A Global Identification of Protein Disulfide Isomerases from ‘duli’ Pear (Pyrus betulaefolia) and Their Expression Profiles under Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Sequence Analysis of PbPDIs
2.2. Phylogenetic and Conserved Motif Analysis of the PbPDIs
2.3. Chromosome Localization, Gene Duplication, and Collinearity Analysis
2.4. Plant Materials, RNA Extraction, cDNA Synthesis, and Real-Time PCR
3. Results
3.1. 24 PDIs Were Identified in the Genome of ‘duli’ Pear
3.2. Phylogeny and Protein Structure Analysis
3.3. Exon and Intron Distribution and Conserved Motif Analysis
3.4. Gene Duplication and Collinearity Analysis
3.5. Analysis of Stress and Hormone Response Cis-Acting Elements in the Promoter Region of PbPDI Genes
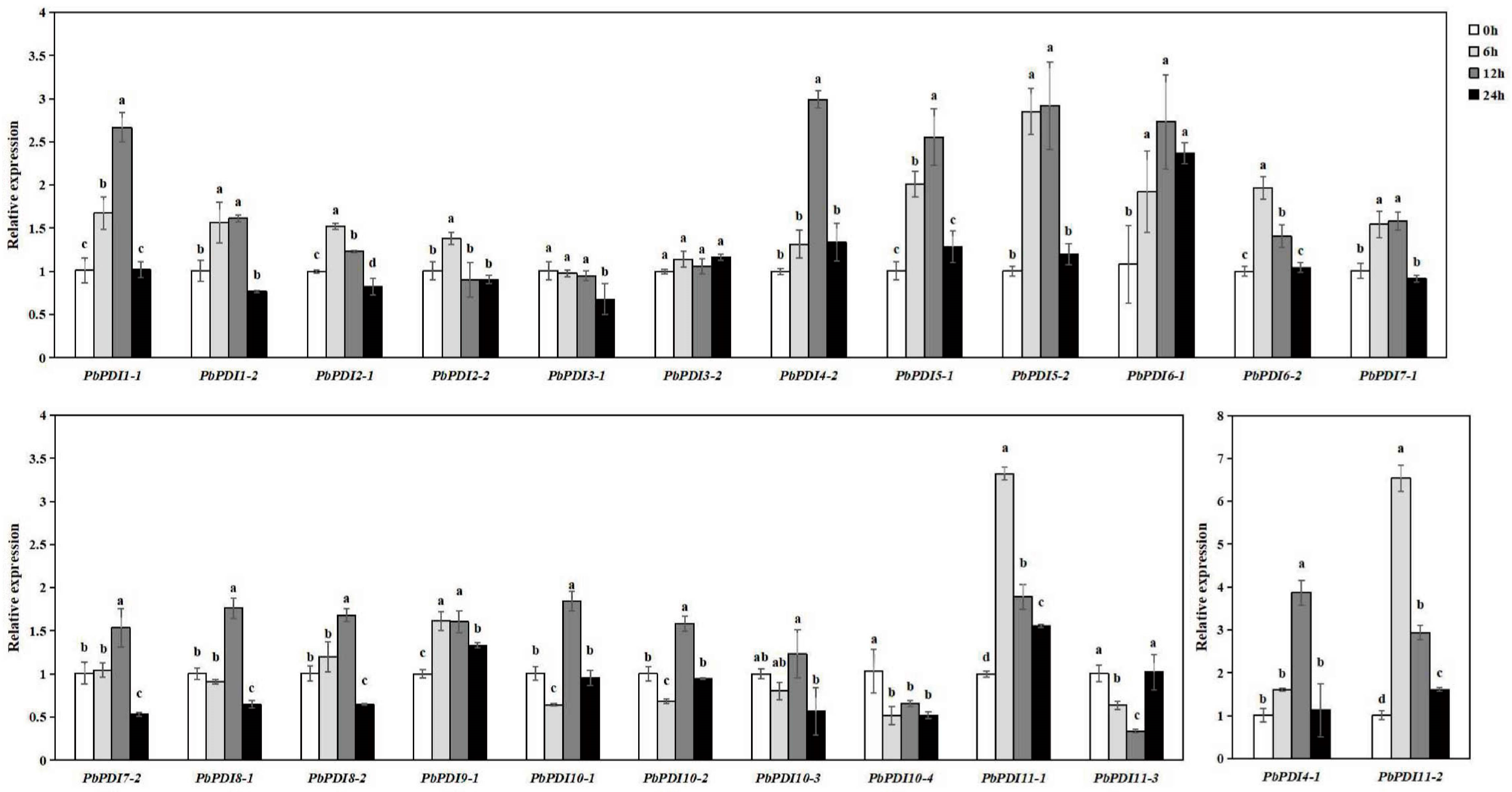
3.6. PbPDIs Expression Profiles under Salt Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tu, B.P.; Ho-Schleyer, S.C.; Travers, K.J.; Weissman, J.S. Biochemical Basis of Oxidative Protein Folding in the Endoplasmic Reticulum. Science 2000, 290, 1571–1574. [Google Scholar] [CrossRef] [PubMed]
- Narayan, M.; Welker, E.; Wedemeyer, W.J.; Scheraga, H.A. Oxidative Folding of Proteins. Acc. Chem. Res. 2000, 33, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Hayano, T.; Hirose, M.; Kikuchi, M. Protein Disulfide Isomerase Mutant Lacking Its Isomerase Activity Accelerates Protein Folding in the Cell. FEBS Lett. 1995, 377, 505–511. [Google Scholar] [PubMed]
- Fu, J.; Gao, J.; Liang, Z.; Yang, D. Pdi-Regulated Disulfide Bond Formation in Protein Folding and Biomolecular Assembly. Molecules 2020, 26, 171. [Google Scholar] [CrossRef]
- Boston, R.S.; Viitanen, P.V.; Vierling, E. Molecular Chaperones and Protein Folding in Plants. Plant Mol. Biol. 1996, 32, 191–222. [Google Scholar] [CrossRef] [PubMed]
- Gruber, C.W.; Cemazar, M.; Heras, B.; Martin, J.L.; Craik, D.J. Protein Disulfide Isomerase: The Structure of Oxidative Folding. Trends Biochem. Sci. 2006, 31, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Anfinsen, C.B. Principles That Govern the Folding of Protein Chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Xiang, S.; Noiva, R.; Lennarz, W.J.; Schindelin, H. The Crystal Structure of Yeast Protein Disulfide Isomerase Suggests Cooperativity between Its Active Sites. Cell 2006, 124, 61–73. [Google Scholar] [CrossRef]
