genes 2024, 15, 1103. https://doi.org/10.3390/genes15081103
Shinkai H, Suzuki K, Itoh T, Yoshioka G, Takenouchi T, Kitazawa H, Uenishi H. Identification of Nonsynonymous SNPs in Immune-Related Genes Associated with Pneumonia Severity in Pigs. Genes. 2024; 15(8):1103. https://doi.org/10.3390/genes15081103
Chicago/Turabian StyleShinkai, Hiroki, Kasumi Suzuki, Tomohito Itoh, Gou Yoshioka, Takato Takenouchi, Haruki Kitazawa, and Hirohide Uenishi. 2024. "Identification of Nonsynonymous SNPs in Immune-Related Genes Associated with Pneumonia Severity in Pigs" Genes 15, no. 8: 1103. https://doi.org/10.3390/genes15081103






