SeqSNP-Based Targeted GBS Provides Insight into the Genetic Relationships among Global Collections of Brassica rapa ssp. oleifera (Turnip Rape)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. SNP Loci Selection, SeqSNP Assay Design and Target Sequencing
2.3. De Novo SNP Discovery, Target and Novel SNP Allele Counting, and Data Filtering
2.4. Converting Allele Counts into Allele Frequencies
2.5. Cluster and Principal Coordinate Analyses, Analysis of Molecular Variance (AMOVA), and Variant Analysis
3. Results
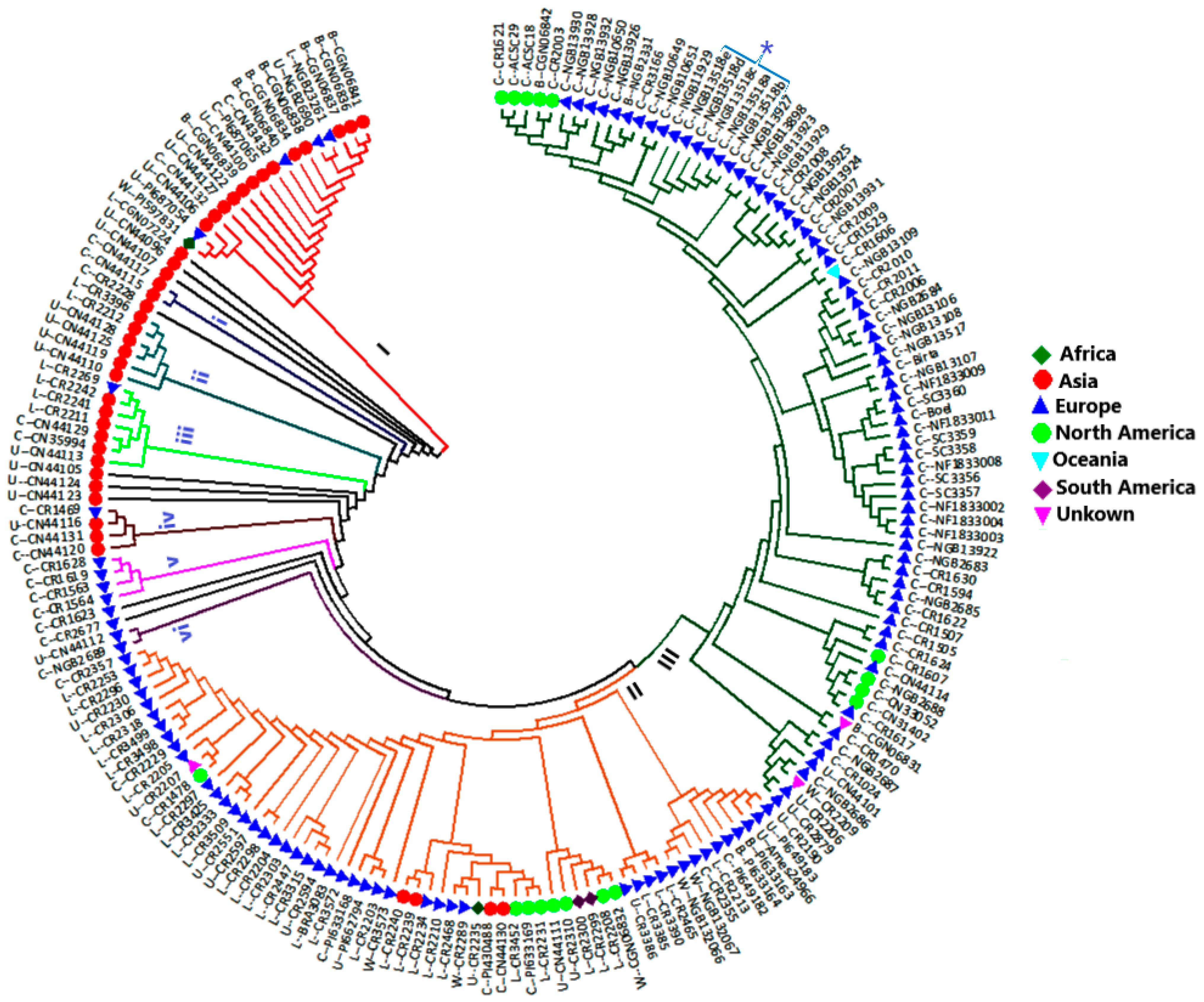
3.1. Cluster Analysis
3.2. Principal Coordinate Analysis
3.3. Analysis of Molecular Variance (AMOVA)
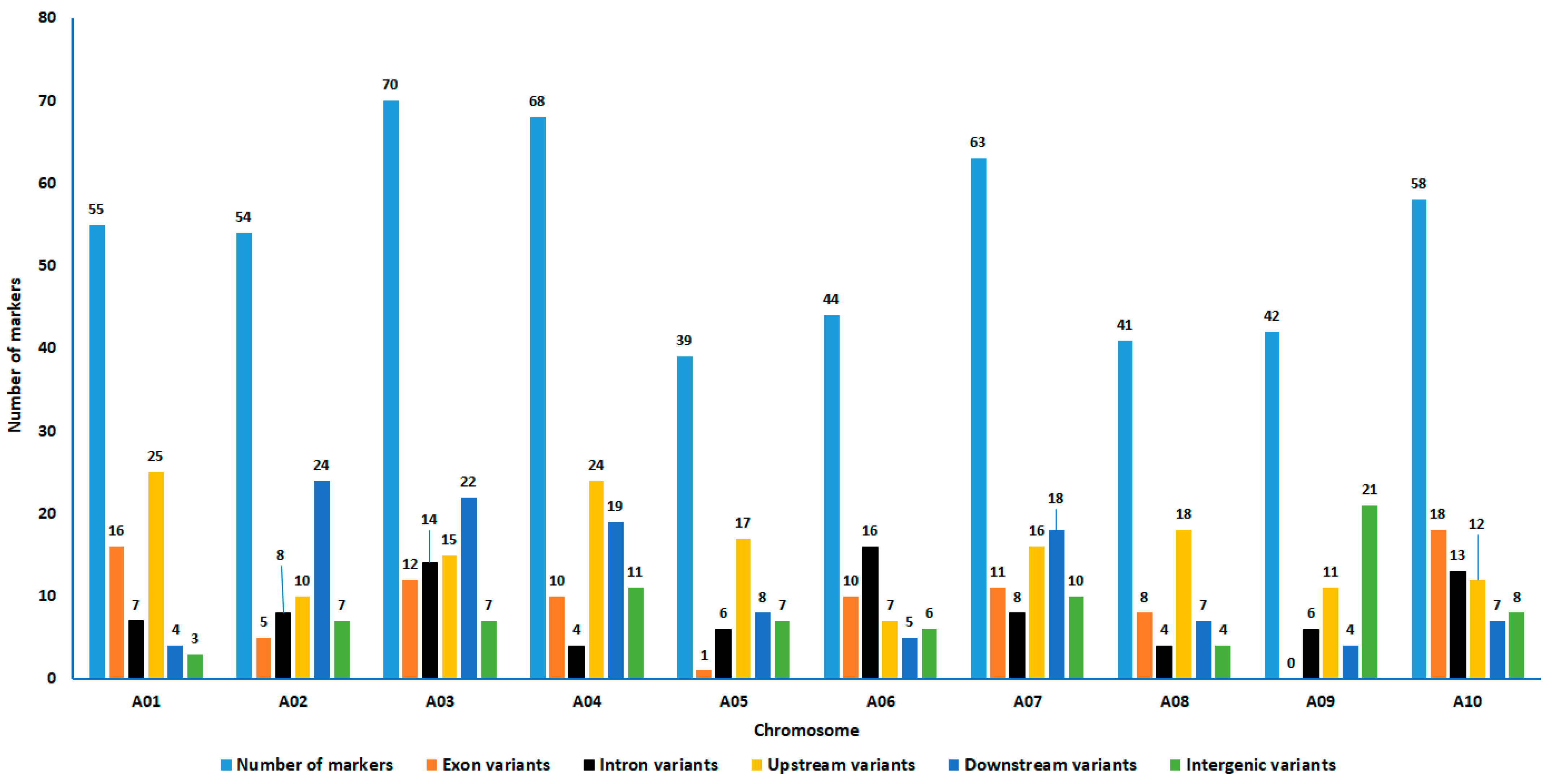
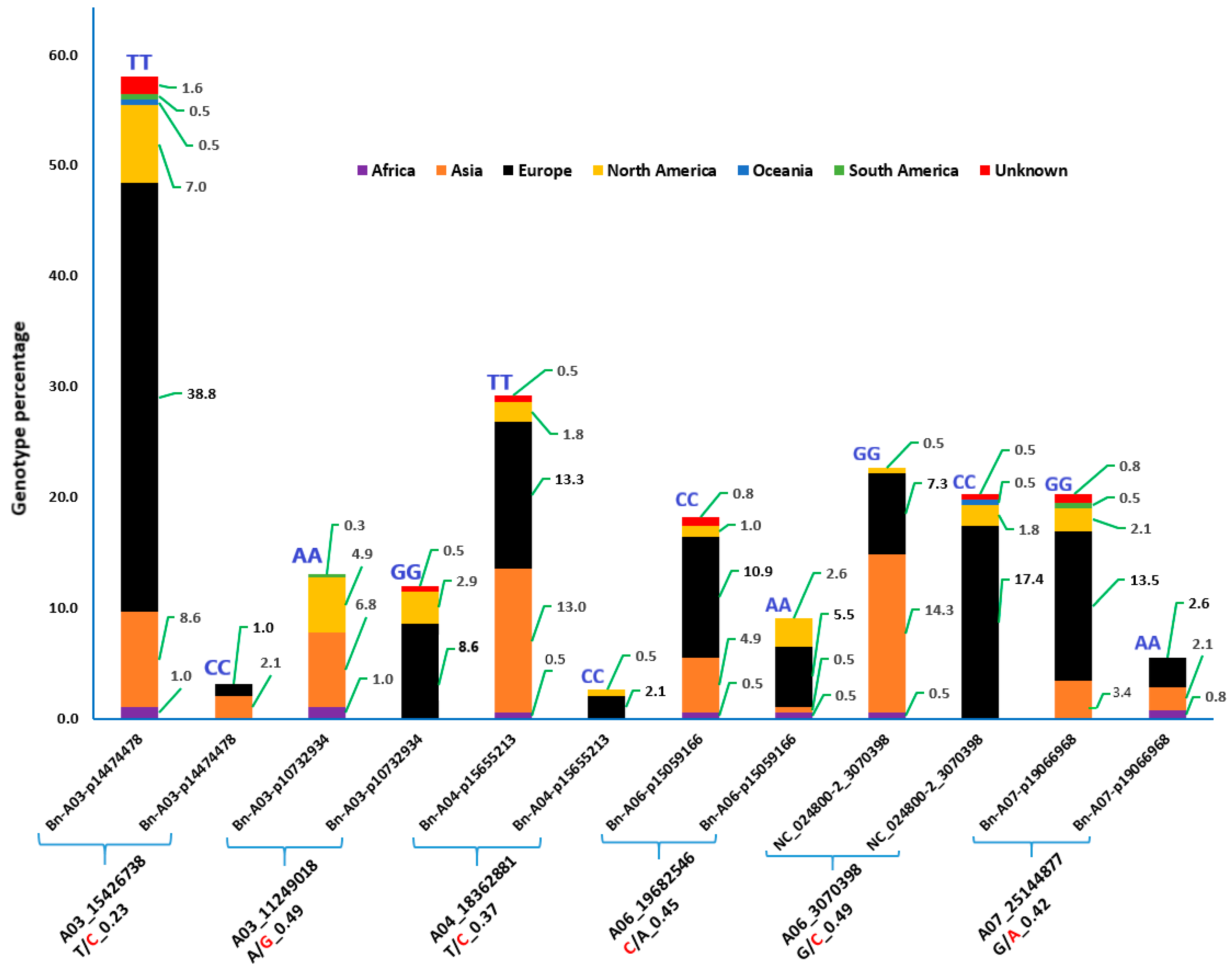
3.4. Region and/or Country-Specific Alleles and the Effects on Their Corresponding Genes
4. Discussion
4.1. Genic SNP Markers with Predicted Moderate to High Effects on Their Associated Genes
4.2. SNP Loci with Region/Country Specific Alleles
4.3. Genetic Relationship between Accessions
4.4. Analysis of Molecular Variance and Genetic Differentiation between Various Groups
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagaharu, U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Gómez-Campo, C.; Prakash, S. 2 Origin and domestication. In Developments in Plant Genetics and Breeding; Elsevier: Amsterdam, The Netherlands, 1999; Volume 4, pp. 33–58. [Google Scholar]
