Association between Complex ACTN3 and ACE Gene Polymorphisms and Elite Endurance Sports in Koreans: A Case–Control Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Blood Sampling and Genomic DNA Extraction
2.3. Genotyping
2.4. Statistical Analysis
3. Results
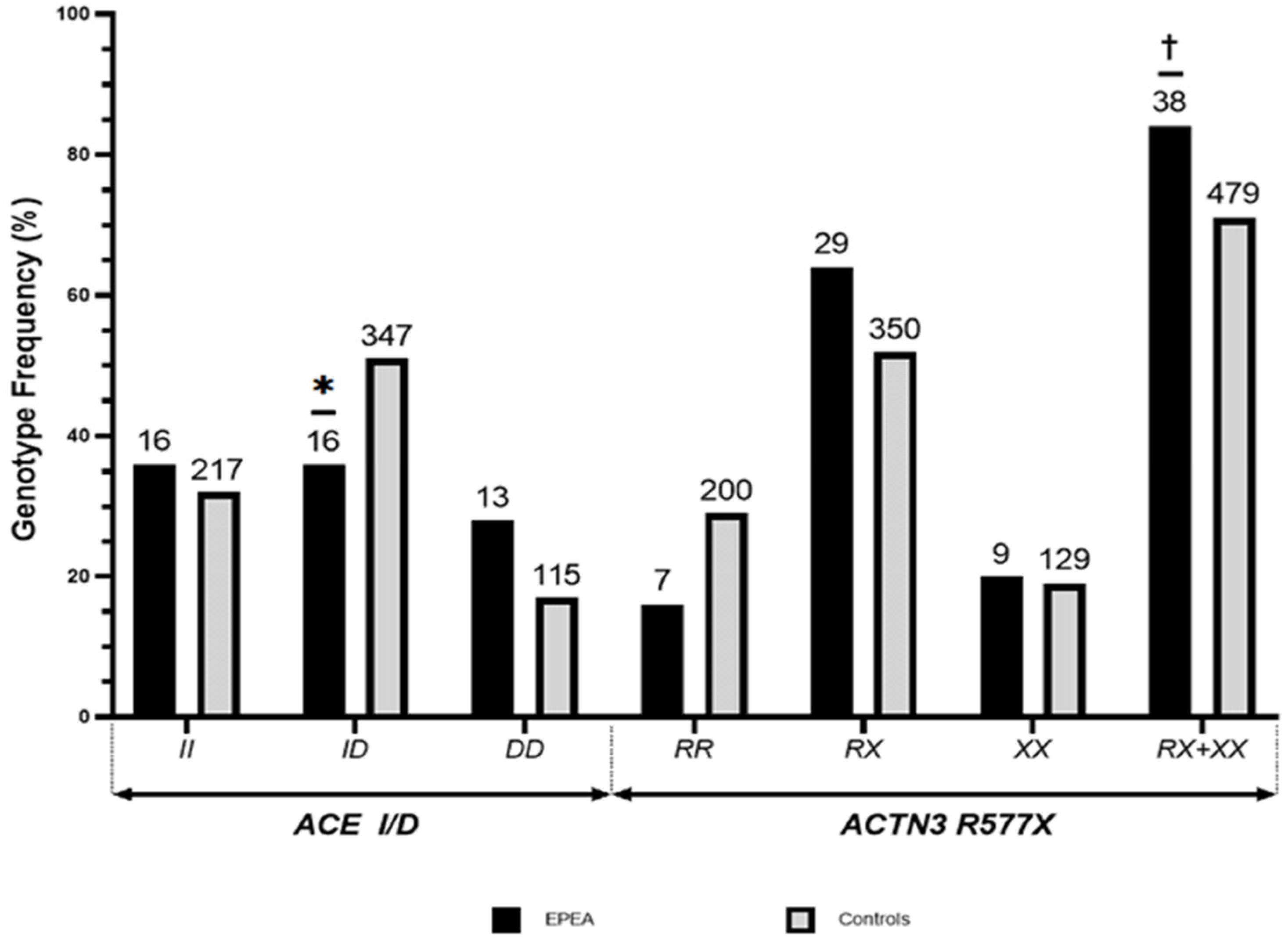
3.1. Distribution of ACTN3 R577X and ACE I/D Polymorphisms among the Non-Athlete Korean Population and EPEA
3.2. Complex ACTN3-ACE Polymorphisms and Pure Elite Endurance in Koreans
3.3. Genetic Relationships between Complex ACTN3-ACE Genotypes and Elite Pure Endurance in Koreans
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmetov, I.I.; Fedotovskaya, O.N. Current Progress in Sports Genomics. Adv. Clin. Chem. 2015, 70, 247–314. [Google Scholar] [CrossRef]
- Takahashi, T.; Tajima, F. The amount of DNA polymorphism when population size changes linearly. Genes. Genet. Syst. 2017, 92, 55–57. [Google Scholar] [CrossRef]
- Semenova, E.A.; Hall, E.C.R.; Ahmetov, I.I. Genes and Athletic Performance: The 2023 Update. Genes 2023, 14, 1235. [Google Scholar] [CrossRef]
- Georgiades, E.; Klissouras, V.; Baulch, J.; Wang, G.; Pitsiladis, Y. Why nature prevails over nurture in the making of the elite athlete. BMC Genom. 2017, 18, 835. [Google Scholar] [CrossRef] [PubMed]
- De Moor, M.H.; Spector, T.D.; Cherkas, L.F.; Falchi, M.; Hottenga, J.J.; Boomsma, D.I.; De Geus, E.J. Genome-wide linkage scan for athlete status in 700 British female DZ twin pairs. Twin Res. Hum. Genet. 2007, 10, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Tanisawa, K.; Wang, G.; Seto, J.; Verdouka, I.; Twycross-Lewis, R.; Karanikolou, A.; Tanaka, M.; Borjesson, M.; Di Luigi, L.; Dohi, M.; et al. Sport and exercise genomics: The FIMS 2019 consensus statement update. Br. J. Sports Med. 2020, 54, 969–975. [Google Scholar] [CrossRef]
- Myburgh, K.H. What makes an endurance athlete world-class? Not simply a physiological conundrum. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2003, 136, 171–190. [Google Scholar] [CrossRef]
- Holloszy, J.O.; Coyle, E.F. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1984, 56, 831–838. [Google Scholar] [CrossRef]
- Davies, C.T.; Thompson, M.W. Aerobic performance of female marathon and male ultramarathon athletes. Eur. J. Appl. Physiol. Occup. Physiol. 1979, 41, 233–245. [Google Scholar] [CrossRef]
- Bouchard, C.; Dionne, F.T.; Simoneau, J.A.; Boulay, M.R. Genetics of aerobic and anaerobic performances. Exerc. Sport. Sci. Rev. 1992, 20, 27–58. [Google Scholar] [PubMed]
- Miyamoto-Mikami, E.; Zempo, H.; Fuku, N.; Kikuchi, N.; Miyachi, M.; Murakami, H. Heritability estimates of endurance-related phenotypes: A systematic review and meta-analysis. Scand. J. Med. Sci. Sports 2018, 28, 834–845. [Google Scholar] [CrossRef]
- Tucker, R.; Collins, M. What makes champions? A review of the relative contribution of genes and training to sporting success. Br. J. Sports Med. 2012, 46, 555–561. [Google Scholar] [CrossRef]
- Naureen, Z.; Perrone, M.; Paolacci, S.; Maltese, P.E.; Dhuli, K.; Kurti, D.; Dautaj, A.; Miotto, R.; Casadei, A.; Fioretti, B.; et al. Genetic test for the personalization of sport training. Acta Biomed. 2020, 91 (Suppl. S13), e2020012. [Google Scholar] [CrossRef]
- Lippi, G.; Longo, U.G.; Maffulli, N. Genetics and sports. Br. Med. Bull. 2010, 93, 27–47. [Google Scholar] [CrossRef] [PubMed]
- McAuley, A.B.T.; Hughes, D.C.; Tsaprouni, L.G.; Varley, I.; Suraci, B.; Roos, T.R.; Herbert, A.J.; Kelly, A.L. Genetic association research in football: A systematic review. Eur. J. Sport. Sci. 2021, 21, 714–752. [Google Scholar] [CrossRef]
