Genetic Etiology in Pelvic Organ Prolapse: Role of Connective Tissue Homeostasis, Hormone Metabolism, and Oxidative Stress
Abstract
:1. Introduction
2. A Familial or Genetic Basis of POP
3. Genetic Variants Associated with Remodeling of Extracellular Matrix (ECM) in Pelvic Floor Connective Tissues
4. Genetic Variants Associated with Hormone Metabolism
5. Genetic Variants Associated with Oxidative Stress That Disturbs Cellular Homeostasis in Pelvic Floor Support Tissues
6. Other Genetic Findings Related to Predisposition of POP
7. Gene Expression of POP-Associated Genes
8. Polygenic Risk Score (PRS) and Interaction
9. Challenges and Prospects
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barber, M.D. Pelvic organ prolapse. BMJ 2016, 354, i3853. [Google Scholar] [CrossRef] [PubMed]
- Ghetti, C.; Skoczylas, L.C.; Oliphant, S.S.; Nikolajski, C.; Lowder, J.L. The emotional burden of pelvic organ prolapse in women seeking treatment: A qualitative study. Urogynecology 2015, 21, 332–338. [Google Scholar] [CrossRef]
- Sanses, T.V.; Schiltz, N.K.; Richter, H.E.; Koroukian, S.M. Trends and factors influencing inpatient prolapse surgical costs and length of stay in the United States. Urogynecology 2016, 22, 103–110. [Google Scholar] [CrossRef]
- Robinson, D.; Prodigalidad, L.T.; Chan, S.; Serati, M.; Lozo, S.; Lowder, J.; Ghetti, C.; Hullfish, K.; Hagen, S.; Dumoulin, C. International Urogynaecology Consultation chapter 1 committee 4: Patients’ perception of disease burden of pelvic organ prolapse. Int. Urogynecol. J. 2022, 33, 189–210. [Google Scholar] [CrossRef]
- Wu, J.M.; Vaughan, C.P.; Goode, P.S.; Redden, D.T.; Burgio, K.L.; Richter, H.E.; Markland, A.D. Prevalence and trends of symptomatic pelvic floor disorders in US women. Obstet. Gynecol. 2014, 123, 141–148. [Google Scholar] [CrossRef]
- Pang, H.; Zhang, L.; Han, S.; Li, Z.; Gong, J.; Liu, Q.; Liu, X.; Wang, J.; Xia, Z.; Lang, J. A nationwide population-based survey on the prevalence and risk factors of symptomatic pelvic organ prolapse in adult women in China–a pelvic organ prolapse quantification system-based study. BJOG Int. J. Obstet. Gynaecol. 2021, 128, 1313–1323. [Google Scholar] [CrossRef]
- Fang, E.F.; Scheibye-Knudsen, M.; Jahn, H.J.; Li, J.; Ling, L.; Guo, H.; Zhu, X.; Preedy, V.; Lu, H.; Bohr, V.A. A research agenda for aging in China in the 21st century. Ageing Res. Rev. 2015, 24, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.M.; Matthews, C.A.; Conover, M.M.; Pate, V.; Funk, M.J. Lifetime risk of stress urinary incontinence or pelvic organ prolapse surgery. Obstet. Gynecol. 2014, 123, 1201–1206. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.M.; Kawasaki, A.; Hundley, A.F.; Dieter, A.A.; Myers, E.R.; Sung, V.W. Predicting the number of women who will undergo incontinence and prolapse surgery, 2010 to 2050. Am. J. Obstet. Gynecol. 2011, 205, 230.e231–230.e235. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- May, M. Biomarkers of aging remain elusive as researchers try to slow the biological clock. Nat. Med. 2023, 29, 2673–2676. [Google Scholar] [CrossRef]
- Jelovsek, J.E.; Maher, C.; Barber, M.D. Pelvic organ prolapse. Lancet 2007, 369, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- DeLancey, J.O.; Low, L.K.; Miller, J.M.; Patel, D.A.; Tumbarello, J.A. Graphic integration of causal factors of pelvic floor disorders: An integrated life span model. Am. J. Obstet. Gynecol. 2008, 199, 610.e611–610.e615. [Google Scholar] [CrossRef]
- Deprest, J.A.; Cartwright, R.; Dietz, H.P.; Brito, L.G.O.; Koch, M.; Allen-Brady, K.; Manonai, J.; Weintraub, A.Y.; Chua, J.W.; Cuffolo, R. International Urogynecological Consultation (IUC): Pathophysiology of pelvic organ prolapse (POP). Int. Urogynecol. J. 2022, 33, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.M.; Wray, N.R.; Stone, J.L.; Visscher, P.M.; O’Donovan, M.C.; Sullivan, P.F.; Sklar, P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009, 460, 748–752. [Google Scholar] [CrossRef]