- Cartea, M.E.; Lema, M.; Francisco, M.; Velasco, P. Basic information on vegetable Brassica crops. In Genetics, Genomics and Breeding of Vegetable Brassicas; CRC Press: Boca Raton, FL, USA, 2011; pp. 1–33. [Google Scholar]
- Bird, K.A.; An, H.; Gazave, E.; Gore, M.A.; Pires, J.C.; Robertson, L.D.; Labate, J.A. Population structure and phylogenetic relationships in a diverse panel of Brassica rapa L. Front. Plant Sci. 2017, 8, 321. [Google Scholar] [CrossRef]
- McAlvay, A.C.; Ragsdale, A.P.; Mabry, M.E.; Qi, X.; Bird, K.A.; Velasco, P.; An, H.; Pires, J.C.; Emshwiller, E. Brassica rapa domestication: Untangling wild and feral forms and convergence of crop morphotypes. Mol. Biol. Evol. 2021, 38, 3358–3372. [Google Scholar] [CrossRef]
- Prakash, S.; Wu, X.; Bhat, S.R. History, evolution, and domestication of Brassica crops. Plant Breed. Rev. 2011, 35, 19–84. [Google Scholar]
- Qi, X.; An, H.; Ragsdale, A.P.; Hall, T.E.; Gutenkunst, R.N.; Chris Pires, J.; Barker, M.S. Genomic inferences of domestication events are corroborated by written records in Brassica rapa. Mol. Ecol. 2017, 26, 3373–3388. [Google Scholar] [CrossRef]
- Reiner, H.; Holzner, W.; Ebermann, R. The development of turnip-type and oilseed-type Brassica rapa crops from the wild-type in Europe–An overview of botanical, historical and linguistic facts. Rapeseed Today Tomorrow 1995, 4, 1066–1069. [Google Scholar]
- Bengtsson, A. Current spring rape and spring turnip rape cultivars. Sven. Frötidning 1992, 61, 8–9. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19930766369 (accessed on 19 August 2024).
