Unambiguous Interpretation of the Pathogenicity of the GLA c.547+3A>G Variant Causing Fabry Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. EMB and Right Heart Catheterization
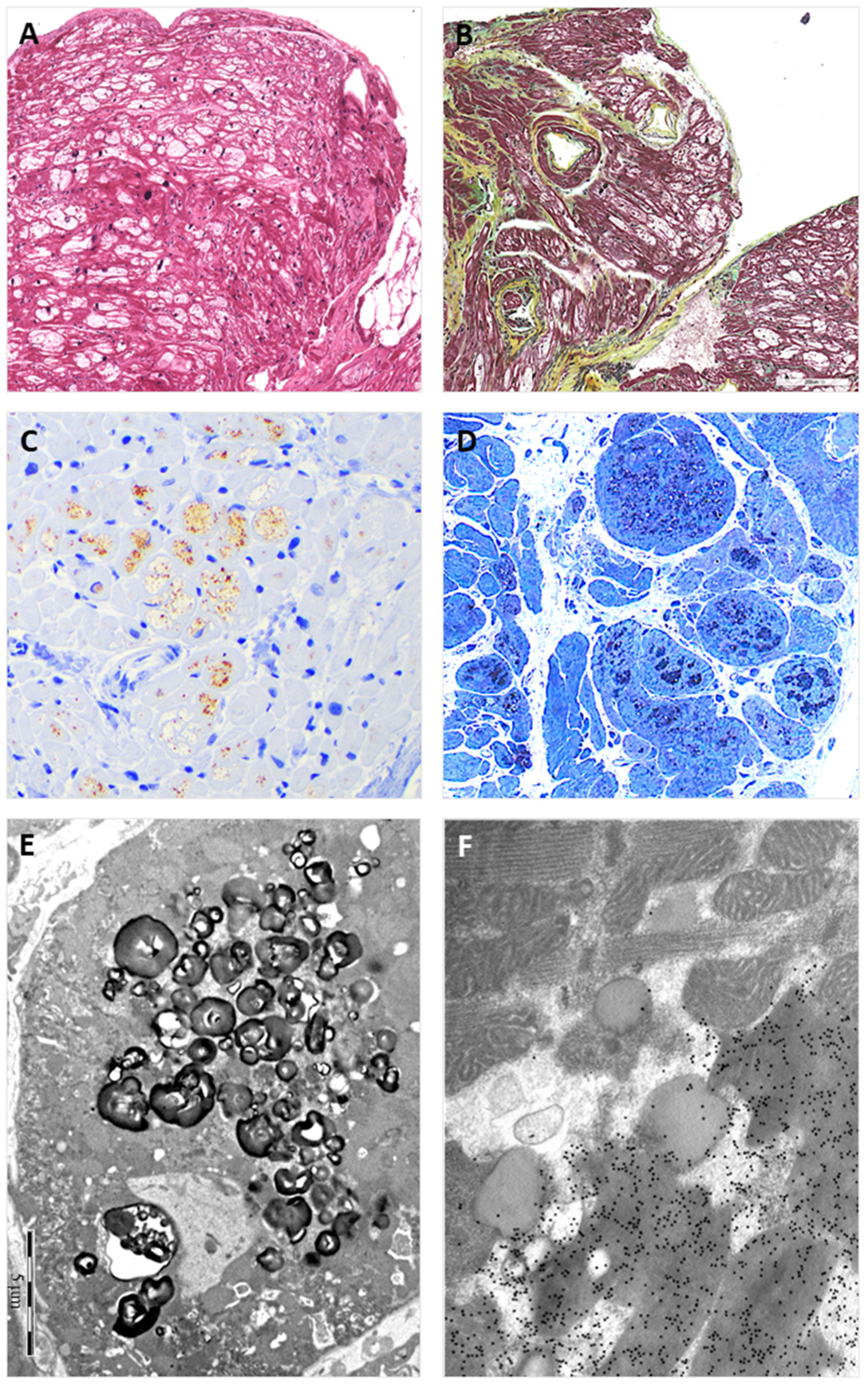
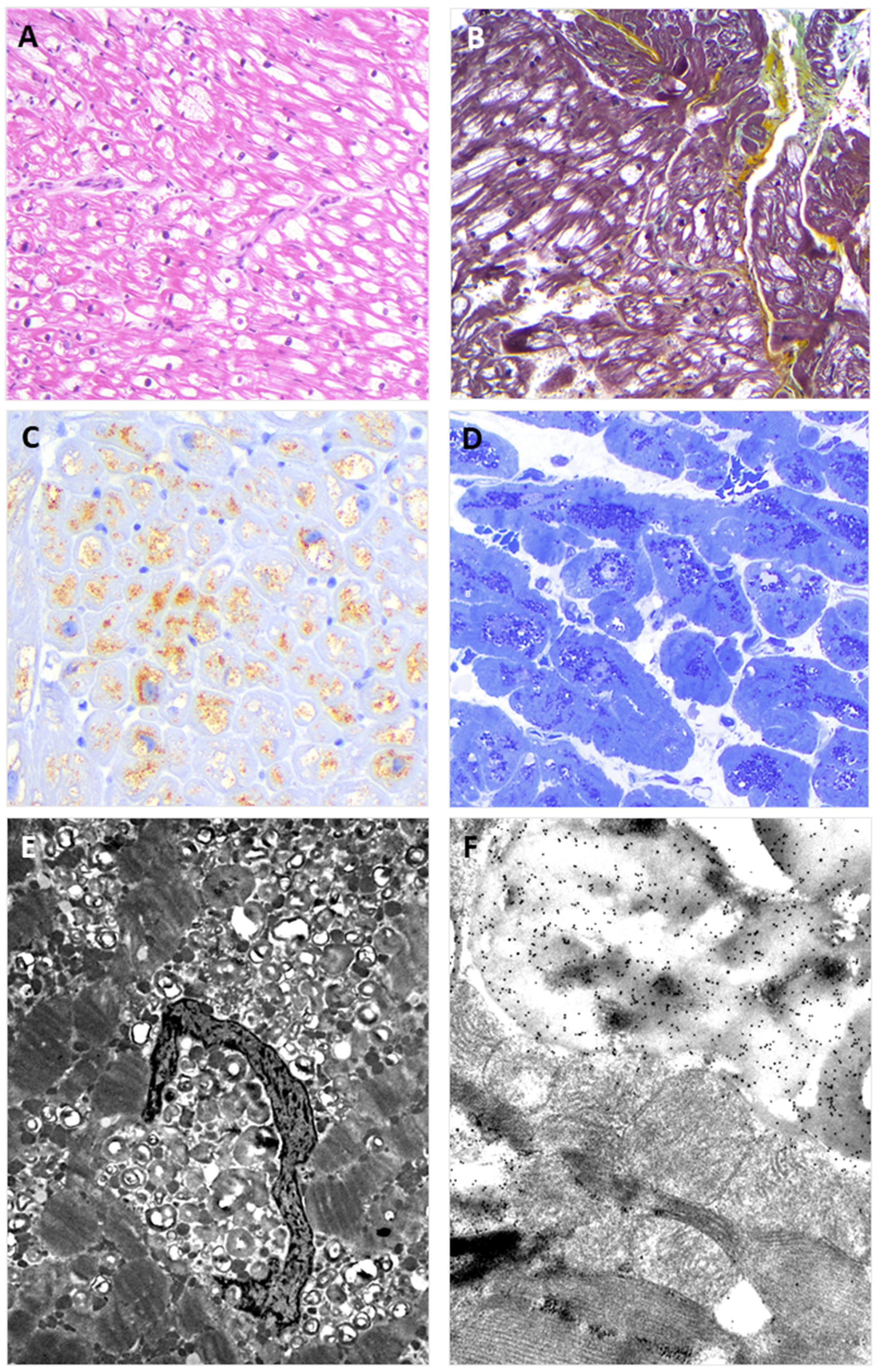
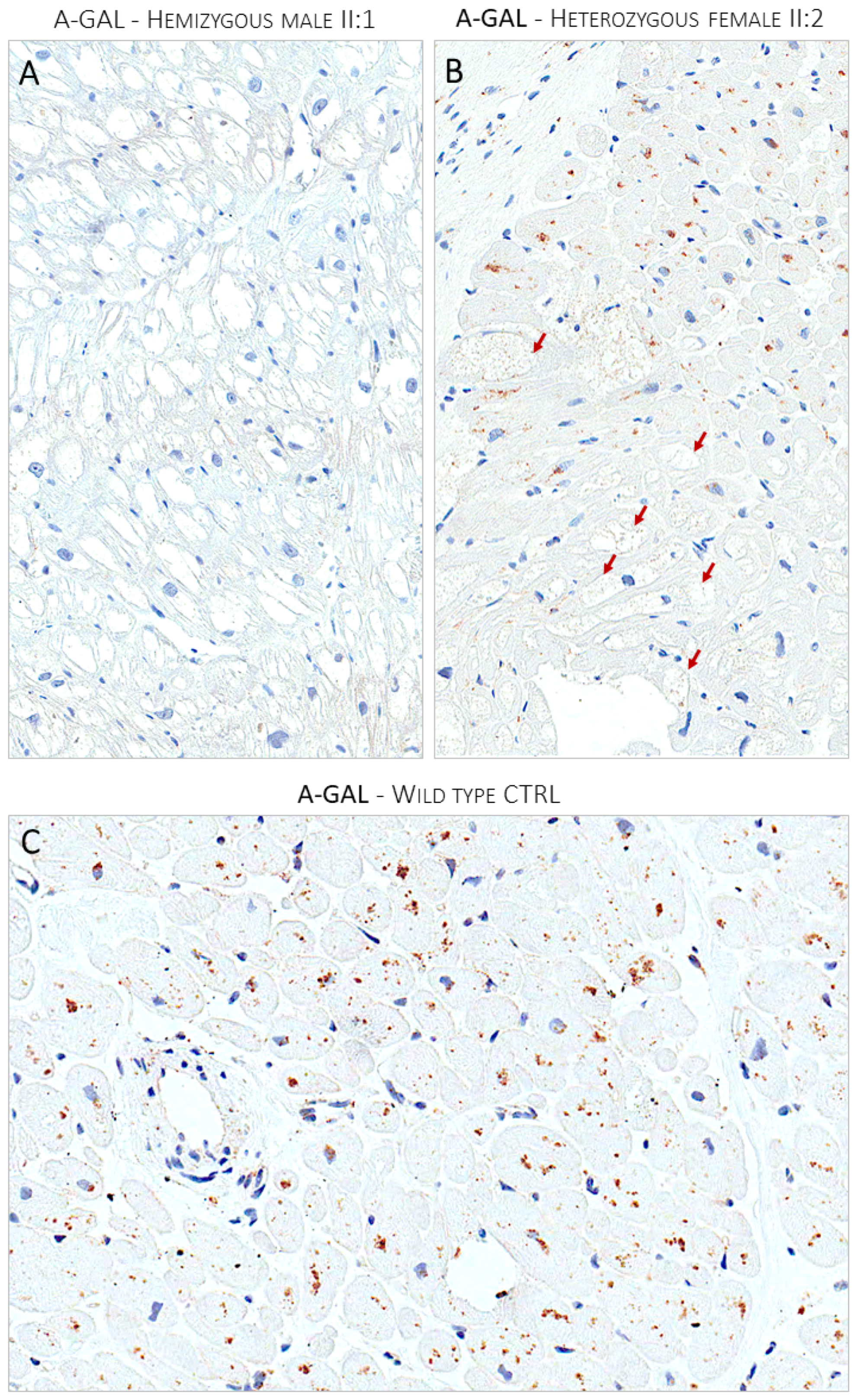
2.2. Light Microscopy Immunohistochemistry
2.3. Electron- and Immuno-Electron Microscopy Study
2.4. Genetic Test, NGS Sequencing and Data Analysis
