GSM1 Requires Hap4 for Expression and Plays a Role in Gluconeogenesis and Utilization of Nonfermentable Carbon Sources
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Media, Growth Conditions, Strains, and Plasmids
2.2. Yeast Transformation and β-Galactosidase Activity Assays
2.3. Serial Dilution of Cells for Growth Analysis
2.4. Cellular Extract Preparation, Immunoblotting, and Immunoprecipitation
3. Results
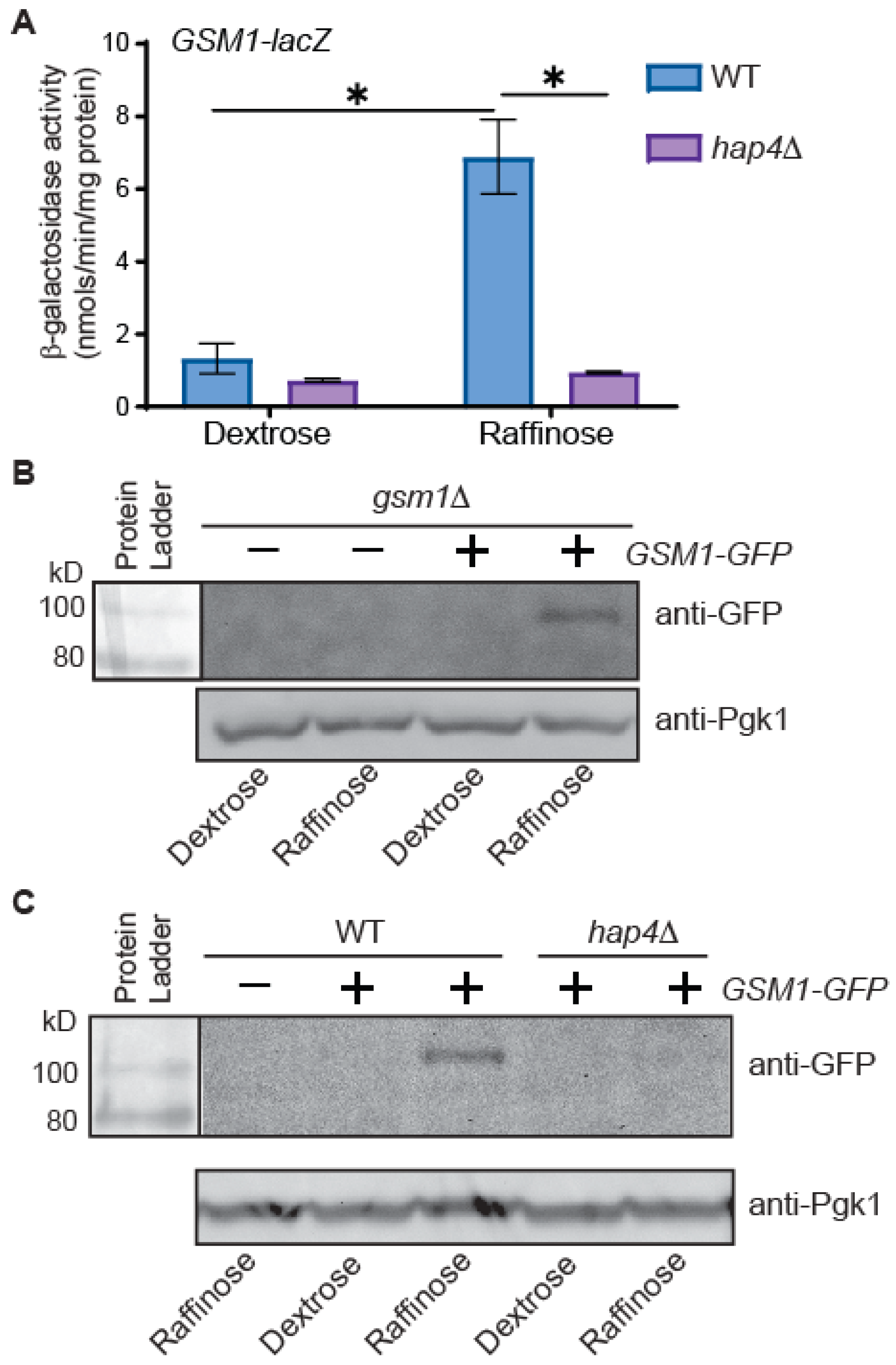
3.1. GSM1 Expression Is Subject to Glucose Repression and Requires Hap4 under Glucose-Derepression Conditions
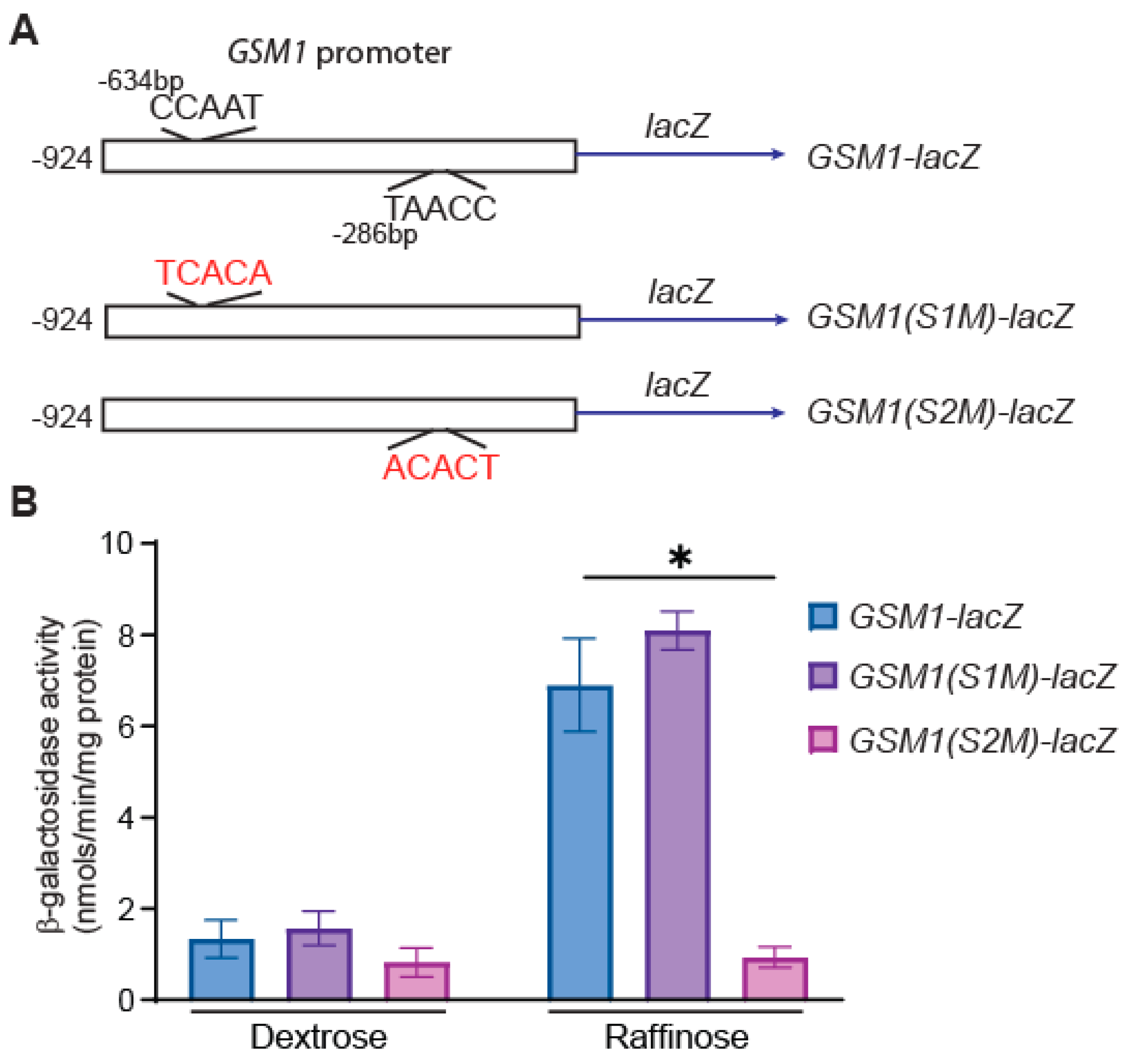
3.2. A Proximal CCAAT Sequence Element Is Required for GSM1 Expression
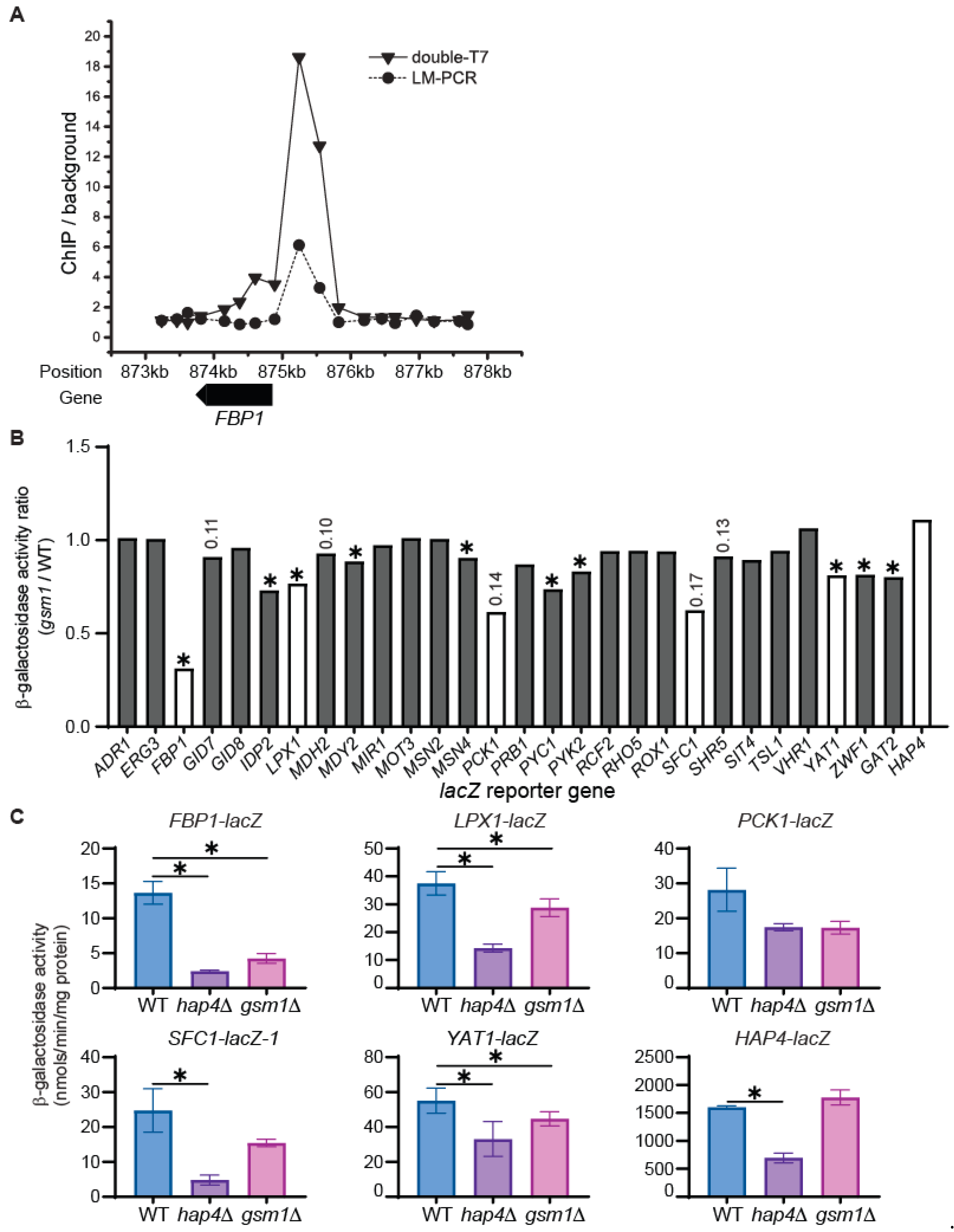
3.3. Identification of Potential Gsm1 Target Genes via Analysis of lacZ Reporter Genes
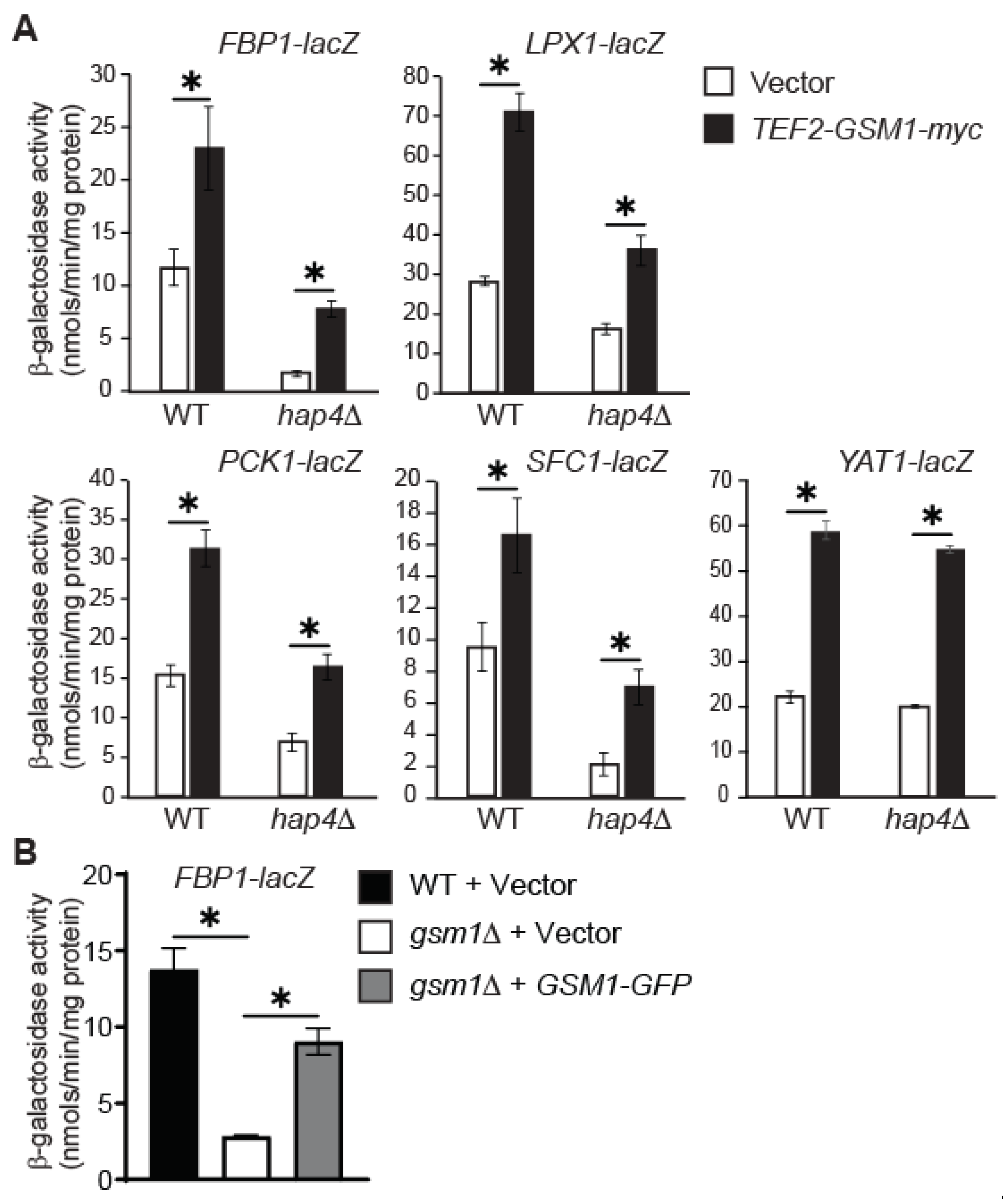
3.4. GSM1 Overexpression Increases Expression of FBP1-, LPX1-, PCK1-, SFC1-, and YAT1-lacZ Reporter Genes in hap4∆ Mutant Cells
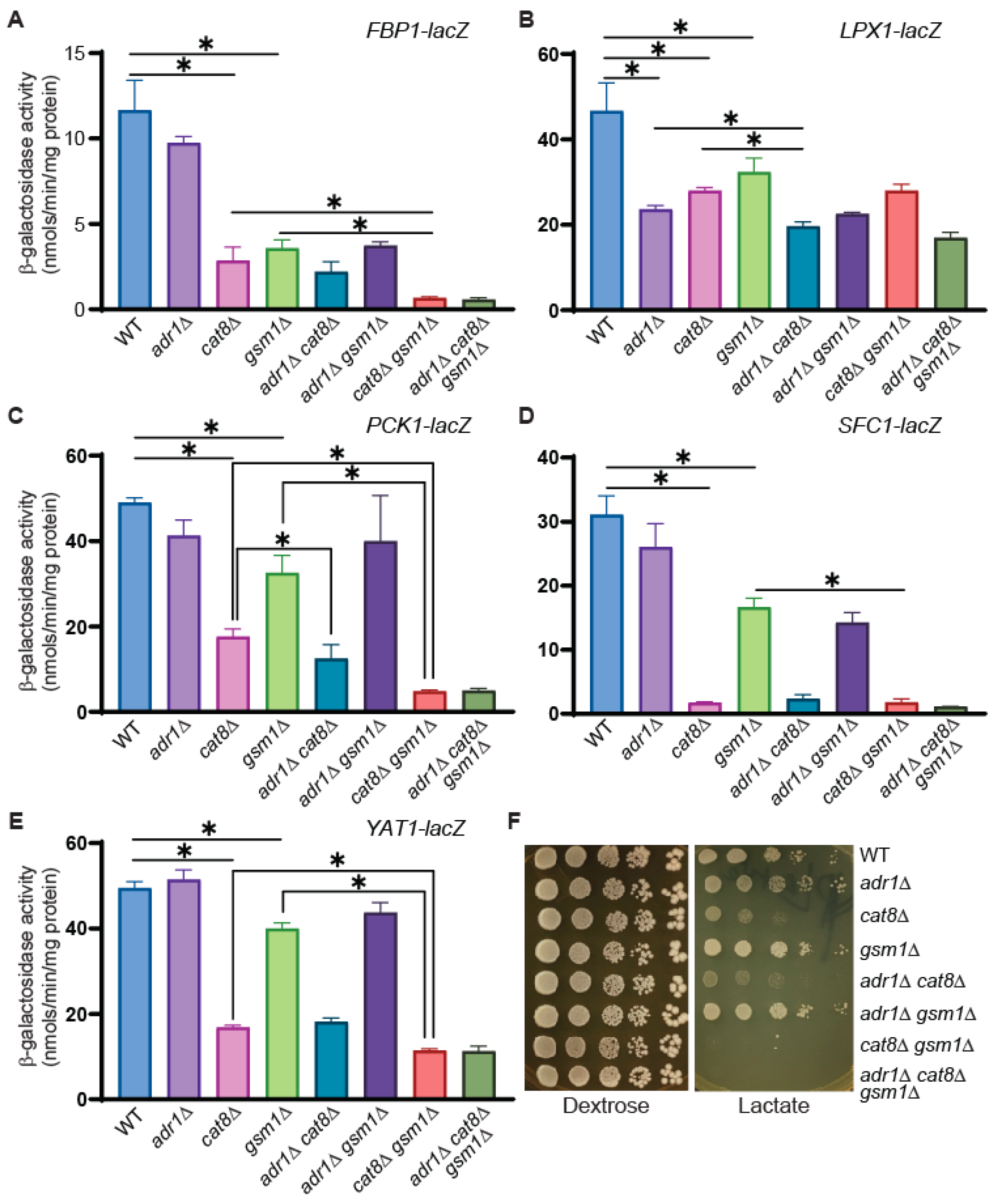
3.5. Cat8 and Gsm1 Are Important in the Transcriptional Regulation of FBP1, PCK1, SFC1, and YAT1 and in the Utilization of Lactate
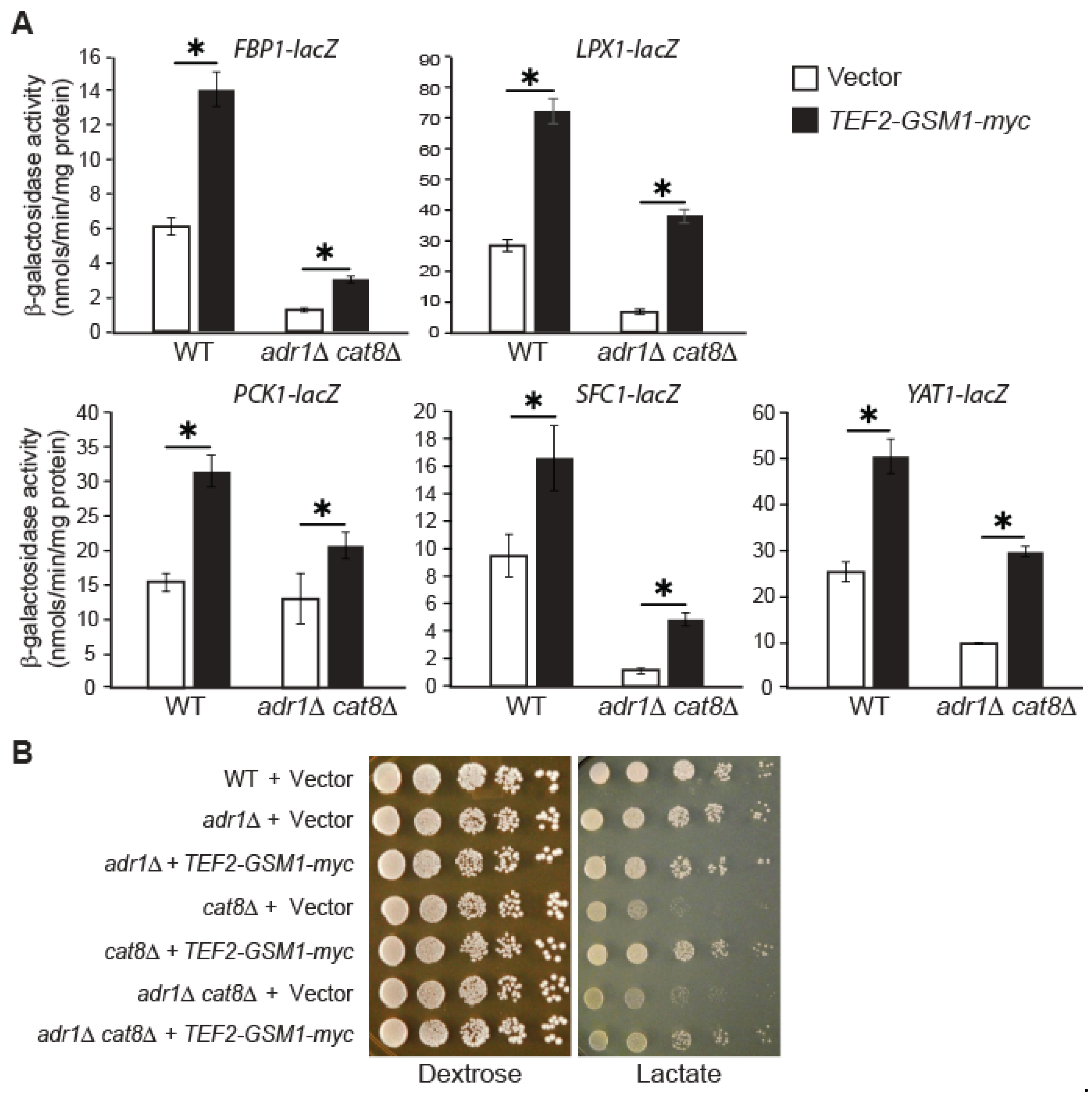
3.6. GSM1 Overexpression Increases Target-Gene Expression in adr1∆ cat8∆ Mutant Cells and Suppresses Growth Defects Associated with cat8Δ
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turcotte, B.; Liang, X.B.; Robert, F.; Soontorngun, N. Transcriptional Regulation of Nonfermentable Carbon Utilization in Budding Yeast. FEMS Yeast Res. 2010, 10, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Schuller, H.J. Transcriptional Control of Nonfermentative Metabolism in the Yeast Saccharomyces Cerevisiae. Curr. Genet. 2003, 43, 139–160. [Google Scholar] [CrossRef]