- Jack, G.S.; Nikolova, G.; Vilain, E.; Raz, S.; Rodríguez, L.V. Familial tranmission of genitovaginal prolapse. Int. Urogynecol. J. 2006, 17, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Buchsbaum, G.M.; Duecy, E.E.; Kerr, L.A.; Huang, L.-S.; Perevich, M.; Guzick, D.S. Pelvic organ prolapse in nulliparous women and their parous sisters. Obstet. Gynecol. 2006, 108, 1388–1393. [Google Scholar] [CrossRef] [PubMed]
- Norton, P.A.; Allen-Brady, K.; Cannon-Albright, L.A. The familiality of pelvic organ prolapse in the Utah Population Database. Int. Urogynecol. J. 2013, 24, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Allen-Brady, K.; Norton, P.A.; Hill, A.J.; Rowe, K.; Cannon-Albright, L.A. Risk of pelvic organ prolapse treatment based on extended family history. Am. J. Obstet. Gynecol. 2020, 223, 105.e101–105.e108. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.; Forsman, M.; Falconer, C.; Lichtenstein, P. Genetic influence on stress urinary incontinence and pelvic organ prolapse. Eur. Urol. 2008, 54, 918–923. [Google Scholar] [CrossRef] [PubMed]
- Lince, S.L.; van Kempen, L.C.; Vierhout, M.E.; Kluivers, K.B. A systematic review of clinical studies on hereditary factors in pelvic organ prolapse. Int. Urogynecol. J. 2012, 23, 1327–1336. [Google Scholar] [CrossRef]
- Friedman, T.; Eslick, G.D.; Dietz, H.P. Risk factors for prolapse recurrence: Systematic review and meta-analysis. Int. Urogynecol. J. 2018, 29, 13–21. [Google Scholar] [CrossRef]
- Samimi, P.; Jones, S.H.; Giri, A. Family history and pelvic organ prolapse: A systematic review and meta-analysis. Int. Urogynecol. J. 2021, 32, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhou, Y.; Wang, K.; Li, Z.; Kong, W. Extracellular Matrix Interactome in Modulating Vascular Homeostasis and Remodeling. Circ. Res. 2024, 134, 931–949. [Google Scholar] [CrossRef] [PubMed]
- del Monte-Nieto, G.; Fischer, J.W.; Gorski, D.J.; Harvey, R.P.; Kovacic, J.C. Basic biology of extracellular matrix in the cardiovascular system, part 1/4: JACC focus seminar. J. Am. Coll. Cardiol. 2020, 75, 2169–2188. [Google Scholar] [CrossRef] [PubMed]
- Ewies, A.A.; Al-Azzawi, F.; Thompson, J. Changes in extracellular matrix proteins in the cardinal ligaments of post-menopausal women with or without prolapse: A computerized immunohistomorphometric analysis. Hum. Reprod. 2003, 18, 2189–2195. [Google Scholar] [CrossRef]
- Ricard-Blum, S.; Vallet, S.D. Fragments generated upon extracellular matrix remodeling: Biological regulators and potential drugs. Matrix Biol. 2019, 75, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Yeh, J. Alterations in connective tissue metabolism in stress incontinence and prolapse. J. Urol. 2011, 186, 1768–1772. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Chung, Y.-W.; Lin, W.-Y.; Wang, J.-C.; Tsai, F.-J.; Tsai, C.-H. Collagen type 3 α 1 polymorphism and risk of pelvic organ prolapse. Int. J. Gynecol. Obstet. 2008, 103, 55–58. [Google Scholar] [CrossRef]
- Kluivers, K.B.; Dijkstra, J.R.; Hendriks, J.C.; Lince, S.L.; Vierhout, M.E.; van Kempen, L.C. COL3A1 2209G> A is a predictor of pelvic organ prolapse. Int. Urogynecol. J. 2009, 20, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sun, Z.; Chen, J.; Zhang, Y.; Shi, H.; Zhu, L. Genetic polymorphisms in collagen-related genes are associated with pelvic organ prolapse. Menopause 2020, 27, 223–229. [Google Scholar] [CrossRef]
- Khadzhieva, M.B.; Kamoeva, S.V.; Chumachenko, A.G.; Ivanova, A.V.; Volodin, I.V.; Vladimirov, I.S.; Abilev, S.K.; Salnikova, L.E. Fibulin-5 (FBLN5) gene polymorphism is associated with pelvic organ prolapse. Maturitas 2014, 78, 287–292. [Google Scholar] [CrossRef]