- Kemmink, J.; Darby, N.J.; Dijkstra, K.; Nilges, M.; Creighton, T.E. The Folding Catalyst Protein Disulfide Isomerase Is Constructed of Active and Inactive Thioredoxin Modules. Curr. Biol. 1997, 7, 239–245. [Google Scholar] [CrossRef]
- Noiva, R.; Lennarz, W.J. Protein Disulfide Isomerase. A Multifunctional Protein Resident in the Lumen of the Endoplasmic Reticulum. J. Biol. Chem. 1992, 267, 3553–3556. [Google Scholar] [CrossRef]
- Klappa, P.; Ruddock, L.W.; Darby, N.J.; Freedman, R.B. The b’ Domain Provides the Principal Peptide-Binding Site of Protein Disulfide Isomerase but All Domains Contribute to Binding of Misfolded Proteins. EMBO J. 1998, 17, 927–935. [Google Scholar] [CrossRef]
- Lucero, H.A.; Kaminer, B. The Role of Calcium on the Activity of Ercalcistorin/Protein-Disulfide Isomerase and the Significance of the C-terminal and Its Calcium Binding. A Comparison with Mammalian Protein-Disulfide Isomerase. J. Biol. Chem. 1999, 274, 3243–3251. [Google Scholar] [CrossRef]
- Denecke, J.; De Rycke, R.; Botterman, J. Plant and Mammalian Sorting Signals for Protein Retention in the Endoplasmic Reticulum Contain a Conserved Epitope. EMBO J. 1992, 11, 2345–2355. [Google Scholar] [CrossRef]
- Houston, N.L.; Fan, C.; Xiang, J.Q.; Schulze, J.M.; Jung, R.; Boston, R.S. Phylogenetic Analyses Identify 10 Classes of the Protein Disulfide Isomerase Family in Plants, Including Single-Domain Protein Disulfide Isomerase-Related Proteins. Plant Physiol. 2005, 137, 762–778. [Google Scholar] [CrossRef]
- Kayum, M.A.; Park, J.I.; Nath, U.K.; Saha, G.; Biswas, M.K.; Kim, H.T.; Nou, I.S. Genome-Wide Characterization and Expression Profiling of Pdi Family Gene Reveals Function as Abiotic and Biotic Stress Tolerance in Chinese Cabbage (Brassica Rapa Ssp. Pekinensis). BMC Genom. 2017, 18, 885. [Google Scholar] [CrossRef]