- Simon, M.; Adam, G.; Rapatz, W.; Spevak, W.; Ruis, H. The Saccharomyces Cerevisiae Adr1 Gene Is a Positive Regulator of Transcription of Genes Encoding Peroxisomal Proteins. Mol. Cell. Biol. 1991, 11, 699–704. [Google Scholar] [PubMed]
- Young, E.T.; Dombek, K.M.; Tachibana, C.; Ideker, T. Multiple Pathways Are Co-Regulated by the Protein Kinase Snf1 and the Transcription Factors Adr1 and Cat8. J. Biol. Chem. 2003, 278, 26146–26158. [Google Scholar] [CrossRef] [PubMed]
- Rahner, A.; Scholer, A.; Martens, E.; Gollwitzer, B.; Schuller, H.J. Dual Influence of the Yeast Cat1p (Snf1p) Protein Kinase on Carbon Source-Dependent Transcriptional Activation of Gluconeogenic Genes by the Regulatory Gene Cat8. Nucleic Acids Res. 1996, 24, 2331–2337. [Google Scholar] [CrossRef] [PubMed]
- Vincent, O.; Carlson, M. Sip4, a Snf1 Kinase-Dependent Transcriptional Activator, Binds to the Carbon Source-Responsive Element of Gluconeogenic Genes. Embo. J. 1998, 17, 7002–7008. [Google Scholar] [CrossRef]
- Hedges, D.; Proft, M.; Entian, K.D. Cat8, a New Zinc Cluster-Encoding Gene Necessary for Derepression of Gluconeogenic Enzymes in the Yeast Saccharomyces Cerevisiae. Mol. Cell. Biol. 1995, 15, 1915–1922. [Google Scholar] [CrossRef]
- Hiesinger, M.; Roth, S.; Meissner, E.; Schuller, H.J. Contribution of Cat8 and Sip4 to the Transcriptional Activation of Yeast Gluconeogenic Genes by Carbon Source-Responsive Elements. Curr. Genet. 2001, 39, 68–76. [Google Scholar] [CrossRef]
- Walther, K.; Schuller, H.J. Adr1 and Cat8 Synergistically Activate the Glucose-Regulated Alcohol Dehydrogenase Gene Adh2 of the Yeast Saccharomyces Cerevisiae. Microbiology (Read.) 2001, 147 Pt 8, 2037–2044. [Google Scholar] [CrossRef]