- Pitsiladis, Y.; Wang, G.; Wolfarth, B.; Scott, R.; Fuku, N.; Mikami, E.; He, Z.; Fiuza-Luces, C.; Eynon, N.; Lucia, A. Genomics of elite sporting performance: What little we know and necessary advances. Br. J. Sports Med. 2013, 47, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Eynon, N.; Ruiz, J.R.; Femia, P.; Pushkarev, V.P.; Cieszczyk, P.; Maciejewska-Karlowska, A.; Sawczuk, M.; Dyatlov, D.A.; Lekontsev, E.V.; Kulikov, L.M.; et al. The ACTN3 R577X polymorphism across three groups of elite male European athletes. PLoS ONE 2012, 7, e43132. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.; Xenophontos, S.L.; Cariolou, M.A.; Mokone, G.G.; Hudson, D.E.; Anastasiades, L.; Noakes, T.D. The ACE gene and endurance performance during the South African Ironman Triathlons. Med. Sci. Sports Exerc. 2004, 36, 1314–1320. [Google Scholar] [CrossRef]
- Beggs, A.H.; Byers, T.J.; Knoll, J.H.; Boyce, F.M.; Bruns, G.A.; Kunkel, L.M. Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J. Biol. Chem. 1992, 267, 9281–9288. [Google Scholar] [CrossRef]
- Takada, F.; Vander Woude, D.L.; Tong, H.Q.; Thompson, T.G.; Watkins, S.C.; Kunkel, L.M.; Beggs, A.H. Myozenin: An alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc. Natl. Acad. Sci. USA 2001, 98, 1595–1600. [Google Scholar] [CrossRef]
- Eynon, N.; Hanson, E.D.; Lucia, A.; Houweling, P.J.; Garton, F.; North, K.N.; Bishop, D.J. Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 2013, 43, 803–817. [Google Scholar] [CrossRef]
- North, K.N.; Beggs, A.H. Deficiency of a skeletal muscle isoform of alpha-actinin (alpha-actinin-3) in merosin-positive congenital muscular dystrophy. Neuromuscul. Disord. 1996, 6, 229–235. [Google Scholar] [CrossRef]
- Blanchard, A.; Ohanian, V.; Critchley, D. The structure and function of alpha-actinin. J. Muscle Res. Cell Motil. 1989, 10, 280–289. [Google Scholar] [CrossRef]
- Endo, T.; Masaki, T. Differential expression and distribution of chicken skeletal- and smooth-muscle-type alpha-actinins during myogenesis in culture. J. Cell Biol. 1984, 99, 2322–2332. [Google Scholar] [CrossRef]
- North, K.N.; Yang, N.; Wattanasirichaigoon, D.; Mills, M.; Easteal, S.; Beggs, A.H. A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat. Genet. 1999, 21, 353–354. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.; Kiely, J. ACTN3: More than Just a Gene for Speed. Front. Physiol. 2017, 8, 1080. [Google Scholar] [CrossRef]
- Vainzof, M.; Costa, C.S.; Marie, S.K.; Moreira, E.S.; Reed, U.; Passos-Bueno, M.R.; Beggs, A.H.; Zatz, M. Deficiency of alpha-actinin-3 (ACTN3) occurs in different forms of muscular dystrophy. Neuropediatrics 1997, 28, 223–228. [Google Scholar] [CrossRef]
- MacArthur, D.G.; Seto, J.T.; Raftery, J.M.; Quinlan, K.G.; Huttley, G.A.; Hook, J.W.; Lemckert, F.A.; Kee, A.J.; Edwards, M.R.; Berman, Y.; et al. Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat. Genet. 2007, 39, 1261–1265. [Google Scholar] [CrossRef]
- Seto, J.T.; Lek, M.; Quinlan, K.G.; Houweling, P.J.; Zheng, X.F.; Garton, F.; MacArthur, D.G.; Raftery, J.M.; Garvey, S.M.; Hauser, M.A.; et al. Deficiency of alpha-actinin-3 is associated with increased susceptibility to contraction-induced damage and skeletal muscle remodeling. Hum. Mol. Genet. 2011, 20, 2914–2927. [Google Scholar] [CrossRef]
- MacArthur, D.G.; North, K.N. A gene for speed? The evolution and function of alpha-actinin-3. Bioessays 2004, 26, 786–795. [Google Scholar] [CrossRef]
- Yang, N.; MacArthur, D.G.; Gulbin, J.P.; Hahn, A.G.; Beggs, A.H.; Easteal, S.; North, K. ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 2003, 73, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, E.M.; Coelho, D.B.; Veneroso, C.E.; Barros Coelho, E.J.; Cruz, I.R.; Morandi, R.F.; De, A.P.G.; Carvalho, M.R.; Garcia, E.S.; De Paz Fernández, J.A. Effect of ACTN3 gene on strength and endurance in soccer players. J. Strength Cond. Res. 2013, 27, 3286–3292. [Google Scholar] [CrossRef]