- Mäkelä, P.S.; Tuulos, A.; Turakainen, M.; Santanen, A.; Stoddard, F.L. Revitalizing the winter turnip rape crop in the northern latitudes. Acta Agric. Scand. B Soil Plant Sci. 2011, 61, 195–201. [Google Scholar] [CrossRef][Green Version]
- Waalen, W.; Øvergaard, S.I.; Åssveen, M.; Eltun, R.; Gusta, L.V. Winter survival of winter rapeseed and winter turnip rapeseed in field trials, as explained by PPLS regression. Eur. J. Agron. 2013, 51, 81–90. [Google Scholar] [CrossRef]
- Jalli, M.; Huusela, E.; Jalli, H.; Kauppi, K.; Niemi, M.; Himanen, S.; Jauhiainen, L. Effects of crop rotation on spring wheat yield and pest occurrence in different tillage systems: A multi-year experiment in Finnish growing conditions. Front. Sustain. Food Syst. 2021, 5, 647335. [Google Scholar] [CrossRef]
- Svanes, E.; Waalen, W.; Uhlen, A.K. Environmental impacts of rapeseed and turnip rapeseed grown in Norway, rape oil and press cake. Sustainability 2020, 12, 10407. [Google Scholar] [CrossRef]
- Peltonen-Sainio, P.; Hakala, K.; Jauhiainen, L.; Ruosteenoja, K. Comparing regional risks in producing turnip rape and oilseed rape–impacts of climate change and breeding. Acta Agric. Scand. B Soil Plant Sci. 2009, 59, 129–138. [Google Scholar] [CrossRef]
- Persson, R. Breeding Brassica rapa in Scandinavia, Sweden, BULLETINS. Global Council for Innovation in Rapeseed and Canola. 2017. Available online: https://www.gcirc.org/fileadmin/documents/Bulletins/B28/2.Poster_Persson_Brassica_rapa_oK.pdf (accessed on 19 August 2024).