- Gasmi, N.; Jacques, P.E.; Klimova, N.; Guo, X.; Ricciardi, A.; Robert, F.; Turcotte, B. The Switch from Fermentation to Respiration in Saccharomyces Cerevisiae Is Regulated by the Ert1 Transcriptional Activator/Repressor. Genetics 2014, 198, 547–560. [Google Scholar] [CrossRef]
- Soontorngun, N.; Larochelle, M.; Drouin, S.; Robert, F.; Turcotte, B. Regulation of Gluconeogenesis in Saccharomyces Cerevisiae Is Mediated by Activator and Repressor Functions of Rds2. Mol. Cell. Biol. 2007, 27, 7895–7905. [Google Scholar] [CrossRef] [PubMed]
- Soontorngun, N.; Baramee, S.; Tangsombatvichit, C.; Thepnok, P.; Cheevadhanarak, S.; Robert, F.; Turcotte, B. Genome-Wide Location Analysis Reveals an Important Overlap between the Targets of the Yeast Transcriptional Regulators Rds2 and Adr1. Biochem. Biophys. Res. Commun. 2012, 423, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Ratnakumar, S.; Kacherovsky, N.; Arms, E.; Young, E.T. Snf1 Controls the Activity of Adr1 through Dephosphorylation of Ser230. Genetics 2009, 182, 735–745. [Google Scholar] [CrossRef]
- Lesage, P.; Yang, X.; Carlson, M. Yeast Snf1 Protein Kinase Interacts with Sip4, a C6 Zinc Cluster Transcriptional Activator: A New Role for Snf1 in the Glucose Response. Mol. Cell. Biol. 1996, 16, 1921–1928. [Google Scholar] [CrossRef]
- Randez-Gil, F.; Bojunga, N.; Proft, M.; Entian, K.D. Glucose Derepression of Gluconeogenic Enzymes in Saccharomyces Cerevisiae Correlates with Phosphorylation of the Gene Activator Cat8p. Mol. Cell. Biol. 1997, 17, 2502–2510. [Google Scholar] [CrossRef]
- Haurie, V.; Perrot, M.; Mini, T.; Jeno, P.; Sagliocco, F.; Boucherie, H. The Transcriptional Activator Cat8p Provides a Major Contribution to the Reprogramming of Carbon Metabolism during the Diauxic Shift in Saccharomyces Cerevisiae. J. Biol. Chem. 2001, 276, 76–85. [Google Scholar] [CrossRef]