- Soubrier, F.; Alhenc-Gelas, F.; Hubert, C.; Allegrini, J.; John, M.; Tregear, G.; Corvol, P. Two putative active centers in human angiotensin I-converting enzyme revealed by molecular cloning. Proc. Natl. Acad. Sci. USA 1988, 85, 9386–9390. [Google Scholar] [CrossRef]
- Erdös, E.G. Angiotensin I converting enzyme. Circ. Res. 1975, 36, 247–255. [Google Scholar] [CrossRef]
- Bánhegyi, V.; Enyedi, A.; Fülöp, G.; Oláh, A.; Siket, I.M.; Váradi, C.; Bottyán, K.; Lódi, M.; Csongrádi, A.; Umar, A.J.; et al. Human Tissue Angiotensin Converting Enzyme (ACE) Activity Is Regulated by Genetic Polymorphisms, Posttranslational Modifications, Endogenous Inhibitors and Secretion in the Serum, Lungs and Heart. Cells 2021, 10, 1708. [Google Scholar] [CrossRef] [PubMed]
- Cambien, F.; Alhenc-Gelas, F.; Herbeth, B.; Andre, J.L.; Rakotovao, R.; Gonzales, M.F.; Allegrini, J.; Bloch, C. Familial resemblance of plasma angiotensin-converting enzyme level: The Nancy Study. Am. J. Hum. Genet. 1988, 43, 774–780. [Google Scholar]
- Rigat, B.; Hubert, C.; Corvol, P.; Soubrier, F. PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCP1) (dipeptidyl carboxypeptidase 1). Nucleic Acids Res. 1992, 20, 1433. [Google Scholar] [CrossRef]
- Gayagay, G.; Yu, B.; Hambly, B.; Boston, T.; Hahn, A.; Celermajer, D.S.; Trent, R.J. Elite endurance athletes and the ACE I allele--the role of genes in athletic performance. Hum. Genet. 1998, 103, 48–50. [Google Scholar] [CrossRef]
- Woods, D. Angiotensin-converting enzyme, renin-angiotensin system and human performance. Med. Sport. Sci. 2009, 54, 72–87. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, R.; Terrados, N.; Ortolano, R.; Iglesias-Cubero, G.; Reguero, J.R.; Batalla, A.; Cortina, A.; Fernández-García, B.; Rodríguez, C.; Braga, S.; et al. Genetic variation in the renin-angiotensin system and athletic performance. Eur. J. Appl. Physiol. 2000, 82, 117–120. [Google Scholar] [CrossRef]
- Scanavini, D.; Bernardi, F.; Castoldi, E.; Conconi, F.; Mazzoni, G. Increased frequency of the homozygous II ACE genotype in Italian Olympic endurance athletes. Eur. J. Hum. Genet. 2002, 10, 576–577. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Yang, Y.; Li, X.; Zhou, F.; Gao, C.; Li, M.; Gao, L. The association of sport performance with ACE and ACTN3 genetic polymorphisms: A systematic review and meta-analysis. PLoS ONE 2013, 8, e54685. [Google Scholar] [CrossRef] [PubMed]
- Rigat, B.; Hubert, C.; Alhenc-Gelas, F.; Cambien, F.; Corvol, P.; Soubrier, F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Investig. 1990, 86, 1343–1346. [Google Scholar] [CrossRef]
- Zhang, B.; Tanaka, H.; Shono, N.; Miura, S.; Kiyonaga, A.; Shindo, M.; Saku, K. The I allele of the angiotensin-converting enzyme gene is associated with an increased percentage of slow-twitch type I fibers in human skeletal muscle. Clin. Genet. 2003, 63, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, D.; Huber-Abel, F.A.; Graber, F.; Hoppeler, H.; Flück, M. The angiotensin converting enzyme insertion/deletion polymorphism alters the response of muscle energy supply lines to exercise. Eur. J. Appl. Physiol. 2013, 113, 1719–1729. [Google Scholar] [CrossRef]
- Santana, H.A.; Moreira, S.R.; Neto, W.B.; Silva, C.B.; Sales, M.M.; Oliveira, V.N.; Asano, R.Y.; Espíndola, F.S.; Nóbrega, O.T.; Campbell, C.S.; et al. The higher exercise intensity and the presence of allele I of ACE gene elicit a higher post-exercise blood pressure reduction and nitric oxide release in elderly women: An experimental study. BMC Cardiovasc. Disord. 2011, 11, 71. [Google Scholar] [CrossRef]