- Swedish Board of Agriculture. 2022. Available online: https://jordbruksverket.se/languages/english/swedish-board-of-agriculture (accessed on 19 August 2024).
- Geleta, M.; Bryngelsson, T.; Bekele, E.; Dagne, K. AFLP and RAPD analyses of genetic diversity of wild and/or weedy Guizotia (Asteraceae) from Ethiopia. Hereditas 2007, 144, 53–62. [Google Scholar] [CrossRef]
- Geleta, M.; Heneen, W.K.; Stoute, A.I.; Muttucumaru, N.; Scott, R.J.; King, G.J.; Kurup, S.; Bryngelsson, T. Assigning Brassica microsatellite markers to the nine C-genome chromosomes using Brassica rapa var. trilocularis–B. oleracea var. alboglabra monosomic alien addition lines. Theor. Appl. Genet. 2012, 125, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Hegay, S.; Geleta, M.; Bryngelsson, T.; Gustavsson, L.; Hovmalm, H.P.; Ortiz, R. Comparing genetic diversity and population structure of common beans grown in Kyrgyzstan using microsatellites. Sci. J. Crop Sci. 2012, 1, 63–75. [Google Scholar]
- Teshome, A.; Bryngelsson, T.; Dagne, K.; Geleta, M. Assessment of genetic diversity in Ethiopian field pea (Pisum sativum L.) accessions with newly developed EST-SSR markers. BMC Genet. 2015, 16, 102. [Google Scholar] [CrossRef] [PubMed]
- Motlhaodi, T.; Geleta, M.; Chite, S.; Fatih, M.; Ortiz, R.; Bryngelsson, T. Genetic diversity in sorghum germplasm from Southern Africa as revealed by microsatellite markers and agromorphological traits. Genet. Resour. Crop Evol. 2017, 64, 599–610. [Google Scholar] [CrossRef]
- Tsehay, S.; Ortiz, R.; Johansson, E.; Bekele, E.; Tesfaye, K.; Hammenhag, C.; Geleta, M. New transcriptome-based SNP markers for noug (Guizotia abyssinica) and their conversion to KASP markers for population genetics analyses. Genes 2020, 11, 1373. [Google Scholar] [CrossRef]
- Enyew, M.; Feyissa, T.; Carlsson, A.S.; Tesfaye, K.; Hammenhag, C.; Geleta, M. Genetic diversity and population structure of sorghum [Sorghum bicolor (L.) Moench] accessions as revealed by single nucleotide polymorphism markers. Front. Plant Sci. 2022, 12, 799482. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.M.; Quiros, C.F. Genetic diversity at isozyme and RFLP loci in Brassica campestris as related to crop type and geographical origin. Theor. Appl. Genet. 1992, 83, 783–790. [Google Scholar] [CrossRef]
- Das, S.; Rajagopal, J.; Bhatia, S.; Srivastava, P.S.; Lakshmikumaran, M. Assessment of genetic variation within Brassica campestris cultivars using amplified fragment length polymorphism and random amplification of polymorphic DNA markers. J. Biosci. 1999, 24, 433–440. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, X.; Deng, B.; Lou, P.; Wu, J.; Sun, R.; Xu, Z.; Vromans, J.; Koornneef, M.; Bonnema, G. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 2005, 110, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Paulo, M.J.; Jamar, D.; Lou, P.; Van Eeuwijk, F.; Bonnema, G.; Vreugdenhil, D.; Koornneef, M. Association mapping of leaf traits, flowering time, and phytate content in Brassica rapa. Genome 2007, 50, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Kulkarni, V.; Liu, N.; Pino Del Carpio, D.; Bucher, J.; Bonnema, G. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 2010, 61, 1817–1825. [Google Scholar] [CrossRef]
- Takuno, S.; Kawahara, T.; Ohnishi, O. Phylogenetic relationships among cultivated types of Brassica rapa L. em. Metzg. as revealed by AFLP analysis. Genet. Resour. Crop Evol. 2007, 54, 279–285. [Google Scholar] [CrossRef]
- Pino Del Carpio, D.; Basnet, R.K.; De Vos, R.C.; Maliepaard, C.; Visser, R.; Bonnema, G. The patterns of population differentiation in a Brassica rapa core collection. Theor. Appl. Genet. 2011, 122, 1105–1118. [Google Scholar] [CrossRef]
- Pino Del Carpio, D.; Basnet, R.K.; De Vos, R.C.; Maliepaard, C.; Paulo, M.J.; Bonnema, G. Comparative methods for association studies: A case study on metabolite variation in a Brassica rapa core collection. PLoS ONE 2011, 6, e19624. [Google Scholar] [CrossRef]
- Guo, N.; Cheng, F.; Wu, J.; Liu, B.; Zheng, S.; Liang, J.; Wang, X. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genom. 2014, 15, 426. [Google Scholar] [CrossRef]
- Cheng, F.; Sun, R.; Hou, X.; Zheng, H.; Zhang, F.; Zhang, Y.; Liu, B.; Liang, J.; Zhuang, M.; Liu, Y.; et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016, 48, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yue, L.; Cai, X.; Li, F.; Zhang, H.; Zhang, S.; Zhang, S.; Sun, R. Fingerprint construction through genotyping by sequencing for applied breeding in Brassica rapa. Genome 2022, 65, 105–113. [Google Scholar] [CrossRef]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Gallegos, J.A.; Kelly, J.D.; Gepts, P. Prebreeding in common bean and use of genetic diversity from wild germplasm. Crop Sci. 2007, 47, S-44–S-59. [Google Scholar] [CrossRef]
- Scheben, A.; Verpaalen, B.; Lawley, C.T.; Chan, C.-K.K.; Bayer, P.E.; Batley, J.; Edwards, D. CropSNPdb: A database of SNP array data for Brassica crops and hexaploid bread wheat. Plant J. 2019, 98, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Rohlf, F.J. NTSYSpc, Version 2.1.; Numerical taxonomy and multivariate analysis system; Exeter Software: Setauket, NY, USA, 2000.