- Tachibana, C.; Yoo, J.Y.; Tagne, J.B.; Kacherovsky, N.; Lee, T.I.; Young, E.T. Combined Global Localization Analysis and Transcriptome Data Identify Genes That Are Directly Coregulated by Adr1 and Cat8. Mol. Cell. Biol. 2005, 25, 2138–2146. [Google Scholar] [CrossRef] [PubMed]
- Buschlen, S.; Amillet, J.M.; Guiard, B.; Fournier, A.; Marcireau, C.; Bolotin-Fukuhara, M. The S. Cerevisiae Hap Complex, a Key Regulator of Mitochondrial Function, Coordinates Nuclear and Mitochondrial Gene Expression. Comp. Funct. Genom. 2003, 4, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Lascaris, R.; Bussemaker, H.J.; Boorsma, A.; Piper, M.; van der Spek, H.; Grivell, L.; Blom, J. Hap4p Overexpression in Glucose-Grown Saccharomyces Cerevisiae Induces Cells to Enter a Novel Metabolic State. Genome Biol. 2003, 4, R3. [Google Scholar] [CrossRef]
- Blom, J.; De Mattos, M.J.; Grivell, L.A. Redirection of the Respiro-Fermentative Flux Distribution in Saccharomyces Cerevisiae by Overexpression of the Transcription Factor Hap4p. Appl. Environ. Microbiol. 2000, 66, 1970–1973. [Google Scholar] [CrossRef]
- Forsburg, S.L.; Guarente, L. Identification and Characterization of Hap4: A Third Component of the Ccaat-Bound Hap2/Hap3 Heteromer. Genes Dev. 1989, 3, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Pinkham, J.L.; Guarente, L. Cloning and Molecular Analysis of the Hap2 Locus: A Global Regulator of Respiratory Genes in Saccharomyces Cerevisiae. Mol. Cell. Biol. 1985, 5, 3410–3416. [Google Scholar] [PubMed]
- Hahn, S.; Guarente, L. Yeast Hap2 and Hap3: Transcriptional Activators in a Heteromeric Complex. Science 1988, 240, 317–321. [Google Scholar] [CrossRef] [PubMed]