- van Ginkel, S.; de Haan, A.; Woerdeman, J.; Vanhees, L.; Serné, E.; de Koning, J.; Flück, M. Exercise intensity modulates capillary perfusion in correspondence with ACE I/D modulated serum angiotensin II levels. Appl. Transl. Genom. 2015, 4, 33–37. [Google Scholar] [CrossRef]
- Halliwill, J.R. Mechanisms and clinical implications of post-exercise hypotension in humans. Exerc. Sport. Sci. Rev. 2001, 29, 65–70. [Google Scholar] [CrossRef]
- Yan, Z.; Okutsu, M.; Akhtar, Y.N.; Lira, V.A. Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J. Appl. Physiol. 2011, 110, 264–274. [Google Scholar] [CrossRef]
- Puthucheary, Z.; Skipworth, J.R.; Rawal, J.; Loosemore, M.; Van Someren, K.; Montgomery, H.E. The ACE gene and human performance: 12 years on. Sports Med. 2011, 41, 433–448. [Google Scholar] [CrossRef]
- Nazarov, I.B.; Woods, D.R.; Montgomery, H.E.; Shneider, O.V.; Kazakov, V.I.; Tomilin, N.V.; Rogozkin, V.A. The angiotensin converting enzyme I/D polymorphism in Russian athletes. Eur. J. Hum. Genet. 2001, 9, 797–801. [Google Scholar] [CrossRef]
- Moran, C.N.; Vassilopoulos, C.; Tsiokanos, A.; Jamurtas, A.Z.; Bailey, M.E.; Montgomery, H.E.; Wilson, R.H.; Pitsiladis, Y.P. The associations of ACE polymorphisms with physical, physiological and skill parameters in adolescents. Eur. J. Hum. Genet. 2006, 14, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Rankinen, T.; Pérusse, L.; Gagnon, J.; Chagnon, Y.C.; Leon, A.S.; Skinner, J.S.; Wilmore, J.H.; Rao, D.C.; Bouchard, C. Angiotensin-converting enzyme ID polymorphism and fitness phenotype in the HERITAGE Family Study. J. Appl. Physiol. 2000, 88, 1029–1035. [Google Scholar] [CrossRef]
- Ruiz, J.R.; Arteta, D.; Buxens, A.; Artieda, M.; Gómez-Gallego, F.; Santiago, C.; Yvert, T.; Morán, M.; Lucia, A. Can we identify a power-oriented polygenic profile? J. Appl. Physiol. 2010, 108, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zaken, S.; Eliakim, A.; Nemet, D.; Meckel, Y. Genetic Variability Among Power Athletes: The Stronger vs. the Faster. J. Strength. Cond. Res. 2019, 33, 1505–1511. [Google Scholar] [CrossRef]
- Maciejewska-Skrendo, A.; Cięszczyk, P.; Chycki, J.; Sawczuk, M.; Smółka, W. Genetic Markers Associated with Power Athlete Status. J. Hum. Kinet. 2019, 68, 17–36. [Google Scholar] [CrossRef]
- Lucía, A.; Gómez-Gallego, F.; Chicharro, J.L.; Hoyos, J.; Celaya, K.; Córdova, A.; Villa, G.; Alonso, J.M.; Barriopedro, M.; Pérez, M.; et al. Is there an association between ACE and CKMM polymorphisms and cycling performance status during 3-week races? Int. J. Sports Med. 2005, 26, 442–447. [Google Scholar] [CrossRef]
- Papadimitriou, I.D.; Lockey, S.J.; Voisin, S.; Herbert, A.J.; Garton, F.; Houweling, P.J.; Cieszczyk, P.; Maciejewska-Skrendo, A.; Sawczuk, M.; Massidda, M.; et al. No association between ACTN3 R577X and ACE I/D polymorphisms and endurance running times in 698 Caucasian athletes. BMC Genom. 2018, 19, 13. [Google Scholar] [CrossRef] [PubMed]
- Ash, G.I.; Scott, R.A.; Deason, M.; Dawson, T.A.; Wolde, B.; Bekele, Z.; Teka, S.; Pitsiladis, Y.P. No association between ACE gene variation and endurance athlete status in Ethiopians. Med. Sci. Sports Exerc. 2011, 43, 590–597. [Google Scholar] [CrossRef]
- Papadimitriou, I.D.; Papadopoulos, C.; Kouvatsi, A.; Triantaphyllidis, C. The ACE I/D polymorphism in elite Greek track and field athletes. J. Sports Med. Phys. Fit. 2009, 49, 459–463. [Google Scholar]