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K.K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Osterman, J.; Hammenhag, C.; Ortiz, R.; Geleta, M. Discovering candidate SNPs for resilience breeding of red clover. Front. Plant Sci. 2022, 13, 997860. [Google Scholar] [CrossRef] [PubMed]
- Zsigmond, L. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 2008, 146, 1721–1737. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Wang, Y.; Wu, M.; Zhu, X.; Teng, X.; Sun, Y.; Zhu, J.; Zhang, Y.; Jing, R.; Lei, J.; et al. The nuclear-localized PPR protein OsNPPR1 is important for mitochondrial function and endosperm development in rice. J. Exp. Bot. 2019, 70, 4705–4720. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, P.; Sun, J.; Zhao, Y.; Zhang, Y.; Yang, Q.; Wang, W.; Chen, Z.; Mai, T.; Zou, Y.; et al. Research progress of PPR proteins in RNA editing, stress response, plant growth and development. Front. Genet. 2021, 12, 765580. [Google Scholar] [CrossRef] [PubMed]
- Turner, W.L.; Knowles, V.L.; Plaxton, W.C. Cytosolic pyruvate kinase: Subunit composition, activity, and amount in developing castor and soybean seeds, and biochemical characterization of the purified castor seed enzyme. Planta 2005, 222, 1051–1062. [Google Scholar] [CrossRef]
- Wulfert, S.; Schilasky, S.; Krueger, S. Transcriptional and biochemical characterization of cytosolic pyruvate kinases in Arabidopsis thaliana. Plants 2020, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Mu, Y.; Chai, X.; Ouyang, M.; Yu, L.J.; Zhang, L.; Meurer, J.; Chi, W. The critical function of the plastid rRNA methyltransferase, CMAL, in ribosome biogenesis and plant development. Nucleic Acids Res. 2020, 48, 3195–3210. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, L.N.T.; Park, S.J.; Cai, J.; Huong, T.T.; Lee, K.; Kang, H. RsmD, a chloroplast rRNA m2G methyltransferase, plays a role in cold stress tolerance by possibly affecting chloroplast translation in Arabidopsis. Plant Cell Physiol. 2021, 62, 948–958. [Google Scholar] [CrossRef]
- Foti, M.; Audhya, A.; Emr, S.D. Sac1 lipid phosphatase and stt4 phosphatidylinositol4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell 2001, 128, 2396–2411. [Google Scholar] [CrossRef] [PubMed]
- Wada, Y.; Kusano, H.; Tsuge, T.; Aoyama, T. Phosphatidylinositol phosphate 5-kinase genes respond to phosphate deficiency for root hair elongation in Arabidopsis thaliana. Plant J. 2015, 81, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, R.; Kato, M.; Tsuge, T.; Aoyama, T. Arabidopsis phosphatidylinositol 4-phosphate 5-kinase genes PIP5K7, PIP5K8, and PIP5K9 are redundantly involved in root growth adaptation to osmotic stress. Plant J. 2021, 106, 913–927. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, Z.; Pandey, G.K.; Tsuchiya, T.; Luan, S. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 2002, 277, 5360–5368. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, Y.; Yao, L.; Li, G.; Wang, L.; Hu, Y.; Wei, H.; Wang, L.; Hammami, R.; Razavi, R.; et al. DetoxiProt: An integrated database for detoxification proteins. BMC Genom. 2011, 12, S2. [Google Scholar] [CrossRef]
- Gong, C.; Yin, X.; Ye, T.; Liu, X.; Yu, M.; Dong, T.; Wu, Y. The F-Box/DUF295 Brassiceae specific 2 is involved in ABA-inhibited seed germination and seedling growth in Arabidopsis. Plant Sci. 2022, 323, 111369. [Google Scholar] [CrossRef]
- Petit, R.J.; El Mousadik, A.; Pons, O. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 1998, 12, 844–855. [Google Scholar] [CrossRef]
- Reyes-Valdés, M.H.; Burgueño, J.; Singh, S.; Martínez, O.; Sansaloni, C.P. An informational view of accession rarity and allele specificity in germplasm banks for management and conservation. PLoS ONE 2018, 13, e0193346. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.M.; Leng, Y.; Li, S.W. Transcriptome profiling provides new insights into ABA-mediated genes and pathways in leaves, stems, and roots of mung bean seedlings. Plant Growth Regul. 2022, 98, 569–587. [Google Scholar] [CrossRef]
- Zheng, Y.; Ma, Q.; Mao, L.; Wu, Z.; Liu, Z.; Zou, X.; Yang, B. Comparative transcriptome analysis identified genes associated with fruit size in pepper (Capsicum annuum L.). Horticulturae 2023, 9, 1009. [Google Scholar] [CrossRef]