- Abulaizi, A.; Abula, A.; Ababaikeli, G.; Wan, X.; Du, R.; Zhakeer, A. Identification of pelvic organ prolapse risk susceptibility gene SNP locus in Xinjiang women. Int. Urogynecol. J. 2020, 31, 123–130. [Google Scholar] [CrossRef]
- Ashikari, A.; Suda, T.; Miyazato, M. Collagen type 1A1, type 3A1, and LOXL1/4 polymorphisms as risk factors of pelvic organ prolapse. BMC Res. Notes 2021, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, L.; Lang, J.; Zhu, L. Common variants in LAMC1 confer risk for pelvic organ prolapse in Chinese population. Hereditas 2020, 157, 26. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M.M.; Rossi, G.; Biondi, M.L.; Vigano, P.; Dell’Utri, C.; Meschia, M. Type I collagen and matrix metalloproteinase 1, 3 and 9 gene polymorphisms in the predisposition to pelvic organ prolapse. Arch. Gynecol. Obstet. 2012, 285, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Chen HueyYi, C.H.; Lin WeiYong, L.W.; Chen YungHsiang, C.Y.; Chen WenChi, C.W.; Tsai FuuJen, T.F.; Tsai ChangHai, T.C. Matrix metalloproteinase-9 polymorphism and risk of pelvic organ prolapse in Taiwanese women. Eur. J. Obstet. Gynecol. Reprod. Biol. 2010, 149, 222–224. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.M.; Visco, A.G.; Grass, E.A.; Craig, D.M.; Fulton, R.G.; Haynes, C.; Weidner, A.C.; Shah, S.H. Matrix metalloproteinase-9 genetic polymorphisms and the risk for advanced pelvic organ prolapse. Obstet. Gynecol. 2012, 120, 587–593. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Z.Q.; Wang, S.Z.; Lu, J.L.; Wang, X.L.; Zhang, Z.Y. Association of matrix metalloproteinase-10 polymorphisms with susceptibility to pelvic organ prolapse. J. Obstet. Gynaecol. Res. 2015, 41, 1972–1981. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, Y.; Yang, H.; Sun, Z.; Chen, J.; Zhu, L. The polymorphisms of extracellular matrix-remodeling genes are associated with pelvic organ prolapse. Int. Urogynecol. J. 2022, 33, 267–274. [Google Scholar] [CrossRef]
- Olafsdottir, T.; Thorleifsson, G.; Sulem, P.; Stefansson, O.A.; Medek, H.; Olafsson, K.; Ingthorsson, O.; Gudmundsson, V.; Jonsdottir, I.; Halldorsson, G.H. Genome-wide association identifies seven loci for pelvic organ prolapse in Iceland and the UK Biobank. Commun. Biol. 2020, 3, 129. [Google Scholar] [CrossRef]
- Pujol-Gualdo, N.; Läll, K.; Lepamets, M.; Estonian Biobank Research Team; Rossi, H.-R.; Arffman, R.K.; Piltonen, T.T.; Mägi, R. Advancing our understanding of genetic risk factors and potential personalized strategies for pelvic organ prolapse. Nat. Commun. 2022, 13, 3584. [Google Scholar] [CrossRef] [PubMed]
- Matsunami, M.; Imamura, M.; Ashikari, A.; Liu, X.; Tomizuka, K.; Hikino, K.; Miwa, K.; Kadekawa, K.; Suda, T. Genome-wide association studies for pelvic organ prolapse in the Japanese population. Commun. Biol. 2024, 7, 1188. [Google Scholar] [CrossRef] [PubMed]
- Allen-Brady, K.; Cannon-Albright, L.; Farnham, J.M.; Teerlink, C.; Vierhout, M.E.; van Kempen, L.C.; Kluivers, K.B.; Norton, P.A. Identification of six loci associated with pelvic organ prolapse using genome-wide association analysis. Obstet. Gynecol. 2011, 118, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, B.; Denschlag, D.; Göbel, H.; Fittkow, C.; Werner, M.; Gitsch, G.; Watermann, D. Uterosacral ligament in postmenopausal women with or without pelvic organ prolapse. Int. Urogynecol. J. 2005, 16, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Skorupski, P.; Król, J.; Staręga, J.; Adamiak, A.; Jankiewicz, K.; Rechberger, T. An α-1 chain of type I collagen Sp1-binding site polymorphism in women suffering from stress urinary incontinence. Am. J. Obstet. Gynecol. 2006, 194, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Steffensen, L.B.; Rasmussen, L.M. A role for collagen type IV in cardiovascular disease? Am. J. Physiol.-Heart Circ. Physiol. 2018, 315, H610–H625. [Google Scholar] [CrossRef]