- Rankinen, T.; Wolfarth, B.; Simoneau, J.A.; Maier-Lenz, D.; Rauramaa, R.; Rivera, M.A.; Boulay, M.R.; Chagnon, Y.C.; Pérusse, L.; Keul, J.; et al. No association between the angiotensin-converting enzyme ID polymorphism and elite endurance athlete status. J. Appl. Physiol. 2000, 88, 1571–1575. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Druzhevskaya, A.M.; Astratenkova, I.V.; Popov, D.V.; Vinogradova, O.L.; Rogozkin, V.A. The ACTN3 R577X polymorphism in Russian endurance athletes. Br. J. Sports Med. 2010, 44, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.A.; Moran, C.; Wilson, R.H.; Onywera, V.; Boit, M.K.; Goodwin, W.H.; Gohlke, P.; Payne, J.; Montgomery, H.; Pitsiladis, Y.P. No association between Angiotensin Converting Enzyme (ACE) gene variation and endurance athlete status in Kenyans. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2005, 141, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Saunders, C.J.; September, A.V.; Xenophontos, S.L.; Cariolou, M.A.; Anastassiades, L.C.; Noakes, T.D.; Collins, M. No association of the ACTN3 gene R577X polymorphism with endurance performance in Ironman Triathlons. Ann. Hum. Genet. 2007, 71 Pt 6, 777–781. [Google Scholar] [CrossRef]
- Cieszczyk, P.; Krupecki, K.; Maciejewska, A.; Sawczuk, M. The angiotensin converting enzyme gene I/D polymorphism in Polish rowers. Int. J. Sports Med. 2009, 30, 624–627. [Google Scholar] [CrossRef]
- Puthucheary, Z.; Skipworth, J.R.; Rawal, J.; Loosemore, M.; Van Someren, K.; Montgomery, H.E. Genetic influences in sport and physical performance. Sports Med. 2011, 41, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Powers, S.K.; Howley, E.T. Exercise Physiology: Theory and Application to Fitness and Performance, 4th ed.; McGraw-Hill College: Boston, MA, USA, 2001; pp. 30–44. [Google Scholar]
- Jo, C.-Y.; Kim, C.-H. Polygenic Association of ACE ID and ACTN3 R577X polymorphisms with Korean endurance status. Korean J. Phys. Educ. 2013, 52, 739–753. [Google Scholar]
- Koch, W.; Latz, W.; Eichinger, M.; Ganser, C.; Schömig, A.; Kastrati, A. Genotyping of the angiotensin I-converting enzyme gene insertion/deletion polymorphism by the TaqMan method. Clin. Chem. 2005, 51, 1547–1549. [Google Scholar] [CrossRef]
- Vincent, B.; De Bock, K.; Ramaekers, M.; Van den Eede, E.; Van Leemputte, M.; Hespel, P.; Thomis, M.A. ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol. Genom. 2007, 32, 58–63. [Google Scholar] [CrossRef]
- Macarthur, D.G.; North, K.N. Genes and human elite athletic performance. Hum. Genet. 2005, 116, 331–339. [Google Scholar] [CrossRef]
- Wang, H.; Meng, L.; Zhao, L.; Wang, J.; Liu, X.; Mi, W. Methylenetetrahydrofolate reductase polymorphism C677T is a protective factor for pediatric acute lymphoblastic leukemia in the Chinese population: A meta-analysis. Genet. Test. Mol. Biomark. 2012, 16, 1401–1407. [Google Scholar] [CrossRef]
- MacKnight, N.J.; Dimos, B.A.; Beavers, K.M.; Muller, E.M.; Brandt, M.E.; Mydlarz, L.D. Disease resistance in coral is mediated by distinct adaptive and plastic gene expression profiles. Sci. Adv. 2022, 8, eabo6153. [Google Scholar] [CrossRef] [PubMed]
- Lavi, E.S.; Lin, Z.P.; Ratner, E.S. Gene expression of non-homologous end-joining pathways in the prognosis of ovarian cancer. iScience 2023, 26, 107934. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zaken, S.; Eliakim, A.; Nemet, D.; Rabinovich, M.; Kassem, E.; Meckel, Y. ACTN3 Polymorphism: Comparison Between Elite Swimmers and Runners. Sports Med. Open 2015, 1, 13. [Google Scholar] [CrossRef]
- MacArthur, D.G.; Seto, J.T.; Chan, S.; Quinlan, K.G.; Raftery, J.M.; Turner, N.; Nicholson, M.D.; Kee, A.J.; Hardeman, E.C.; Gunning, P.W.; et al. An Actn3 knockout mouse provides mechanistic insights into the association between alpha-actinin-3 deficiency and human athletic performance. Hum. Mol. Genet. 2008, 17, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Eynon, N.; Duarte, J.A.; Oliveira, J.; Sagiv, M.; Yamin, C.; Meckel, Y.; Sagiv, M.; Goldhammer, E. ACTN3 R577X polymorphism and Israeli top-level athletes. Int. J. Sports Med. 2009, 30, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Huang, C.; Chang, Q.; Zhang, L.; Huang, T. Association between the ACTN3 R577X polymorphism and female endurance athletes in China. Int. J. Sports Med. 2010, 31, 913–916. [Google Scholar] [CrossRef]