- Wai, A.H.; Waseem, M.; Khan, A.; Nath, U.K.; Lee, D.J.; Kim, S.T.; Kim, C.K.; Chung, M.Y. Genome-Wide Identification and Expression Profiling of the Pdi Gene Family Reveals Their Probable Involvement in Abiotic Stress Tolerance in Tomato (Solanum lycopersicum L.). Genes 2020, 12, 23. [Google Scholar] [CrossRef]
- Meng, L.; Du, X. Genome-Wide Characterization of the Pdi Gene Family in Medicago Truncatula and Their Roles in Response to Endoplasmic Reticulum Stress. Genome 2021, 64, 599–614. [Google Scholar] [CrossRef]
- Parveen, K.; Saddique, M.A.B.; Waqas, M.U.; Attia, K.A.; Rizwan, M.; Abushady, A.M.; Shamsi, I.H. Genome-Wide Analysis and Expression Divergence of Protein Disulfide Isomerase (Pdi) Gene Family Members in Chickpea (Cicer Arietinum) under Salt Stress. Funct. Plant Biol. 2024, 51, FP23253. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yuan, L.; Zhou, Z.; Wang, M.; Wang, X.; Zhang, S.; Sun, Q. The Thiol-Disulfide Exchange Activity of Atpdi1 Is Involved in the Response to Abiotic Stresses. BMC Plant Biol. 2021, 21, 557. [Google Scholar] [CrossRef]
- Feldeverd, E.; Porter, B.W.; Yuen, C.Y.L.; Iwai, K.; Carrillo, R.; Smith, T.; Barela, C.; Wong, K.; Wang, P.; Kang, B.H.; et al. The Arabidopsis Protein Disulfide Isomerase Subfamily M Isoform, Pdi9, Localizes to the Endoplasmic Reticulum and Influences Pollen Viability and Proper Formation of the Pollen Exine During Heat Stress. Front. Plant Sci. 2020, 11, 610052. [Google Scholar] [CrossRef]
- Wittenberg, G.; Levitan, A.; Klein, T.; Dangoor, I.; Keren, N.; Danon, A. Knockdown of the Arabidopsis thaliana Chloroplast Protein Disulfide Isomerase 6 Results in Reduced Levels of Photoinhibition and Increased D1 Synthesis in High Light. Plant J. 2014, 78, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Boavida, L.C.; Ron, M.; McCormick, S. Truncation of a Protein Disulfide Isomerase, Pdil2-1, Delays Embryo Sac Maturation and Disrupts Pollen Tube Guidance in Arabidopsis thaliana. Plant Cell 2008, 20, 3300–3311. [Google Scholar] [CrossRef]