- Seppinen, L.; Sormunen, R.; Soini, Y.; Elamaa, H.; Heljasvaara, R.; Pihlajaniemi, T. Lack of collagen XVIII accelerates cutaneous wound healing, while overexpression of its endostatin domain leads to delayed healing. Matrix Biol. 2008, 27, 535–546. [Google Scholar] [CrossRef]
- Ward, R.M.; Edwards, D.R.V.; Edwards, T.; Giri, A.; Jerome, R.N.; Wu, J.M. Genetic epidemiology of pelvic organ prolapse: A systematic review. Am. J. Obstet. Gynecol. 2014, 211, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Chen, X.; Lu, Y. COL3A1 rs1800255 polymorphism is associated with pelvic organ prolapse susceptibility in Caucasian individuals: Evidence from a meta-analysis. PLoS ONE 2021, 16, e0250943. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, R.G.; Pepicelli, F.C.; Batista, N.C.; de Carvalho, C.V.; Bortolini, M.A.; Castro, R.A. Collagen XVIII and LOXL-4 polymorphisms in women with and without advanced pelvic organ prolapse. Int. Urogynecol. J. 2018, 29, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Giri, A.; Wu, J.M.; Ward, R.M.; Hartmann, K.E.; Park, A.J.; North, K.E.; Graff, M.; Wallace, R.B.; Bareh, G.; Qi, L. Genetic determinants of pelvic organ prolapse among African American and Hispanic women in the Women’s Health Initiative. PLoS ONE 2015, 10, e0141647. [Google Scholar] [CrossRef] [PubMed]
- Mecham, R.P. Elastin synthesis and fiber assembly. Ann. N. Y. Acad. Sci. 1991, 624, 137–146. [Google Scholar] [CrossRef]
- Ozsvar, J.; Yang, C.; Cain, S.A.; Baldock, C.; Tarakanova, A.; Weiss, A.S. Tropoelastin and elastin assembly. Front. Bioeng. Biotechnol. 2021, 9, 643110. [Google Scholar] [CrossRef]
- Wise, S.G.; Weiss, A.S. Tropoelastin. Int. J. Biochem. Cell Biol. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Drewes, P.G.; Yanagisawa, H.; Starcher, B.; Hornstra, I.; Csiszar, K.; Marinis, S.I.; Keller, P.; Word, R.A. Pelvic organ prolapse in fibulin-5 knockout mice: Pregnancy-induced changes in elastic fiber homeostasis in mouse vagina. Am. J. Pathol. 2007, 170, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.K.; Pandit, A.; Zawistowski, M.; Dutta, D.; Narla, G.; Swenson, C.W. Genome-Wide Association Study of Pelvic Organ Prolapse Using the Michigan Genomics Initiative. Urogynecology 2021, 27, 502–506. [Google Scholar] [CrossRef]
- Zhao, B.h.; Zhou, J.h. Decreased expression of elastin, fibulin-5 and lysyl oxidase-like 1 in the uterosacral ligaments of postmenopausal women with pelvic organ prolapse. J. Obstet. Gynaecol. Res. 2012, 38, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Allen-Brady, K.; Cannon-Albright, L.A.; Farnham, J.M.; Norton, P.A. Evidence for pelvic organ prolapse predisposition genes on chromosomes 10 and 17. Am. J. Obs. Gynecol. 2015, 212, e771–e777. [Google Scholar] [CrossRef]
- Pulst, S.M. Genetic linkage analysis. Arch. Neurol. 1999, 56, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, Y.; Gao, J.; Pawlyk, B.; Starcher, B.; Spencer, J.A.; Yanagisawa, H.; Zuo, J.; Li, T. Elastic fiber homeostasis requires lysyl oxidase–like 1 protein. Nat. Genet. 2004, 36, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, G.; Lee, H.; Berkovitz, S.; Nelson, S.; Sinsheimer, J.; Vilain, E.; Rodríguez, L.V. Sequence variant in the laminin γ1 (LAMC1) gene associated with familial pelvic organ prolapse. Hum. Genet. 2007, 120, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Hill, L.D.; Schubert, C.M.; Strauss III, J.F.; Matthews, C.A. Is laminin γ-1 a candidate gene for advanced pelvic organ prolapse? Am. J. Obstet. Gynecol. 2010, 202, 505.e501–505.e505. [Google Scholar] [CrossRef]
- Wu, J.M.; Visco, A.G.; Grass, E.A.; Craig, D.M.; Fulton, R.G.; Haynes, C.; Amundsen, C.L.; Shah, S.H. Comprehensive analysis of LAMC1 genetic variants in advanced pelvic organ prolapse. Am. J. Obstet. Gynecol. 2012, 206, 447.e441–447.e446. [Google Scholar] [CrossRef] [PubMed]