- Kikuchi, N.; Miyamoto-Mikami, E.; Murakami, H.; Nakamura, T.; Min, S.K.; Mizuno, M.; Naito, H.; Miyachi, M.; Nakazato, K.; Fuku, N. ACTN3 R577X genotype and athletic performance in a large cohort of Japanese athletes. Eur. J. Sport. Sci. 2016, 16, 694–701. [Google Scholar] [CrossRef]
- Anomasiri, W.; Sanguanrungsirikul, S.; Saichandee, P. Low dose creatine supplementation enhances sprint phase of 400 m swimming performance. J. Med. Assoc. Thail. 2004, 87 (Suppl. S2), S228–S232. [Google Scholar]
- Hogarth, M.W.; Garton, F.C.; Houweling, P.J.; Tukiainen, T.; Lek, M.; Macarthur, D.G.; Seto, J.T.; Quinlan, K.G.; Yang, N.; Head, S.I.; et al. Analysis of the ACTN3 heterozygous genotype suggests that alpha-actinin-3 controls sarcomeric composition and muscle function in a dose-dependent fashion. Hum. Mol. Genet. 2016, 25, 866–877. [Google Scholar] [CrossRef]
- Garton, F.C.; Seto, J.T.; Quinlan, K.G.R.; Yang, N.; Houweling, P.J.; North, K.N. alpha-Actinin-3 deficiency alters muscle adaptation in response to denervation and immobilization. Hum. Mol. Genet. 2014, 23, 1879–1893. [Google Scholar] [CrossRef]
- Chan, S.; Seto, J.T.; MacArthur, D.G.; Yang, N.; North, K.N.; Head, S.I. A gene for speed: Contractile properties of isolated whole EDL muscle from an alpha-actinin-3 knockout mouse. Am. J. Physiol. Cell Physiol. 2008, 295, C897–C9042008. [Google Scholar] [CrossRef] [PubMed]
- Berman, Y.; North, K.N. A gene for speed: The emerging role of alpha-actinin-3 in muscle metabolism. Physiology 2010, 25, 250–259. [Google Scholar] [CrossRef]
- Quinlan, K.G.; Seto, J.T.; Turner, N.; Vandebrouck, A.; Floetenmeyer, M.; Macarthur, D.G.; Raftery, J.M.; Lek, M.; Yang, N.; Parton, R.G.; et al. alpha-actinin-3 deficiency results in reduced glycogen phosphorylase activity and altered calcium handling in skeletal muscle. Hum. Mol. Genet. 2010, 19, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.; Seto, J.T.; Houweling, P.J.; Yang, N.; North, K.N.; Head, S.I. Properties of extensor digitorum longus muscle and skinned fibers from adult and aged male and female Actn3 knockout mice. Muscle Nerve 2011, 43, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, M.H.; Roseguini, B. Mechanisms for exercise training-induced increases in skeletal muscle blood flow capacity: Differences with interval sprint training versus aerobic endurance training. J. Physiol. Pharmacol. 2008, 59 (Suppl. S7), 71–88. [Google Scholar]
- Broos, S.; Malisoux, L.; Theisen, D.; Francaux, M.; Deldicque, L.; Thomis, M.A. Role of alpha-actinin-3 in contractile properties of human single muscle fibers: A case series study in paraplegics. PLoS ONE 2012, 7, e49281. [Google Scholar] [CrossRef]
- Norman, B.; Esbjörnsson, M.; Rundqvist, H.; Osterlund, T.; von Walden, F.; Tesch, P.A. Strength, power, fiber types, and mRNA expression in trained men and women with different ACTN3 R577X genotypes. J. Appl. Physiol. 2009, 106, 959–965. [Google Scholar] [CrossRef]
- Montgomery, H.E.; Marshall, R.; Hemingway, H.; Myerson, S.; Clarkson, P.; Dollery, C.; Hayward, M.; Holliman, D.E.; Jubb, M.; World, M.; et al. Human gene for physical performance. Nature 1998, 393, 221–222. [Google Scholar] [CrossRef]
- Myerson, S.; Hemingway, H.; Budget, R.; Martin, J.; Humphries, S.; Montgomery, H. Human angiotensin I-converting enzyme gene and endurance performance. J. Appl. Physiol. 1999, 87, 1313–1316. [Google Scholar] [CrossRef]
- Shahmoradi, S.; Ahmadalipour, A.; Salehi, M. Evaluation of ACE gene I/D polymorphism in Iranian elite athletes. Adv. Biomed. Res. 2014, 3, 207. [Google Scholar] [CrossRef]