2.5. Variant Interpretation
3. Results
3.1. Clinical History and Phenotype in the Proband
3.2. Clinical Family Screening
3.3. Genetic Test in the Proband and Relatives
3.4. Endomyocardial Biopsy
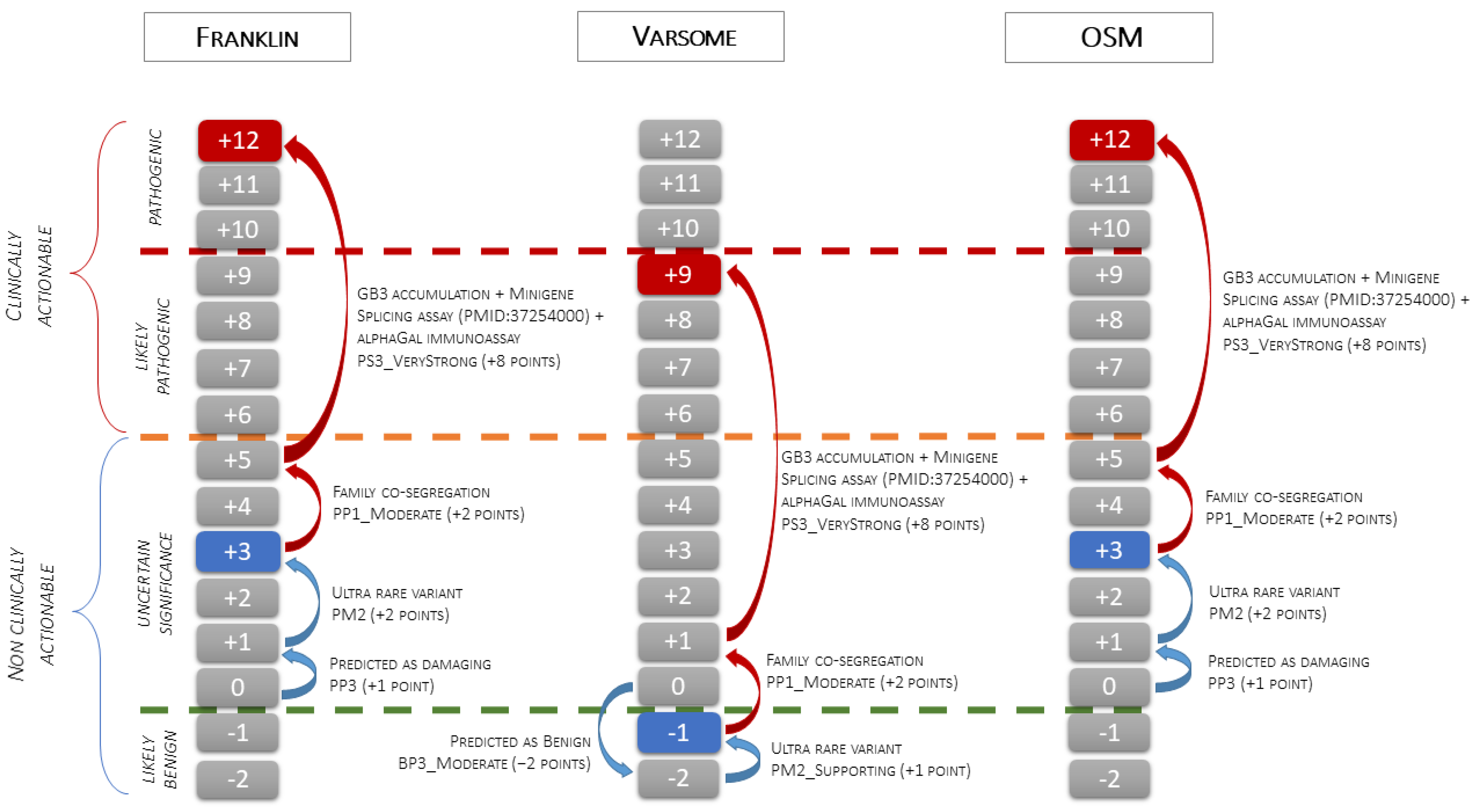
3.5. Variant Classification
- -
- VUS, when using Franklin (v.2022.6) software that activates the PM2 criterion with moderate strength (extremely low frequency in gnomAD population databases), and PP3 (for a missense or a splicing region variant, computational prediction tools unanimously support a deleterious effect on the gene, based on the latest recommendations for PP3/BP4 rules) with supporting strength “https://franklin.genoox.com/clinical-db/variant/snp/chrX-100656617-T-C (accessed on 13 August 2024)”;
- -
- Likely Benign, when using Varsome (v.11.10.0), which activates the PM2 criterion with supporting strength and the BP4 criterion with moderate strength (multiple lines of computational evidence suggest no impact of gene or gene product) “https://varsome.com/variant/hg19/chrX-100656617-T-C (accessed on 13 August 2024)”.
- -
- VUS, when using OSM, which activates the PM2 criterion with moderate strength, and the PP3 criterion (MaxEntScan splice effect variation of −75.82% and SpliceAI Donor Loss of 0.81; High impact).