- Nakad, B.; Fares, F.; Azzam, N.; Feiner, B.; Zilberlicht, A.; Abramov, Y. Estrogen receptor and laminin genetic polymorphism among women with pelvic organ prolapse. Taiwan. J. Obstet. Gynecol. 2017, 56, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G. Tissue inhibitors of metalloproteinases. Genome Biol. 2011, 12, 233. [Google Scholar] [CrossRef] [PubMed]
- Fadista, J.; Skotte, L.; Karjalainen, J.; Abner, E.; Sørensen, E.; Ullum, H.; Werge, T.; iPSYCH Group; Esko, T.; Milani, L.; et al. Comprehensive genome-wide association study of different forms of hernia identifies more than 80 associated loci. Nat. Commun. 2022, 13, 3200. [Google Scholar] [CrossRef]
- Xu, K.; Sun, Y.; Al-Ani, M.K.; Wang, C.; Sha, Y.; Sung, K.P.; Dong, N.; Qiu, X.; Yang, L. Synergistic promoting effects of bone morphogenetic protein 12/connective tissue growth factor on functional differentiation of tendon derived stem cells and patellar tendon window defect regeneration. J. Biomech. 2018, 66, 95–102. [Google Scholar] [CrossRef]
- Haddad-Weber, M.; Prager, P.; Kunz, M.; Seefried, L.; Jakob, F.; Murray, M.M.; Evans, C.H.; Nöth, U.; Steinert, A.F. BMP12 and BMP13 gene transfer induce ligamentogenic differentiation in mesenchymal progenitor and anterior cruciate ligament cells. Cytotherapy 2010, 12, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Attaar, M.; Shi, Z.; Na, R.; Resurreccion, W.; Haggerty, S.; Zheng, S.; Helfand, B.; Ujiki, M.; Xu, J. Identification of fifty-seven novel loci for abdominal wall hernia development and their biological and clinical implications: Results from the UK Biobank. Hernia 2022, 26, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Connell, K.A.; Guess, M.K.; Chen, H.; Andikyan, V.; Bercik, R.; Taylor, H.S. HOXA11 is critical for development and maintenance of uterosacral ligaments and deficient in pelvic prolapse. J. Clin. Investig. 2008, 118, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Guess, M.; Datar, A.; Hennessey, A.; Cardenas, I.; Johnson, J.; Connell, K.A. Knockdown of Hoxa11 in vivo in the uterosacral ligament and uterus of mice results in altered collagen and matrix metalloproteinase activity. Biol. Reprod. 2012, 86, 100. [Google Scholar] [CrossRef] [PubMed]
- Castelán, F.; Cuevas-Romero, E.; Martínez-Gómez, M. The expression of hormone receptors as a gateway toward understanding endocrine actions in female pelvic floor muscles. Endocr. Metab. Immune Disord.-Drug Targets (Former. Curr. Drug Targets-Immune Endocr. Metab. Disord.) 2020, 20, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Koehler, K.F.; Helguero, L.A.; Haldosén, L.-A.; Warner, M.; Gustafsson, J.-A.k. Reflections on the discovery and significance of estrogen receptor β. Endocr. Rev. 2005, 26, 465–478. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Chung, Y.-W.; Lin, W.-Y.; Chen, W.-C.; Tsai, F.-J.; Tsai, C.-H. Estrogen receptor α polymorphism is associated with pelvic organ prolapse risk. Int. Urogynecol. J. 2008, 19, 1159–1163. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Chung, Y.-W.; Lin, W.-Y.; Chen, W.-C.; Tsai, F.-J.; Tsai, C.-H. Progesterone receptor polymorphism is associated with pelvic organ prolapse risk. Acta Obstet. Gynecol. Scand. 2009, 88, 835–838. [Google Scholar] [CrossRef]
- Hall, J.M.; Couse, J.F.; Korach, K.S. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J. Biol. Chem. 2001, 276, 36869–36872. [Google Scholar] [CrossRef]
- Alperin, M.; Burnett, L.; Lukacz, E.; Brubaker, L. The mysteries of menopause and urogynecologic health: Clinical and scientific gaps. Menopause 2019, 26, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Wan, L.; Chung, Y.-W.; Chen, W.-C.; Tsai, F.-J.; Tsai, C.-H. Estrogen receptor β gene haplotype is associated with pelvic organ prolapse. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008, 138, 105–109. [Google Scholar] [CrossRef]