- Oh, S.D. The distribution of I/D polymorphism in the ACE gene among Korean male elite athletes. J. Sports Med. Phys. Fit. 2007, 47, 250–254. [Google Scholar]
- Taylor, R.R.; Mamotte, C.D.; Fallon, K.; van Bockxmeer, F.M. Elite athletes and the gene for angiotensin-converting enzyme. J. Appl. Physiol. 1999, 87, 1035–1037. [Google Scholar] [CrossRef] [PubMed]
- Tobina, T.; Michishita, R.; Yamasawa, F.; Zhang, B.; Sasaki, H.; Tanaka, H.; Saku, K.; Kiyonaga, A. Association between the angiotensin I-converting enzyme gene insertion/deletion polymorphism and endurance running speed in Japanese runners. J. Physiol. Sci. 2010, 60, 325–330. [Google Scholar] [CrossRef]
- Pranckeviciene, E.; Gineviciene, V.; Jakaitiene, A.; Januska, L.; Utkus, A. Total Genotype Score Modelling of Polygenic Endurance-Power Profiles in Lithuanian Elite Athletes. Genes 2021, 12, 1067. [Google Scholar] [CrossRef]
- Eynon, N.; Ruiz, J.R.; Oliveira, J.; Duarte, J.A.; Birk, R.; Lucia, A. Genes and elite athletes: A roadmap for future research. J. Physiol. 2011, 589 Pt 13, 3063–3070. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, J.; Lancha, A.H., Jr. Total genotype score and athletic status: An exploratory cross-sectional study of a Brazilian athlete cohort. Ann. Hum. Genet. 2020, 84, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.G.; Rayson, M.P.; Jubb, M.; World, M.; Woods, D.R.; Hayward, M.; Martin, J.; Humphries, S.E.; Montgomery, H.E. The ACE gene and muscle performance. Nature 2000, 403, 614. [Google Scholar] [CrossRef]
- Brink, M.; Wellen, J.; Delafontaine, P. Angiotensin II causes weight loss and decreases circulating insulin-like growth factor I in rats through a pressor-independent mechanism. J. Clin. Investig. 1996, 97, 2509–2516. [Google Scholar] [CrossRef]
- Zhao, G.; Bernstein, R.D.; Hintze, T.H. Nitric oxide and oxygen utilization: Exercise, heart failure and diabetes. Coron. Artery Dis. 1999, 10, 315–320. [Google Scholar] [CrossRef]
- Valdivieso, P.; Vaughan, D.; Laczko, E.; Brogioli, M.; Waldron, S.; Rittweger, J.; Flück, M. The Metabolic Response of Skeletal Muscle to Endurance Exercise Is Modified by the ACE-I/D Gene Polymorphism and Training State. Front. Physiol. 2017, 8, 993. [Google Scholar] [CrossRef]
- McAuley, A.B.T.; Hughes, D.C.; Tsaprouni, L.G.; Varley, I.; Suraci, B.; Roos, T.R.; Herbert, A.J.; Kelly, A.L. The association of the ACTN3 R577X and ACE I/D polymorphisms with athlete status in football: A systematic review and meta-analysis. J. Sports Sci. 2021, 39, 200–211. [Google Scholar] [CrossRef]
- Vaughan, D.; Brogioli, M.; Maier, T.; White, A.; Waldron, S.; Rittweger, J.; Toigo, M.; Wettstein, J.; Laczko, E.; Flück, M. The Angiotensin Converting Enzyme Insertion/Deletion Polymorphism Modifies Exercise-Induced Muscle Metabolism. PLoS ONE 2016, 11, e0149046. [Google Scholar] [CrossRef]
- Mägi, A.; Unt, E.; Prans, E.; Raus, L.; Eha, J.; Veraksitš, A.; Kingo, K.; Kõks, S. The Association Analysis between ACE and ACTN3 Genes Polymorphisms and Endurance Capacity in Young Cross-Country Skiers: Longitudinal Study. J. Sports Sci. Med. 2016, 15, 287–294. [Google Scholar] [PubMed]
- Booth, F.W.; Ruegsegger, G.N.; Toedebusch, R.G.; Yan, Z. Endurance Exercise and the Regulation of Skeletal Muscle Metabolism. Prog. Mol. Biol. Transl. Sci. 2015, 135, 129–151. [Google Scholar] [CrossRef] [PubMed]
- Layec, G.; Haseler, L.J.; Hoff, J.; Hart, C.R.; Liu, X.; Le Fur, Y.; Jeong, E.K.; Richardson, R.S. Short-term training alters the control of mitochondrial respiration rate before maximal oxidative ATP synthesis. Acta Physiol. 2013, 208, 376–386. [Google Scholar] [CrossRef]