- Hu, J.; Yu, M.; Chang, Y.; Tang, H.; Wang, W.; Du, L.; Wang, K.; Yan, Y.; Ye, X. Functional Analysis of Tapdi Genes on Storage Protein Accumulation by Crispr/Cas9 Edited Wheat Mutants. Int. J. Biol. Macromol. 2022, 196, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ghatak, A.; Bazargani, M.M.; Bajaj, P.; Varshney, R.K.; Chaturvedi, P.; Jiang, D.; Weckwerth, W. Spatial Distribution of Proteins and Metabolites in Developing Wheat Grain and Their Differential Regulatory Response During the Grain Filling Process. Plant J. 2021, 107, 669–687. [Google Scholar] [CrossRef] [PubMed]
- d’Aloisio, E.; Paolacci, A.R.; Dhanapal, A.P.; Tanzarella, O.A.; Porceddu, E.; Ciaffi, M. The Protein Disulfide Isomerase Gene Family in Bread Wheat (T. aestivum L.). BMC Plant Biol. 2010, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zheng, Y.; Fu, Y.; Wang, J.; Yuan, S.; Wang, Y.; Zhu, Q.; Ou, X.; Li, G.; Kang, G. Pdil1-2 Can Indirectly and Negatively Regulate Expression of the Agpl1 Gene in Bread Wheat. Biol. Res. 2019, 52, 56. [Google Scholar] [CrossRef]
- Li, T.; Wang, Y.H.; Huang, Y.; Liu, J.X.; Xing, G.M.; Sun, S.; Li, S.; Xu, Z.S.; Xiong, A.S. A Novel Plant Protein-Disulfide Isomerase Participates in Resistance Response against the Tylcv in Tomato. Planta 2020, 252, 25. [Google Scholar] [CrossRef]
- Akhtar, M.S. Salt Stress, Microbes, and Plant Interactions: Mechanisms and Molecular Approaches; Springer: Singapore, 2019; Volume 2. [Google Scholar]
- Poole, R.L. The Tair Database. Methods Mol. Biol. 2007, 406, 179–212. [Google Scholar]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. Cdd: Ncbi’s Conserved Domain Database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. Plantcare, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the Expasy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. Tbtools-Ii: A One for All, All for One Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Lv, W.; Jiang, S.; Zhang, D.; Cai, G.; Pan, J.; Li, D. Genome-Wide Identification and Expression Analysis of Calcium-Dependent Protein Kinase in Maize. BMC Genom. 2013, 14, 433. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple Methods for Estimating the Numbers of Synonymous and Nonsynonymous Nucleotide Substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [PubMed]