- Sollis, E.; Mosaku, A.; Abid, A.; Buniello, A.; Cerezo, M.; Gil, L.; Groza, T.; Güneş, O.; Hall, P.; Hayhurst, J. The NHGRI-EBI GWAS Catalog: Knowledgebase and deposition resource. Nucleic Acids Res. 2023, 51, D977–D985. [Google Scholar] [CrossRef]
- Sapkota, Y.; Steinthorsdottir, V.; Morris, A.P.; Fassbender, A.; Rahmioglu, N.; De Vivo, I.; Buring, J.E.; Zhang, F.; Edwards, T.L.; Jones, S. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat. Commun. 2017, 8, 15539. [Google Scholar] [CrossRef] [PubMed]
- Biason-Lauber, A.; Konrad, D. WNT4 and sex development. Sex. Dev. 2008, 2, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Mandel, H.; Shemer, R.; Borochowitz, Z.U.; Okopnik, M.; Knopf, C.; Indelman, M.; Drugan, A.; Tiosano, D.; Gershoni-Baruch, R.; Choder, M. SERKAL syndrome: An autosomal-recessive disorder caused by a loss-of-function mutation in WNT4. Am. J. Hum. Genet. 2008, 82, 39–47. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.-H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative stress and autophagy: The clash between damage and metabolic needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef]
- Hong, S.; Hong, L.; Wu, D.; Li, B.; Liu, C.; Guo, W.; Min, J.; Hu, M.; Zhao, Y.; Yang, Q. Oxidative damage to human parametrial ligament fibroblasts induced by mechanical stress. Mol. Med. Rep. 2015, 12, 5342–5348. [Google Scholar] [CrossRef] [PubMed]
- Kim JiYoung, K.J.; Kim EunJae, K.E.; Jeon MyungJae, J.M.; Kim Ran, K.R.; Lee MinWoo, L.M.; Kim SuhngWook, K.S. Association between susceptibility to advanced pelvic organ prolapse and glutathione S-transferase P1 Ile105Val polymorphism. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014, 175, 205–208. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, E.J.; Jeon, M.J.; Kim, H.; Moon, Y.J.; Bai, S.W. Association between the poly (ADP-ribose) polymerase-1 gene polymorphism and advanced pelvic organ prolapse. Menopause 2014, 21, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular stress signaling through poly (ADP-ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Von Scheidt, M.; Zhao, Y.; de Aguiar Vallim, T.Q.; Che, N.; Wierer, M.; Seldin, M.M.; Franzén, O.; Kurt, Z.; Pang, S.; Bongiovanni, D. Transcription factor MAFF (MAF basic leucine zipper transcription factor F) regulates an atherosclerosis relevant network connecting inflammation and cholesterol metabolism. Circulation 2021, 143, 1809–1823. [Google Scholar] [CrossRef]
- Darwich, R.; Li, W.; Yamak, A.; Komati, H.; Andelfinger, G.; Sun, K.; Nemer, M. KLF13 is a genetic modifier of the Holt-Oram syndrome gene TBX5. Hum. Mol. Genet. 2017, 26, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, R.; Gunawan, F.; Ramadass, R.; Beisaw, A.; Konzer, A.; Mullapudi, S.T.; Gentile, A.; Maischein, H.-M.; Graumann, J.; Stainier, D.Y. Mechanical forces regulate cardiomyocyte myofilament maturation via the VCL-SSH1-CFL axis. Dev. Cell 2019, 51, 62–77.e5. [Google Scholar] [CrossRef] [PubMed]
- Fitz, F.F.; Bortolini, M.A.T.; Pereira, G.M.V.; Salerno, G.R.F.; Castro, R.A. PEOPLE: Lifestyle and comorbidities as risk factors for pelvic organ prolapse—A systematic review and meta-analysis PEOPLE: PElvic Organ Prolapse Lifestyle comorbiditiEs. Int. Urogynecol. J. 2023, 34, 2007–2032. [Google Scholar] [CrossRef] [PubMed]
- Isık, H.; Aynıoglu, O.; Sahbaz, A.; Selimoglu, R.; Timur, H.; Harma, M. Are hypertension and diabetes mellitus risk factors for pelvic organ prolapse? Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 197, 59–62. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; Girão, M.J.B.C.; da Silva, I.D.C.G.; Sartori, M.G.F.; Martins, K.d.F.; Castro, R.d.A. COL1A1 Sp1-binding site polymorphism as a risk factor for genital prolapse. Int. Urogynecol. J. 2008, 19, 1471–1475. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Jung, H.J.; Kim, S.K.; Choi, J.R.; Cho, N.H.; Bai, S.W. Polymorphism of a COLIA1 gene Sp1 binding site in Korean women with pelvic organ prolapse. Yonsei Med. J. 2009, 50, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Feiner, B.; Fares, F.; Azam, N.; Auslender, R.; David, M.; Abramov, Y. Does COLIA1 SP1-binding site polymorphism predispose women to pelvic organ prolapse? Int. Urogynecol. J. 2009, 20, 1061–1065. [Google Scholar] [CrossRef]