- McNabb, D.S.; Xing, Y.; Guarente, L. Cloning of Yeast Hap5: A Novel Subunit of a Heterotrimeric Complex Required for Ccaat Binding. Genes Dev. 1995, 9, 47–58. [Google Scholar] [CrossRef]
- Mao, Y.; Chen, C. The Hap Complex in Yeasts: Structure, Assembly Mode, and Gene Regulation. Front. Microbiol. 2019, 10, 1645. [Google Scholar] [CrossRef]
- Mattoon, J.R.; Caravajal, E.; Guthrie, D. Effects of Hap Mutations on Heme and Cytochrome Formation in Yeast. Curr. Genet. 1990, 17, 179–183. [Google Scholar] [CrossRef]
- Zhang, T.; Bu, P.; Zeng, J.; Vancura, A. Increased Heme Synthesis in Yeast Induces a Metabolic Switch from Fermentation to Respiration Even under Conditions of Glucose Repression. J. Biol. Chem. 2017, 292, 16942–16954. [Google Scholar] [CrossRef]
- McNabb, D.S.; Pinto, I. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA Complex in Saccharomyces Cerevisiae. Eukaryot. Cell 2005, 4, 1829–1839. [Google Scholar] [CrossRef]
- Roberts, G.G.; Hudson, A.P. Transcriptome Profiling of Saccharomyces Cerevisiae during a Transition from Fermentative to Glycerol-Based Respiratory Growth Reveals Extensive Metabolic and Structural Remodeling. Mol. Genet. Genom. 2006, 276, 170–186. [Google Scholar] [CrossRef]
- Martinez, K.P.; Gasmi, N.; Jeronimo, C.; Klimova, N.; Robert, F.; Turcotte, B. Yeast Zinc Cluster Transcription Factors Involved in the Switch from Fermentation to Respiration Show Interdependency for DNA Binding Revealing a Novel Type of DNA Recognition. Nucleic Acids Res. 2024, 52, 2242–2259. [Google Scholar] [CrossRef]
- van Bakel, H.; van Werven, F.J.; Radonjic, M.; Brok, M.O.; van Leenen, D.; Holstege, F.C.; Timmers, H.T. Improved Genome-Wide Localization by Chip-Chip Using Double-Round T7 RNA Polymerase-Based Amplification. Nucleic Acids Res. 2008, 36, e21. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.W.; Jona, G.; Chen, C.T.; Johnston, M.; Snyder, M. Linking DNA-Binding Proteins to Their Recognition Sequences by Using Protein Microarrays. Proc. Natl. Acad. Sci. USA 2006, 103, 9940–9945. [Google Scholar] [CrossRef] [PubMed]