- Nekrutenko, A.; Makova, K.D.; Li, W.H. The K(a)/K(S) Ratio Test for Assessing the Protein-Coding Potential of Genomic Regions: An Empirical and Simulation Study. Genome Res. 2002, 12, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Nørgaard, P.; Winther, J.R. Mutation of Yeast Eug1p Cxxs Active Sites to Cxxc Results in a Dramatic Increase in Protein Disulphide Isomerase Activity. Biochem. J. 2001, 358 Pt 1, 269–274. [Google Scholar] [CrossRef]
- Chivers, P.T.; Laboissière, M.C.; Raines, R.T. The Cxxc Motif: Imperatives for the Formation of Native Disulfide Bonds in the Cell. EMBO J. 1996, 15, 2659–2667. [Google Scholar] [CrossRef]
- Cuervo, N.Z.; Grandvaux, N. Redox Proteomics and Structural Analyses Provide Insightful Implications for Additional Non-Catalytic Thiol-Disulfide Motifs in Pdis. Redox Biol. 2023, 59, 102583. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of Duplicate Genes in Exon-Intron Structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Ma, Z.; Mao, H. Duplicate Genes Contribute to Variability in Abiotic Stress Resistance in Allopolyploid Wheat. Plants 2023, 12, 2465. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Cantizano, N.; Ko, D.K.; Angelos, E.; Pu, Y.; Brandizzi, F. Functional Diversification of Er Stress Responses in Arabidopsis. Trends Biochem. Sci. 2020, 45, 123–136. [Google Scholar] [CrossRef]
- Liu, J.X.; Howell, S.H. Endoplasmic Reticulum Protein Quality Control and Its Relationship to Environmental Stress Responses in Plants. Plant Cell 2010, 22, 2930–2942. [Google Scholar] [CrossRef]
- Reyes-Impellizzeri, S.; Moreno, A.A. The Endoplasmic Reticulum Role in the Plant Response to Abiotic Stress. Front. Plant Sci. 2021, 12, 755447. [Google Scholar] [CrossRef]

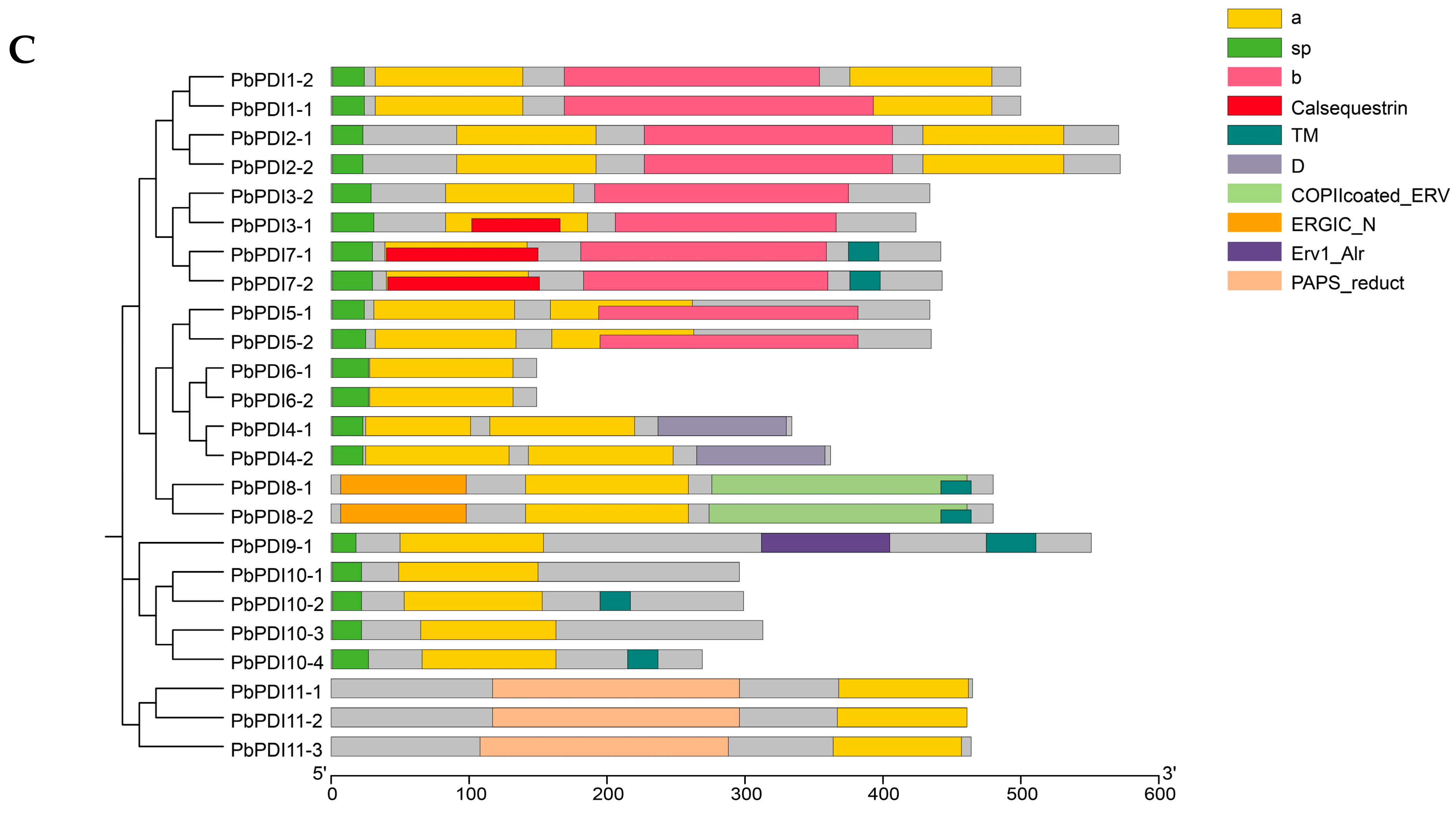
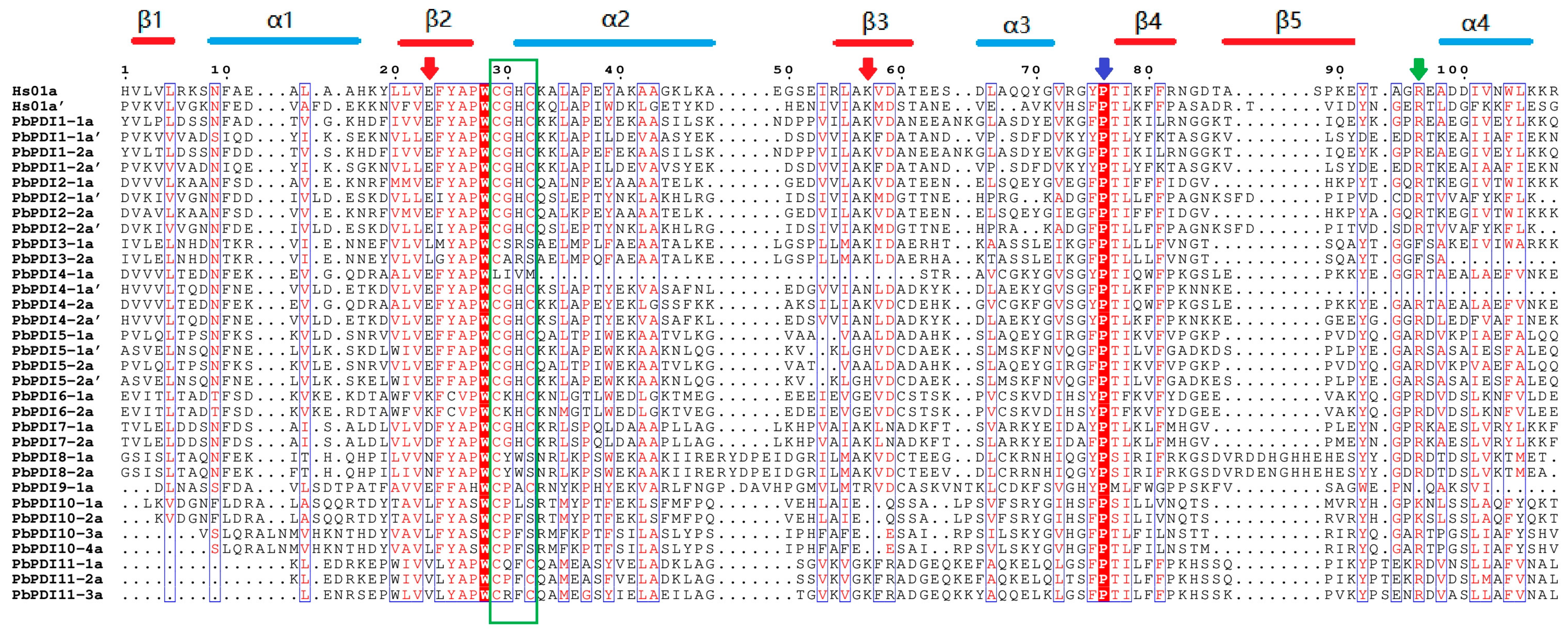
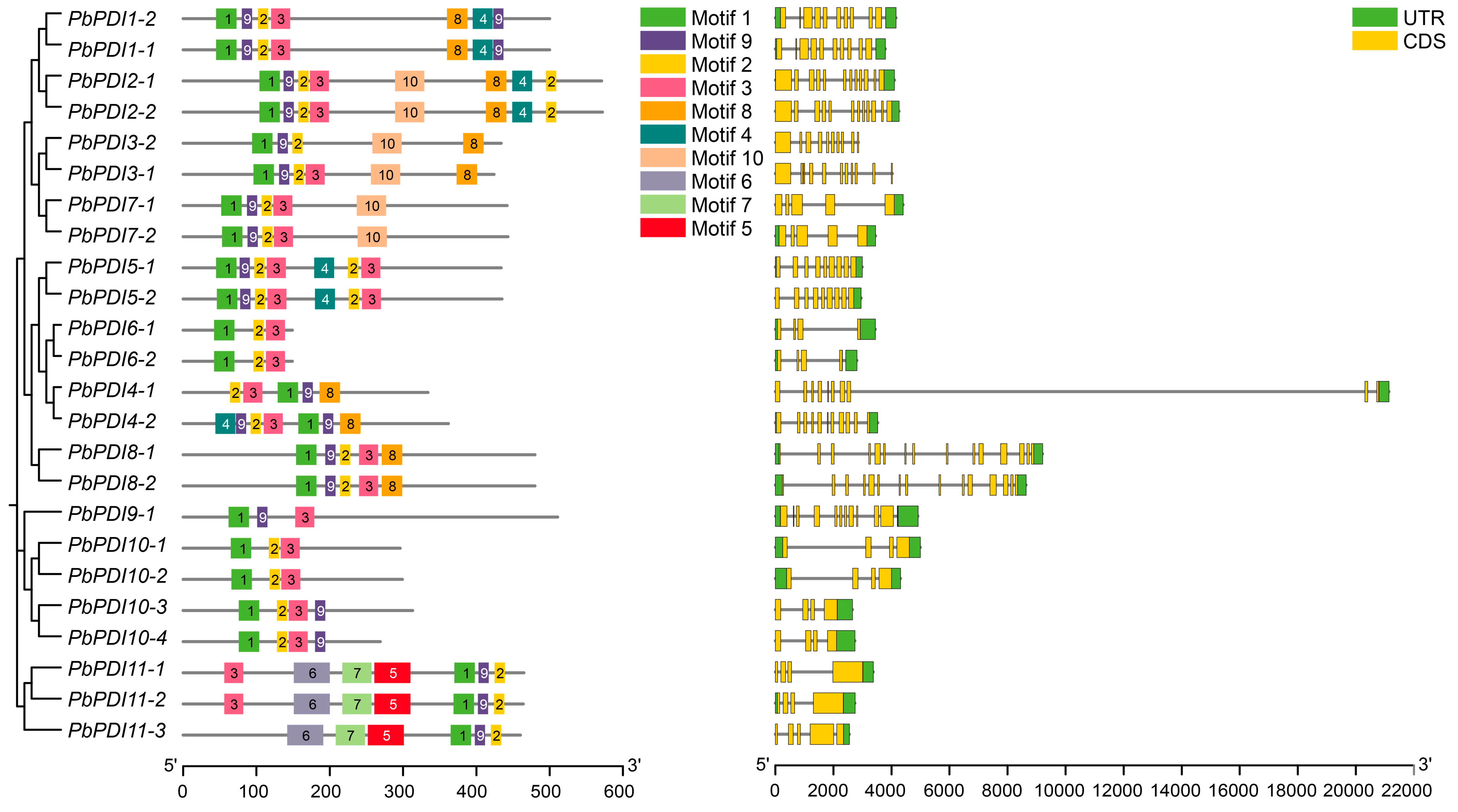
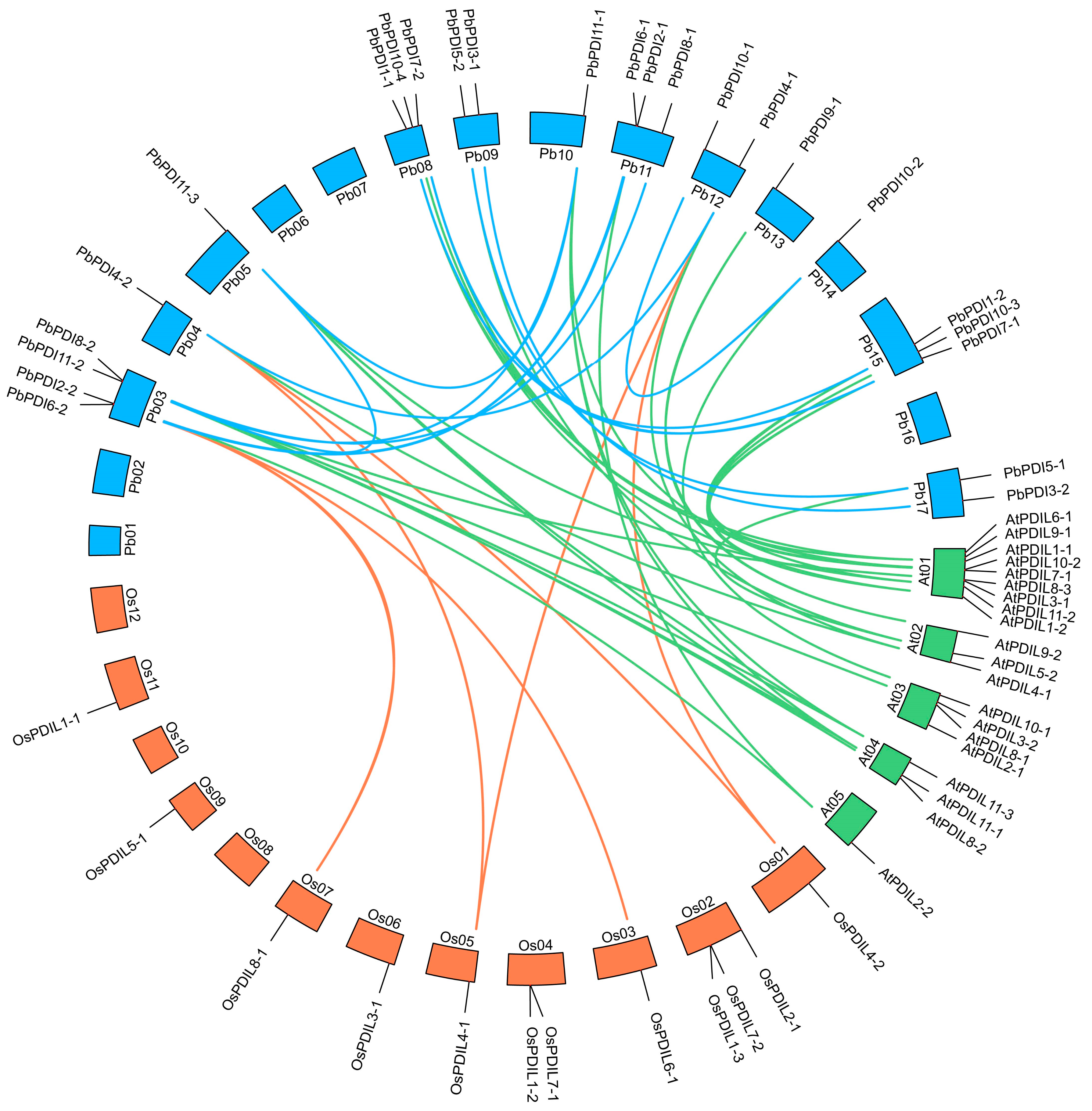

| Gene Name | Locus Name | Chromosome | No. of Exons | Proteins | Subcellular Localization | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. (Strand) | Length (bp) | ORF (bp) | Length (aa) | MW (kDa) | PI | ||||
| PbPDI1-1 | GWHPAAYT052317 | Chr8+ | 3792 | 1503 | 10 | 500 | 56,1 | 4.92 | ER |