- Brandt, R.; Cabedo, M.; Xie, Y.; Wenkel, S. Homeodomain leucine-zipper proteins and their role in synchronizing growth and development with the environment. J. Integr. Plant Biol. 2014, 56, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Xue, Y.; Cao, D.; Luan, X.; Zhao, K.; Liu, Q.; Ren, Y.; Zhu, Z.; Li, Y.; Liu, X. Identification of candidate genes for drought resistance during soybean seed development. Agriculture 2023, 13, 949. [Google Scholar] [CrossRef]
- Xiang, L.; Wang, M.; Pan, F.; Wang, G.; Jiang, W.; Wang, Y.; Chen, X.; Yin, C.; Mao, Z. Transcriptome analysis Malus domestica ‘M9T337’root molecular responses to Fusarium solani infection. Physiol. Mol. Plant Pathol. 2021, 113, 101567. [Google Scholar] [CrossRef]
- Chen, L.; Huang, X.X.; Zhao, S.-M.; Xiao, D.-W.; Xiao, L.-T.; Tong, J.-H.; Wang, W.-S.; Li, Y.-J.; Ding, Z.; Hou, B.-K. IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 6910–6917. [Google Scholar] [CrossRef] [PubMed]
- Duckney, P.; Deeks, M.J.; Dixon, M.R.; Kroon, J.; Hawkins, T.J.; Hussey, P.J. Actin–membrane interactions mediated by NETWORKED 2 in Arabidopsis pollen tubes through associations with Pollen Receptor-Like Kinase 4 and 5. New Phytol. 2017, 216, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Khouider, S.; Borges, F.; LeBlanc, C.; Ungru, A.; Schnittger, A.; Martienssen, R.; Colot, V.; Bouyer, D. Male fertility in Arabidopsis requires active DNA demethylation of genes that control pollen tube function. Nat. Commun. 2021, 12, 410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.G.; Ofori, A.; Lu, C.M. Genetic diversity of European and Chinese oilseed Brassica rapa cultivars from different breeding periods. Agric. Sci. China 2009, 8, 931–938. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. The Second Report on the State of the World’s Plant Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome Italy, 2010. [Google Scholar]
- Anglin, N.L.; Amri, A.; Kehel, Z.; Ellis, D. A case of need: Linking traits to genebank accessions. Biopreserv. Biobank 2018, 16, 337–349. [Google Scholar] [CrossRef]
- Singh, N.; Wu, S.; Raupp, W.J.; Sehgal, S.; Arora, S.; Tiwari, V.; Vikram, P.; Singh, S.; Chhuneja, P.; Gill, B.S.; et al. Efficient curation of gene banks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 2019, 9, 650. [Google Scholar]
- Sahu, T.K.; Singh, A.K.; Mittal, S.; Jha, S.K.; Kumar, S.; Jacob, S.R.; Singh, K. G-DIRT: A web server for identification and removal of duplicate germplasms based on identity-by-state analysis using single nucleotide polymorphism genotyping data. Brief. Bioinform. 2022, 23, bbac348. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Takahashi, Y.; Yokoi, S.; Takahata, Y. Genetic divergence of turnip (Brassica rapa L. em. Metzg. subsp. rapa) inferred from simple sequence repeats in chloroplast and nuclear genomes and morphology. Genet. Resour. Crop Evol. 2016, 63, 869–879. [Google Scholar] [CrossRef]
- Jarvis, D.I.; Hodgkin, T. Wild relatives and crop cultivars: Detecting natural introgression and farmer selection of new genetic combinations in agroecosystems. Mol. Ecol. 1999, 8, S159–S173. [Google Scholar] [CrossRef]
- Ofori, A.; Becker, H.C.; Kopisch-Obuch, F.J. Effect of crop improvement on genetic diversity in oilseed Brassica rapa (turnip-rape) cultivars, detected by SSR markers. J. Appl. Genet. 2008, 49, 207–212. [Google Scholar] [CrossRef]

| GenBank | Africa | Asia | Europe | North America | Oceania | South America | Unknown | Total |
|---|---|---|---|---|---|---|---|---|
| NordGen | 0 | 0 | 33 | 1 | 0 | 0 | 0 | 34 |
| Jerrestad AB | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 13 |
| PGRC | 0 | 24 | 3 | 3 | 0 | 0 | 0 | 30 |
| CGN | 0 | 7 | 1 | 2 | 0 | 0 | 1 | 11 |
| IPK | 1 | 8 | 64 | 8 | 1 | 2 | 2 | 86 |
| NPGS | 1 | 2 | 8 | 1 | 0 | 0 | 0 | 12 |
| A&A Canada | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| Total | 2 | 41 | 122 | 17 | 1 | 2 | 3 | 188 |
| Consequence_Effect | Chromosome | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A01 | A02 | A03 | A04 | A05 | A06 | A07 | A08 | A09 | A10 | ||