- Amberg, D.C.; Burke, D.J.; Strathern, J.N. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2005. [Google Scholar]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional Profiling of the Saccharomyces Cerevisiae Genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Chelstowska, A.; Liu, Z.; Jia, Y.; Amberg, D.; Butow, R.A. Signalling between Mitochondria and the Nucleus Regulates the Expression of a New D-Lactate Dehydrogenase Activity in Yeast. Yeast 1999, 15, 1377–1391. [Google Scholar] [CrossRef]
- Capps, D.; Hunter, A.; Chiang, M.; Pracheil, T.; Liu, Z. Ubiquitin-Conjugating Enzymes Ubc1 and Ubc4 Mediate the Turnover of Hap4, a Master Regulator of Mitochondrial Biogenesis in Saccharomyces Cerevisiae. Microorganisms 2022, 10, 2370. [Google Scholar] [CrossRef] [PubMed]
- Mumberg, D.; Muller, R.; Funk, M. Yeast Vectors for the Controlled Expression of Heterologous Proteins in Different Genetic Backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Gietz, D.; Jean, A.S.; Woods, R.A.; Schiestl, R.H. Improved Method for High Efficiency Transformation of Intact Yeast Cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef]
- Yaffe, M.P.; Schatz, G. Two Nuclear Mutations That Block Mitochondrial Protein Import in Yeast. Proc. Natl. Acad. Sci. USA 1984, 81, 4819–4823. [Google Scholar] [CrossRef]
- Deng, Y.; He, T.; Wu, Y.; Vanka, P.; Yang, G.; Huang, Y.; Yao, H.; Brown, S.J. Computationally Analyzing the Possible Biological Function of Yjl103c--an Orf Potentially Involved in the Regulation of Energy Process in Yeast. Int. J. Mol. Med. 2005, 15, 123–127. [Google Scholar] [CrossRef]