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Germain, D.P. Fabry disease. Orphanet J. Rare Dis. 2010, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, A.; Germain, D.P.; Desnick, R.J.; Politei, J.; Mauer, M.; Burlina, A.; Eng, C.; Hopkin, R.J.; Laney, D.; Linhart, A.; et al. Fabry disease revisited: Management and treatment recommendations for adult patients. Mol. Genet. Metab. 2018, 123, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Izhar, R.; Borriello, M.; La Russa, A.; Di Paola, R.; De, A.; Capasso, G.; Ingrosso, D.; Perna, A.F.; Simeoni, M. Fabry Disease in Women: Genetic Basis, Available Biomarkers, and Clinical Manifestations. Genes 2023, 15, 37. [Google Scholar] [CrossRef] [PubMed]
- Pieroni, M.; Moon, J.C.; Arbustini, E.; Barriales-Villa, R.; Camporeale, A.; Vujkovac, A.C.; Elliott, P.M.; Hagege, A.; Kuusisto, J.; Linhart, A.; et al. Cardiac Involvement in Fabry Disease: JACC Review Topic of the Week. J. Am. Coll. Cardiol. 2021, 77, 922–936. [Google Scholar] [CrossRef]
- Doheny, D.; Srinivasan, R.; Pagant, S.; Chen, B.; Yasuda, M.; Desnick, R.J. Fabry Disease: Prevalence of affected males and heterozygotes with pathogenic. J. Med. Genet. 2018, 55, 261–268. [Google Scholar] [CrossRef]
- Favalli, V.; Disabella, E.; Molinaro, M.; Tagliani, M.; Scarabotto, A.; Serio, A.; Grasso, M.; Narula, N.; Giorgianni, C.; Caspani, C.; et al. Genetic Screening of Anderson-Fabry Disease in Probands Referred from Multispecialty Clinics. J. Am. Coll. Cardiol. 2016, 68, 1037–1050. [Google Scholar] [CrossRef]
- Monserrat, L.; Gimeno-Blanes, J.R.; Marín, F.; Hermida-Prieto, M.; García-Honrubia, A.; Pérez, I.; Fernández, X.; de Nicolas, R.; de la Morena, G.; Payá, E.; et al. Prevalence of fabry disease in a cohort of 508 unrelated patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2007, 50, 2399–2403. [Google Scholar] [CrossRef]
- Shi, Q.; Chen, J.; Pongmoragot, J.; Lanthier, S.; Saposnik, G. Prevalence of Fabry disease in stroke patients—A systematic review and meta-analysis. J. Stroke Cerebrovasc. Dis. 2014, 23, 985–992. [Google Scholar] [CrossRef]
- Capuano, I.; Garofalo, C.; Buonanno, P.; Pinelli, M.; Di Risi, T.; Feriozzi, S.; Riccio, E.; Pisani, A. Identifying Fabry patients in dialysis population: Prevalence of GLA mutations by renal clinic screening, 1995–2019. J. Nephrol. 2020, 33, 569–581. [Google Scholar] [CrossRef]
- Nagai-Sangawa, M.; Fukunaga, A.; Takeuchi, C.; Nishiyama, S.; Horikawa, T.; Nagano, C.; Nozu, K.; Fujii, H.; Nishigori, C. Beneficial screening of Fabry disease in patients with hypohidrosis. J. Dermatol. 2022, 49, 308–312. [Google Scholar] [CrossRef]
- Fanlo, P.; Garralda, A.; Gómez-Cerezo, J.F.; Echeverria, M.; López, M.; Heras, H.; Riera-Mestre, A. Results of the screening program for Fabry disease in patients with cornea verticillata at the University Hospital of Navarre. Rev. Clin. Esp. 2023, 223, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Sahyoun, J.Y.; Sabeti, S.; Robert, M.C. Drug-induced corneal deposits: An up-to-date review. BMJ Open Ophthalmol. 2022, 7, e000943. [Google Scholar] [CrossRef] [PubMed]