| Missense variant_Moderate | 12 | 5 | 9 | 5 | 0 | 9 | 7 | 6 | 0 | 13 | 66 |
| Synonymous variant_Low | 4 | 0 | 1 | 4 | 1 | 0 | 4 | 2 | 0 | 5 | 21 |
| Splice polypyrimidine tract variant_Low | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 4 |
| Splice donor variant_High | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 3 |
| Splice polypyrimidine tract variant/Intron variant_Low | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 3 |
| Splice acceptor variant_High | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| Splice donor fifth base variant/Intron variant_Low | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| Splice donor region variant/Intron variant_Low | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| Splice region variant/Synonymous variant_Low | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| Missense variant/stop retained variant_Moderate | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Splice region variant/Intron variant_Low | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Stop lost_High | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 16 | 6 | 16 | 12 | 1 | 14 | 13 | 8 | 2 | 20 | 108 |
| Source of Variation | Degrees of Freedom | Sum of Squares | Variance Components | Percentage of Variation | Fixation Index | p-Value |
|---|---|---|---|---|---|---|
| Among accession groups a | 5 | 2.311 | 0.015 | 19.0 | PhiPT = 0.194 | 0.001 |
| Within accession groups a | 180 | 11.230 | 0.062 | 81.0 | ||
| Total | 185 | 13.542 | 0.077 | |||
| Among accession groups b | 2 | 0.954 | 0.012 | 17.5 | PhiPT = 0.175 | 0.001 |
| Within accession groups b | 139 | 8.108 | 0.058 | 82.5 | ||
| Total | 141 | 9.063 | 0.071 | |||
| Among accession groups c | 4 | 2.227 | 0.025 | 38.0 | PhiPT = 0.378 | 0.001 |
| Within accession groups c | 112 | 4.590 | 0.041 | 62.0 | ||
| Total | 116 | 6.817 | 0.066 | |||
| Among accession groups d | 4 | 2.332 | 0.023 | 27.0 | PhiPT = 0.268 | 0.005 |
| Within accession groups d | 179 | 11.025 | 0.062 | 73.0 | ||
| Total | 183 | 13.357 | 0.084 |
| Source of Variation | Degrees of Freedom | Sum of Squares | Variance Components | Percentage of Variation | Fixation Index | p-Value |
|---|---|---|---|---|---|---|
| Among accession groups a | 6 | 58.4 | 0.38 Va | 23.49 | FST = 0.24 | 0.003 |
| Within accession groups a | 175 | 217.0 | 1.24 Vb | 76.51 | ||
| Total | 181 | 275.4 | 1.62 | |||
| Among accession groups b | 2 | 9.0 | 0.15 Va | 17.08 | FST = 0.17 | 0.03 |
| Within accession groups b | 145 | 105.6 | 0.73 Vb | 82.92 | ||
| Total | 147 | 114.7 | 0.88 | |||
| Among accession groups c | 7 | 100.3 | 0.85 Va | 51.07 | FST = 0.51 | 0 |
| Within accession groups c | 150 | 122.8 | 0.82 Vb | 48.93 | ||
| Total | 157 | 223.1 | 1.67 | |||
| Among accession groups d | 3 | 96.3 | 1.47 Va | 58.97 | FST = 0.59 | 0 |
| Within accession groups d | 170 | 174.1 | 1.02 Vb | 41.03 | ||
| Total | 173 | 270.4 | 2.49 |
| Accession-group a | Cultivar | Landrace | Wild | |||
|---|---|---|---|---|---|---|
| Cultivar | - | |||||
| Landrace | 0.188 * | - | ||||
| Wild | 0.208 * | 0.040 | - | |||
| Accession-group b | China | Germany | India | Italy | Sweden | |
| China | - | |||||
| Germany | 0.188 * | - | ||||
| India | 0.243 * | 0.368 * | - | |||
| Italy | 0.340 * | 0.234 * | 0.475 * | - | ||
| Sweden | 0.466 * | 0.133 * | 0.558 * | 0.388 * | - | |
| Accession-group c | Africa | Asia | Europe | North America | South America | |
| Africa | - | |||||
| Asia | 0.276 * | - | ||||
| Europe | 0.487 * | 0.308 * | - | |||
| North America | 0.459 * | 0.334 * | 0.072 * | - | ||
| South America | 0.198 | 0.317 * | 0.174 | 0.146 | - | |
| Accession-group d | Jerrestad AB | CGN | IPK | NordGen | NPGS | PGRC |
| Jerrestad AB | - | |||||
| CGN | 0.538 * | - | ||||
| IPK | 0.169 * | 0.363 * | - | |||
| NordGen | 0.088 | 0.416 * | 0.105 * | - | ||
| NPGS | 0.281 * | 0.194 * | 0.072 * | 0.187 * | - | |
| PGRC | 0.407 * | 0.172 * | 0.167 * | 0.243 * | 0.076 * | - |
| Accession Group a | Cultivar | Landrace | Wild | |||||
|---|---|---|---|---|---|---|---|---|
| Cultivar | - | |||||||