- Forsburg, S.L.; Guarente, L. Communication between Mitochondria and the Nucleus in Regulation of Cytochrome Genes in the Yeast Saccharomyces Cerevisiae. Annu. Rev. Cell Biol. 1989, 5, 153–180. [Google Scholar] [CrossRef]
| Strain | Genotype | Source |
|---|---|---|
| BY4741 (WT) | MATa ura3 leu2 his3 met15 | Lab stock |
| ZLY2811 (hap4) | BY4741 hap4::kanMX4 | [34] |
| MBY123 (gsm1) | BY4741 gsm1::kanMX4 | This study |
| ZLY3707 (adr1) | BY4741 adr1::kanMX4 | [34] |
| ZLY3701 (cat∆) | BY4741 cat8::kanMX4 | [34] |
| ZLY5048 (adr1 cat8) | BY4741 adr1::kanMX4 cat8::HIS3 | This study |
| ZLY5103 (adr1 gsm1) | MATa ura3 leu2 his3 lys2 adr1::kanMX4 gsm1::kanMX4 | This study |
| ZLY5081 (cat8 gsm1) | BY4741 gsm1::kanMX4 cat8::HIS3 | This study |
| ZLY5109 (adr1 cat8 gsm1) | MATa ura3 leu2 his3 lys2 adr1::kanMX4 gsm1::kanMX4 cat8::HIS3 | This study |
| Plasmid | Description | Source |
|---|---|---|
| pZL3454 | pGSM1-lacZ expressing lacZ under the control of a 924 bp GSM1 promoter in the centromeric plasmid WEJ derived from pRS416 [35]. | This study |
| pZL3462 | pRS416-GSM1-GFP expressing Gsm1 from its own promoter with a GFP tag at the C-terminus. | This study |
| pMB165 | pGSM1(S1M)-lacZ expressing lacZ under the control of a GSM1 promoter with a mutation in the CCAAT sequence at position –634 in relation to the ATG start codon. | This study |
| pMB168 | pGSM1(S2M)-lacZ expressing lacZ under the control of a GSM1 promoter with a mutation in the CCAAT sequence at position −286 in relation to the ATG start codon. | This study |
| pMB179 | pFBP1-lacZ expressing lacZ under the control of a 924 bp FBP1 promoter. | This study |