- Hilz, M.J.; Arbustini, E.; Dagna, L.; Gasbarrini, A.; Goizet, C.; Lacombe, D.; Liguori, R.; Manna, R.; Politei, J.; Spada, M.; et al. Non-specific gastrointestinal features: Could it be Fabry disease? Dig. Liver Dis. 2018, 50, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Di Toro, A.; Narula, N.; Giuliani, L.; Concardi, M.; Smirnova, A.; Favalli, V.; Urtis, M.; Alvisi, C.; Antoniazzi, E.; Arbustini, E. Pathologic substrate of gastropathy in Anderson-Fabry disease. Orphanet J. Rare Dis. 2020, 15, 156. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Arbustini, E.; Behr, E.R.; Carrier, L.; van Duijn, C.; Evans, P.; Favalli, V.; van der Harst, P.; Haugaa, K.H.; Jondeau, G.; Kääb, S.; et al. Interpretation and actionability of genetic variants in cardiomyopathies: A position statement from the European Society of Cardiology Council on cardiovascular genomics. Eur. Heart J. 2022, 43, 1901–1916. [Google Scholar]
- Tavtigian, S.V.; Harrison, S.M.; Boucher, K.M.; Biesecker, L.G. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum. Mutat. 2020, 41, 1734–1737. [Google Scholar] [CrossRef]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef]
- Okada, E.; Horinouchi, T.; Yamamura, T.; Aoto, Y.; Suzuki, R.; Ichikawa, Y.; Tanaka, Y.; Masuda, C.; Kitakado, H.; Kondo, A.; et al. All reported non-canonical splice site variants in GLA cause aberrant splicing. Clin. Exp. Nephrol. 2023, 27, 737–746. [Google Scholar] [CrossRef]
- Oliveira, J.P.; Ferreira, S. Multiple phenotypic domains of Fabry disease and their relevance for establishing genotype-phenotype correlations. Appl. Clin. Genet. 2019, 12, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, M.; Bazan-Socha, S.; Błażejewska-Hyżorek, B.; Kłopotowski, M.M.; Komar, M.; Kusztal, M.A.; Liberek, T.; Małyszko, J.; Mizia-Stec, K.; Oko-Sarnowska, Z.; et al. A review and recommendations for oral chaperone therapy in adult patients with Fabry disease. Orphanet J. Rare Dis. 2024, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://fabry-database.org/mutants/ (accessed on 14 August 2024).

| Likely Pathogenic/Pathogenic | Variants of Uncertain Significance | |
|---|---|---|
| c.1000−10G>A c.19_369+582del c.194+1222_998dup c.195−89_712del c.370−530_1279del c.370−532_1278del c.639+4A>T c.640−801G>A c.640−814T>C c.640−859C>T c.801+3A>G c.802−3_802-2del | c. −1A>C c. −1A>G c. −2C>T c. −79G>A c. −7C>T c.195−13_195-12delinsGC c.195−9C>A c.369+3G>A c.369+4A>T c.369+5G>T c.547+397G>A c.547+3A>G | c.547+4A>C c.548−10T>A c.548−3C>A c.639+3G>A c.639+5G>A c.639+852_639+855del c.640−336T>C c.640−4A>C c.802−14A>T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urtis, M.; Cavaliere, C.; Vilardo, V.; Paganini, C.; Smirnova, A.; Giorgianni, C.; Di Toro, A.; Chiapparini, L.; Pellegrini, C.; Grasso, M.; et al. Unambiguous Interpretation of the Pathogenicity of the GLA c.547+3A>G Variant Causing Fabry Disease. Genes 2024, 15, 1212. https://doi.org/10.3390/genes15091212
Urtis M, Cavaliere C, Vilardo V, Paganini C, Smirnova A, Giorgianni C, Di Toro A, Chiapparini L, Pellegrini C, Grasso M, et al. Unambiguous Interpretation of the Pathogenicity of the GLA c.547+3A>G Variant Causing Fabry Disease. Genes. 2024; 15(9):1212. https://doi.org/10.3390/genes15091212
Chicago/Turabian StyleUrtis, Mario, Claudia Cavaliere, Viviana Vilardo, Chiara Paganini, Alexandra Smirnova, Carmelina Giorgianni, Alessandro Di Toro, Luisa Chiapparini, Carlo Pellegrini, Maurizia Grasso, and et al. 2024. "Unambiguous Interpretation of the Pathogenicity of the GLA c.547+3A>G Variant Causing Fabry Disease" Genes 15, no. 9: 1212. https://doi.org/10.3390/genes15091212
APA StyleUrtis, M., Cavaliere, C., Vilardo, V., Paganini, C., Smirnova, A., Giorgianni, C., Di Toro, A., Chiapparini, L., Pellegrini, C., Grasso, M., & Arbustini, E. (2024). Unambiguous Interpretation of the Pathogenicity of the GLA c.547+3A>G Variant Causing Fabry Disease. Genes, 15(9), 1212. https://doi.org/10.3390/genes15091212