- Batista, N.C.; Bortolini, M.A.; Silva, R.S.; Teixeira, J.B.; Melo, N.C.; Santos, R.G.; Pepicelli, F.A.; Castro, R.A. Collagen I and collagen III polymorphisms in women with pelvic organ prolapse. Neurourol. Urodyn. 2020, 39, 1977–1984. [Google Scholar] [CrossRef]
- Palos, C.C.; Timm, B.F.; de Souza Paulo, D.; Fernandes, C.E.; de Souto, R.P.; Oliveira, E. Evaluation of COLIA1-1997 G/T polymorphism as a related factor to genital prolapse. Int. Urogynecol. J. 2020, 31, 133–137. [Google Scholar] [CrossRef] [PubMed]
- de Freitas Rosa, J.P.; Haddad, R.F.; Maeda, F.G.R.; Souto, R.P.; Fernandes, C.E.; de Oliveira, E. Association between col1a2 polymorphism and the occurrence of pelvic organ prolapse in Brazilian women. Rev. Bras. De Ginecol. E Obs./RBGO Gynecol. Obstet. 2019, 41, 031–036. [Google Scholar] [CrossRef]
- Teixeira, F.H.; Fernandes, C.E.; do Souto, R.P.; de Oliveira, E. Polymorphism rs1800255 from COL3A1 gene and the risk for pelvic organ prolapse. Int. Urogynecol. J. 2020, 31, 73–78. [Google Scholar] [CrossRef]
- Paula, M.V.B.d.; Lira Júnior, M.A.d.F.; Souto, R.P.; Fernandes, C.E.; Oliveira, E.d. Evaluation of the fibulin 5 gene polymorphism as a factor related to the occurrence of pelvic organ prolapse. Rev. Da Assoc. Médica Bras. 2020, 66, 680–686. [Google Scholar] [CrossRef]
- Neupane, R.; Sadeghi, Z.; Fu, R.; Hagstrom, S.A.; Moore, C.K.; Daneshgari, F. Mutation screen of LOXL1 in patients with female pelvic organ prolapse. Urogynecology 2014, 20, 316–321. [Google Scholar] [CrossRef]
- Skorupski, P.; Jankiewicz, K.; Miotła, P.; Marczak, M.; Kulik-Rechberger, B.; Rechberger, T. The polymorphisms of the MMP-1 and the MMP-3 genes and the risk of pelvic organ prolapse. Int. Urogynecol. J. 2013, 24, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Karachalios, C.; Bakas, P.; Kaparos, G.; Demeridou, S.; Liapis, I.; Grigoriadis, C.; Liapis, A. Matrix metalloproteinase-3 gene promoter polymorphisms: A potential risk factor for pelvic organ prolapse. Biomed. Rep. 2016, 5, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Maeda, P.M.; Bicudo, A.P.S.; Watanabe, R.T.; Fonseca, T.S.; do Souto, R.P.; Fernandes, C.E.; de Oliveira, E. Study of the polymorphism rs3025058 of the MMP-3 gene and risk of pelvic organ prolapse in Brazilian women. Eur. J. Obstet. Gynecol. Reprod. Biol. X 2019, 3, 100031. [Google Scholar] [CrossRef]
- Ghersel, F.R.; Souto, R.P.; Gonzales, E.W.P.; Paulo, D.S.; Fernandes, C.E.; Oliveira, E. Assessment of metalloproteinase matrix 9 (MMP9) gene polymorphisms risk factors for pelvic organ prolapse in the Brazilian population. Rev. Bras. De Ginecol. E Obs. 2019, 41, 164–169. [Google Scholar] [CrossRef]
- Singh, K.P.; Miaskowski, C.; Dhruva, A.A.; Flowers, E.; Kober, K.M. Mechanisms and measurement of changes in gene expression. Biol. Res. Nurs. 2018, 20, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Battle, A.; Brown, C.D.; Engelhardt, B.E.; Montgomery, S.B. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [PubMed]
- DeLancey, J.O. Anatomie aspects of vaginal eversion after hysterectomy. Am. J. Obstet. Gynecol. 1992, 166, 1717–1728. [Google Scholar] [CrossRef]
- Zhao, Y.; Xia, Z.; Lin, T.; Qin, M. Transforming growth factor β 1 and p44/42 expression in cardinal ligament tissues of patients with pelvic organ prolapse. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2021, 27, e930433-1. [Google Scholar] [CrossRef]