| PbPDI1-2 | GWHPAAYT023135 | Chr15+ | 4157 | 1503 | 10 | 500 | 56.1 | 4.9 | ER |
| PbPDI2-1 | GWHPAAYT007464 | Chr11- | 4107 | 1716 | 12 | 571 | 63.5 | 4.71 | Vacuole membrane |
| PbPDI2-2 | GWHPAAYT035228 | Chr3- | 4261 | 1719 | 12 | 572 | 63.7 | 4.6 | ER |
| PbPDI3-1 | GWHPAAYT055473 | Chr9- | 4031 | 1275 | 11 | 424 | 47.2 | 5.17 | Chloroplast |
| PbPDI3-2 | GWHPAAYT029967 | Chr17- | 2865 | 1305 | 10 | 434 | 48.4 | 4.96 | Chloroplast |
| PbPDI4-1 | GWHPAAYT012267 | Chr12+ | 21,142 | 1005 | 10 | 334 | 36.9 | 5.26 | Extracellular |
| PbPDI4-2 | GWHPAAYT039863 | Chr4+ | 3528 | 1089 | 11 | 362 | 39.8 | 5.69 | Extracellular |
| PbPDI5-1 | GWHPAAYT028894 | Chr17+ | 2992 | 1305 | 9 | 434 | 47.4 | 5.29 | Vacuole membrane |
| PbPDI5-2 | GWHPAAYT054427 | Chr9+ | 2957 | 1308 | 9 | 435 | 47.7 | 5.38 | ER |
| PbPDI6-1 | GWHPAAYT007378 | Chr11+ | 3446 | 450 | 4 | 149 | 16.7 | 4.66 | Extracellular |
| PbPDI6-2 | GWHPAAYT035173 | Chr3+ | 2815 | 450 | 4 | 149 | 16.8 | 4.89 | Extracellular |
| PbPDI7-1 | GWHPAAYT024090 | Chr15+ | 4405 | 1329 | 5 | 442 | 49.8 | 5.01 | PM |
| PbPDI7-2 | GWHPAAYT053038 | Chr8+ | 3456 | 1332 | 5 | 443 | 49.8 | 4.9 | PM |
| PbPDI8-1 | GWHPAAYT008875 | Chr11+ | 9376 | 1443 | 15 | 480 | 53.5 | 7.36 | PM |
| PbPDI8-2 | GWHPAAYT036573 | Chr3+ | 8640 | 1443 | 15 | 480 | 53.7 | 7.06 | PM |
| PbPDI9-1 | GWHPAAYT013244 | Chr13- | 4914 | 1536 | 12 | 511 | 57.1 | 8.24 | Chloroplast |
| PbPDI10-1 | GWHPAAYT009923 | Chr12+ | 4996 | 891 | 4 | 296 | 33.7 | 9.8 | PM |
| PbPDI10-2 | GWHPAAYT016488 | Chr14+ | 4312 | 900 | 4 | 299 | 33.9 | 9.73 | PM |
| PbPDI10-3 | GWHPAAYT023589 | Chr15- | 2655 | 942 | 4 | 313 | 34.9 | 9.07 | Cytoplasm |
| PbPDI10-4 | GWHPAAYT052662 | Chr8- | 2739 | 810 | 4 | 269 | 30.0 | 8.21 | PM |
| PbPDI11-1 | GWHPAAYT005858 | Chr10+ | 3378 | 1398 | 4 | 465 | 51.6 | 6.36 | Chloroplast |
| PbPDI11-2 | GWHPAAYT044219 | Chr3- | 2551 | 1395 | 5 | 464 | 51.6 | 6.74 | Chloroplast |
| PbPDI11-3 | GWHPAAYT036474 | Chr5+ | 2744 | 1383 | 4 | 460 | 50.8 | 8.9 | Chloroplast |
| Name | Signal Peptide | Trans- Membrane | Domain Organization | Active Site Motif | Conserved Charge Pair Sequence | Conserved Arginine | C-Terminal Signal |