| pZL3628 | pPCK1-lacZ expressing lacZ under the control of a 1484 bp PCK1 promoter. | This study |
| pMB209 | pSFC1-lacZ expressing lacZ under the control of a 1454 bp SFC1 promoter. | This study |
| pMB181 | pLPX1-lacZ expressing lacZ under the control of a 1039 bp LPX1 promoter. | This study |
| pZL3625 | pYAT1-lacZ expressing lacZ under the control of a 1555 bp YAT1 promoter. | This study |
| pDC124 | pHAP4-lacZ expressing lacZ under the control of a 1.8 kbp HAP4 promoter. | [36] |
| pZL3613 | pRS415-GSM1-GFP expressing Gsm1 from its own promoter with a GFP tag at the C-terminus. | This study |
| pZL3459 | pRS415-TEF2-GSM1-myc, expressing Gsm1 with a C-terminal 3x myc epitope tag under the control of the strong TEF2 promoter. The plasmid was constructed from pRS415TEF [37]. | This study |
| pRS415TEF | pRS415-TEF2p-CYC1 terminator. | [37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhondeley, M.; Liu, Z. GSM1 Requires Hap4 for Expression and Plays a Role in Gluconeogenesis and Utilization of Nonfermentable Carbon Sources. Genes 2024, 15, 1128. https://doi.org/10.3390/genes15091128
Bhondeley M, Liu Z. GSM1 Requires Hap4 for Expression and Plays a Role in Gluconeogenesis and Utilization of Nonfermentable Carbon Sources. Genes. 2024; 15(9):1128. https://doi.org/10.3390/genes15091128
Chicago/Turabian StyleBhondeley, Manika, and Zhengchang Liu. 2024. "GSM1 Requires Hap4 for Expression and Plays a Role in Gluconeogenesis and Utilization of Nonfermentable Carbon Sources" Genes 15, no. 9: 1128. https://doi.org/10.3390/genes15091128
APA StyleBhondeley, M., & Liu, Z. (2024). GSM1 Requires Hap4 for Expression and Plays a Role in Gluconeogenesis and Utilization of Nonfermentable Carbon Sources. Genes, 15(9), 1128. https://doi.org/10.3390/genes15091128