- Mortensen, S.P.; Saltin, B. Regulation of the skeletal muscle blood flow in humans. Exp. Physiol. 2014, 99, 1552–1558. [Google Scholar] [CrossRef]
- Gouzi, F.; Préfaut, C.; Abdellaoui, A.; Roudier, E.; de Rigal, P.; Molinari, N.; Laoudj-Chenivesse, D.; Mercier, J.; Birot, O.; Hayot, M. Blunted muscle angiogenic training-response in COPD patients versus sedentary controls. Eur. Respir. J. 2013, 41, 806–814. [Google Scholar] [CrossRef]
- Lewis, M.I.; Fournier, M.; Wang, H.; Storer, T.W.; Casaburi, R.; Kopple, J.D. Effect of endurance and/or strength training on muscle fiber size, oxidative capacity, and capillarity in hemodialysis patients. J. Appl. Physiol. 2015, 119, 865–871. [Google Scholar] [CrossRef]
- Messonnier, L.; Freund, H.; Féasson, L.; Prieur, F.; Castells, J.; Denis, C.; Linossier, M.T.; Geyssant, A.; Lacour, J.R. Blood lactate exchange and removal abilities after relative high-intensity exercise: Effects of training in normoxia and hypoxia. Eur. J. Appl. Physiol. 2001, 84, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, J.M.; Rankinen, T.; Loos, R.J.; Pérusse, L.; Roth, S.M.; Wolfarth, B.; Bouchard, C. Advances in exercise, fitness, and performance genomics in 2010. Med. Sci. Sports Exerc. 2011, 43, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Siska, V.; Jones, E.R.; Jeon, S.; Bhak, Y.; Kim, H.M.; Cho, Y.S.; Kim, H.; Lee, K.; Veselovskaya, E.; Balueva, T.; et al. Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago. Sci. Adv. 2017, 3, e1601877. [Google Scholar] [CrossRef] [PubMed]
| EPEA | Controls | ||
|---|---|---|---|
| (n = 45) | (n = 679) | ||
| Sex | |||
| Male | 36 (80.0%) | 361 (53.2%) | |
| Female | 9 (20.0%) | 318 (46.8%) | |
| Age | 20.6 ± 4.4 | 32.6 ± 4.8 | |
| Sport events | |||
| Marathon | 15 (33.3%) | ||
| 10,000 m run | 10 (22.2%) | ||
| 5000 m run | 18 (40.0%) | ||
| 10~20-kmW | 2 (4.4%) |
| Material | Designation | Sequence |
|---|---|---|
| Primers | ACE 111 a | CCC-ATC-CTT-TCT-CCC-ATT-TCT-C |
| ACE 112 b | AGC-TGG-AAT-AAA-ATT-GGC-GAA-AC | |
| ACE 113 a | CCT-CCC-AAA-GTG-CTG-GGA-TTA | |
| Probes | I allele-specific c | AGG-CGT-GAT-ACA-GTC-A |
| D allele-specific d | TGC-TGC-CTA-TAC-AGT-CA |
| Complex Genotypes | |||
|---|---|---|---|
| EDCGs | ENCGs | ERCGs | |
| RX+XX/II+DD | RX+XX/ID RR/II+DD | RR/ID | |
| EPEA | 23 * (51.1%) | 21 (46.7%) | 1 † (2.2%) |
| Controls | 227 (33.4%) | 358 (52.7%) | 94 (13.8%) |
| Complex Genotypes | EPEA (n = 45) | Controls (n = 679) | p (Linear by Linear Association) | EPEA vs. Controls | |
|---|---|---|---|---|---|
| ORs (90% CI) | |||||
| RX+XX | II (n = 165) | 15 (33.3%) | 150 (22.1%) | 0.007 | 1.763 (1.037–3.089) |
| DD (n = 85) | 8 (17.8%) | 77 (11.3%) | 1.690 (0.863–3.221) | ||
| ID (n = 267) | 15 (33.3%) | 252 (37.1%) | 0.847 (0.504–1.468) | ||
| RR | II (n = 69) | 1 (2.2%) | 68 (10.0%) | 0.264 (0.051–1.235) | |
| DD (n = 43) | 5 (11.1%) | 38 (5.6%) | 2.109 (0.945–4.742) | ||
| ID (n = 95) | 1 (2.2%) | 94 (13.8%) | 0.183 (0.036–0.847) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chae, J.H.; Eom, S.-H.; Lee, S.-K.; Jung, J.-H.; Kim, C.-H. Association between Complex ACTN3 and ACE Gene Polymorphisms and Elite Endurance Sports in Koreans: A Case–Control Study. Genes 2024, 15, 1110. https://doi.org/10.3390/genes15091110
Chae JH, Eom S-H, Lee S-K, Jung J-H, Kim C-H. Association between Complex ACTN3 and ACE Gene Polymorphisms and Elite Endurance Sports in Koreans: A Case–Control Study. Genes. 2024; 15(9):1110. https://doi.org/10.3390/genes15091110
Chicago/Turabian StyleChae, Ji Heon, Seon-Ho Eom, Sang-Ki Lee, Joo-Ha Jung, and Chul-Hyun Kim. 2024. "Association between Complex ACTN3 and ACE Gene Polymorphisms and Elite Endurance Sports in Koreans: A Case–Control Study" Genes 15, no. 9: 1110. https://doi.org/10.3390/genes15091110