|---|---|---|---|---|---|---|---|
| PbPDI1-1 | 1-24 | No | a-b-a’ | CGHC, CGHC | E54-K88, E398-K431′ | R128, R468 | KDEL |
| PbPDI1-2 | 1-24 | No | a-b-a’ | CGHC, CGHC | E54-K88, E398-K431 | R128, R468 | KDEL |
| PbPDI2-1 | 1-23 | No | a-b-a’ | CGHC, CGHC | E113-K145, E452-K485 | R181, R523 | RFEG |
| PbPDI2-2 | 1-23 | No | a-b-a’ | CGHC, CGHC | E113-K145, E452-K485 | R181, R523 | KDEL |
| PbPDI3-1 | 1-31 | No | a-c-b | CSRS | L105-K139 | F175 | SSAQ |
| PbPDI3-2 | 1-29 | No | a-b | CARS | L103-K137 | F173 | ACIL |
| PbPDI4-1 | 1-23 | No | ao-a-D | CGHC | E138-N171 | R90 | TSSS |
| PbPDI4-2 | 1-23 | No | ao-a-D | CGHC, CGHC | E47-K80, E166-N199 | R118, R237 | ASSS |
| PbPDI5-1 | 1-24 | No | ao-a-b | CGHC, CGHC | E54-A85, E182-H213 | R122, R251 | KEEL |
| PbPDI5-2 | 1-25 | No | ao-a-b | CGHC, CGHC | E55-A86, E183-H214 | R123, R252 | KDEL |
| PbPDI6-1 | 1-27 | No | a | CKHC | K51-E84 | R121 | DKEL |
| PbPDI6-2 | 1-27 | No | a | CKHC | K51-E84 | R121 | DKEL |
| PbPDI7-1 | 1-30 | 375-397 | a-c-b-t | CGHC | D61-K95 | R131 | EKED |
| PbPDI7-2 | 1-30 | 376-398 | a-c-b-t | CGHC | D62-K96 | R132 | EKED |
| PbPDI8-1 | No | 442-464 | g-a-f-t | CYWS | N163-K202 | R248 | GKNF |
| PbPDI8-2 | No | 442-464 | g-a-f-t | CYWS | N163-K202 | R248 | GKNF |
| PbPDI9-1 | 1-18 | 475-511 | a-g-t | CPAC | K71-R109 | Q149 | RSWS |
| PbPDI10-1 | 1-22 | No | a | CPLS | L93-I120 | K152 | SSTN |
| PbPDI10-2 | 1-22 | 195-217 | a-t | CPFS | L94-I121 | K153 | SLSS |
| PbPDI10-3 | 1-27 | No | a | CPFS | L85-F116 | R152 | YSSD |
| PbPDI10-4 | 1-22 | 215-237 | a-t | CPFS | L85-F116 | R152 | SAVV |
| PbPDI11-1 | No | No | h-a′ | CQFC | V379-K411 | R452 | NALR |
| PbPDI11-2 | No | No | h-a′ | CPFC | V377-K410 | R451 | NALR |
| PbPDI11-3 | No | No | h-a′ | CRFC | V374-K406 | R447 | NALR |
| Duplicated Gene Pairs | Ka | Ks | Ka_Ks | Duplication Type | Types of Selection | Time (MYA) | |
|---|---|---|---|---|---|---|---|
| PbPDI1-1 | PbPDI1-2 | 0.030656727 | 0.228689663 | 0.134053839 | Segmental | Purifying selection | 7.62 |