| Landrace | 0.215 * | - | ||||||
| Wild | 0.109 | 0.121 | - | |||||
| Accession group b | Africa | Asia | Europe | North America | ||||
| Africa | - | |||||||
| Asia | 0.599 | - | ||||||
| Europe | 0.521 | 0.835 | - | |||||
| North America | 0.674 | 0.873 | −0.007 | - | ||||
| Accession group c | Jerrestad AB | A&A Canada | CGN | IPK | NordGen | NPGS | PGRC | |
| Jerrestad AB | - | |||||||
| A&A Canada | −0.151 | - | ||||||
| CGN | 0.495 * | 0.271 | - | |||||
| IPK | −0.021 | −0.097 | 0.635 * | - | ||||
| NordGen | 0.010 | −0.077 | 0.461 * | 0.021 | - | |||
| NPGS | 0.441 | 0.234 | 0.221 | 0.500 * | 0.268 * | - | ||
| PGRC | 0.041 | −0.113 | 0.310 | 0.088 | 0.020 | 0.153 | - | |
| Accession group d | Austria | Bangladesh | Canada | Finland | Germany | Italy | Sweden | USA |
| Austria | - | |||||||
| Bangladesh | 0.953 | - | ||||||
| Canada | −0.125 | 0.950 * | - | |||||
| Finland | −0.045 | 0.792 * | 0.038 | - | ||||
| Germany | 0.261 | 0.987 * | 0.196 | 0.028 | - | |||
| Italy | 0.096 | 0.931 * | 0.157 | 0.133 | 0.480 | - | ||
| Sweden | −0.089 | 0.908 * | −0.027 | 0.036 | 0.054 | 0.143 | - | |
| USA | 0.040 | 0.951 | 0.106 | 0.049 | 0.650 | −0.078 | 0.078 | - |
| Region Specific Allele | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Europe/France | Europe/France | Europe/ ** | Europe/Italy | Europe/ ** | Europe/ ** | Europe/ ** | Europe/ ** | North America/Cuba | |
| Locus | Bn-A01-p1001022 | Bn-A02-p20845905 | NC_024796-2_ 3347226 | NC_024800-2_ 3070370 | NC_024803-2_ 35208073 | NC_024804-2_15703515 | NC_024803-2_33076115 | NC_024803-2_30767803 | NC_024795-2_25991653 |
| Chromosome | A01 | A02 | A02 | A06 | A09 | A10 | A09 | A09 | A01 |
| SNP position | 1061243 | 24397768 | 3347226 | 3070370 | 35208073 | 15703515 | 33076115 | 30767803 | 25991653 |
| Ref/Alt alleles | A/C | A/C | T/A | T/C | C/T | C/T | T/C | G/T | C/T |
| PA | C | C | A | C | T | T | C | T | T |
| PA freq | 0.004 | 0.004 | 0.021 | 0.017 | 0.030 | 0.017 | 0.025 | 0.012 | 0.176 |
| PA description | Upstream gene variant | Intergenic variant | Missense variant | Missense variant | Upstream gene variant | Downstream gene variant | Intergenic variant | Intergenicvariant | Intergenic variant |
| PA effect | Modifier | Modifier | Moderate | Moderate | Modifier | Modifier | Modifier | Modifier | Modifier |
| PA-associated gene | A01p002320.1_ BraROA | A02g509130.1_ BraROA * | A02p008020.1_ BraROA | A06p008790.1_ BraROA | A09g511940.1_ BraROA | A10p024240.1_ BraROA | A09g510750.1_BraROA * | A09g510320.1_BraROA * | A01p046190.1_BraROA * |
| PA-associated Protein | homeobox-leucine zipper protein ATHB-40 | - | uncharacterized | phosphatidylinositol 4-phosphate 5-kinase 7 | UDP-glycosyltransferase 76F1 | uncharacterized | uncharacterized | uncharacterized | Pollen receptor-like kinase 4 |
| Coding strand | Forward | Forward | Forward | Reverse | Forward | Reverse | Forward | Reverse | Reverse |
| Codon change | - | - | Ttc/Atc | gAg/gGg | - | - | - | - | - |
| AA change | - | - | Phe/Ile | Glu/Gly | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geleta, M.; Sundaramoorthy, J.; Carlsson, A.S. SeqSNP-Based Targeted GBS Provides Insight into the Genetic Relationships among Global Collections of Brassica rapa ssp. oleifera (Turnip Rape). Genes 2024, 15, 1187. https://doi.org/10.3390/genes15091187
Geleta M, Sundaramoorthy J, Carlsson AS. SeqSNP-Based Targeted GBS Provides Insight into the Genetic Relationships among Global Collections of Brassica rapa ssp. oleifera (Turnip Rape). Genes. 2024; 15(9):1187. https://doi.org/10.3390/genes15091187
Chicago/Turabian StyleGeleta, Mulatu, Jagadeesh Sundaramoorthy, and Anders S. Carlsson. 2024. "SeqSNP-Based Targeted GBS Provides Insight into the Genetic Relationships among Global Collections of Brassica rapa ssp. oleifera (Turnip Rape)" Genes 15, no. 9: 1187. https://doi.org/10.3390/genes15091187
APA StyleGeleta, M., Sundaramoorthy, J., & Carlsson, A. S. (2024). SeqSNP-Based Targeted GBS Provides Insight into the Genetic Relationships among Global Collections of Brassica rapa ssp. oleifera (Turnip Rape). Genes, 15(9), 1187. https://doi.org/10.3390/genes15091187