| PbPDI2-1 | PbPDI2-2 | 0.037174043 | 0.264643094 | 0.140468594 | Segmental | Purifying selection | 8.82 |
| PbPDI3-1 | PbPDI3-2 | 0.080273354 | 0.281013353 | 0.285656725 | Segmental | Purifying selection | 9.37 |
| PbPDI4-1 | PbPDI4-2 | 0.053035942 | 0.128765197 | 0.411881031 | Segmental | Purifying selection | 4.29 |
| PbPDI5-1 | PbPDI5-2 | 0.017175359 | 0.240972829 | 0.071275086 | Segmental | Purifying selection | 8.03 |
| PbPDI6-1 | PbPDI6-2 | 0.041379614 | 0.306937951 | 0.134814266 | Segmental | Purifying selection | 10.23 |
| PbPDI7-1 | PbPDI7-2 | 0.05256445 | 0.158091931 | 0.332492932 | Segmental | Purifying selection | 5.27 |
| PbPDI8-1 | PbPDI8-2 | 0.02186113 | 0.223403215 | 0.097855039 | Segmental | Purifying selection | 7.45 |
| PbPDI10-1 | PbPDI10-2 | 0.044270767 | 0.116345503 | 0.380511196 | Segmental | Purifying selection | 3.88 |
| PbPDI11-1 | PbPDI11-2 | 0.022953656 | 0.301454658 | 0.076142981 | Segmental | Purifying selection | 10.05 |
| PbPDI11-1 | PbPDI11-3 | 0.163012376 | 2.493442125 | 0.065376443 | Segmental | Purifying selection | 83.11 |
| PbPDI11-2 | PbPDI11-3 | 0.154988455 | 2.21329614 | 0.070026081 | Segmental | Purifying selection | 73.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhang, Y.; Cui, K.; Liu, C.; Chen, M.; Fu, Y.; Li, Z.; Ma, H.; Zhang, H.; Qi, B.; et al. A Global Identification of Protein Disulfide Isomerases from ‘duli’ Pear (Pyrus betulaefolia) and Their Expression Profiles under Salt Stress. Genes 2024, 15, 968. https://doi.org/10.3390/genes15080968
Zhang H, Zhang Y, Cui K, Liu C, Chen M, Fu Y, Li Z, Ma H, Zhang H, Qi B, et al. A Global Identification of Protein Disulfide Isomerases from ‘duli’ Pear (Pyrus betulaefolia) and Their Expression Profiles under Salt Stress. Genes. 2024; 15(8):968. https://doi.org/10.3390/genes15080968
Chicago/Turabian StyleZhang, Hao, Yuyue Zhang, Kexin Cui, Chang Liu, Mengya Chen, Yufan Fu, Zhenjie Li, Hui Ma, Haixia Zhang, Baoxiu Qi, and et al. 2024. "A Global Identification of Protein Disulfide Isomerases from ‘duli’ Pear (Pyrus betulaefolia) and Their Expression Profiles under Salt Stress" Genes 15, no. 8: 968. https://doi.org/10.3390/genes15080968