- Yucel, N.; Usta, A.; Guzin, K.; Kanter, M.; Bilgic, E.; Ozel, N.O.; Ozgul, M. Immunohistochemical analysis of connective tissue in patients with pelvic organ prolapse. J. Mol. Histol. 2013, 44, 97–102. [Google Scholar] [CrossRef]
- Dökmeci, F.; Tekşen, F.; Çetinkaya, Ş.E.; Özkan, T.; Kaplan, F.; Köse, K. Expressions of homeobox, collagen and estrogen genes in women with uterine prolapse. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 233, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-p.; Xie, T.; Guo, T.; Sun, Z.-j.; Zhu, L.; Lang, J.-h. Evaluation of extracellular matrix protein expression and apoptosis in the uterosacral ligaments of patients with or without pelvic organ prolapse. Int. Urogynecol. J. 2021, 32, 2273–2281. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zheng, Q.; Zhang, L.; Chen, L.; Wang, J. Effect of Vaginal Microecological Alterations on Female Pelvic Organ Prolapse. Int. Urogynecol. J. 2024, 35, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Söderberg, M.W.; Byström, B.; Kalamajski, S.; Malmström, A.; Ekman-Ordeberg, G. Gene expressions of small leucine-rich repeat proteoglycans and fibulin-5 are decreased in pelvic organ prolapse. MHR Basic Sci. Reprod. Med. 2009, 15, 251–257. [Google Scholar] [CrossRef]
- Moalli, P.A.; Shand, S.H.; Zyczynski, H.M.; Gordy, S.C.; Meyn, L.A. Remodeling of vaginal connective tissue in patients with prolapse. Obstet. Gynecol. 2005, 106, 953–963. [Google Scholar] [CrossRef]
- Lin, S.-Y.; Tee, Y.-T.; Ng, S.-C.; Chang, H.; Lin, P.; Chen, G.-D. Changes in the extracellular matrix in the anterior vagina of women with or without prolapse. Int. Urogynecol. J. 2007, 18, 43–48. [Google Scholar] [CrossRef]
- Mosier, E.; Lin, V.K.; Zimmern, P. Extracellular matrix expression of human prolapsed vaginal wall. Neurourol. Urodyn. Off. J. Int. Cont. Soc. 2010, 29, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Lee, J.H.; Wen, Y.; Constantinou, C.; Yoshinobu, M.; Omata, S.; Chen, B. Biomechanical properties and associated collagen composition in vaginal tissue of women with pelvic organ prolapse. J. Urol. 2012, 188, 875–880. [Google Scholar] [CrossRef]
- Kerkhof, M.H.; Ruiz-Zapata, A.M.; Bril, H.; Bleeker, M.C.; Belien, J.A.; Stoop, R.; Helder, M.N. Changes in tissue composition of the vaginal wall of premenopausal women with prolapse. Am. J. Obstet. Gynecol. 2014, 210, 168.e161–168.e169. [Google Scholar] [CrossRef] [PubMed]
- Vetuschi, A.; D’Alfonso, A.; Sferra, R.; Zanelli, D.; Pompili, S.; Patacchiola, F.; Gaudio, E.; Carta, G. Changes in muscularis propria of anterior vaginal wall in women with pelvic organ prolapse. Eur. J. Histochem. EJH 2016, 60, 2604. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wu, R.; Li, H.; Gu, Y.; Wei, W. Expression and significance of metalloproteinase and collagen in vaginal wall tissues of patients with pelvic organ prolapse. Ann. Clin. Lab. Sci. 2017, 47, 698–705. [Google Scholar]
- Vetuschi, A.; Pompili, S.; Gallone, A.; D’Alfonso, A.; Carbone, M.G.; Carta, G.; Festuccia, C.; Gaudio, E.; Colapietro, A.; Sferra, R. Immunolocalization of advanced glycation end products, mitogen activated protein kinases, and transforming growth factor-β/Smads in pelvic organ prolapse. J. Histochem. Cytochem. 2018, 66, 673–686. [Google Scholar] [CrossRef]
- Lin, T.; Ji, Y.; Zhao, Y.; Xia, Z. Expression of COX-2 and Nrf2/GPx3 in the anterior vaginal wall tissues of women with pelvic organ prolapse. Arch. Gynecol. Obstet. 2021, 303, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Goepel, C. Differential elastin and tenascin immunolabeling in the uterosacral ligaments in postmenopausal women with and without pelvic organ prolapse. Acta Histochem. 2008, 110, 204–209. [Google Scholar] [CrossRef]
- de Landsheere, L.; Blacher, S.; Munaut, C.; Nusgens, B.; Rubod, C.; Noel, A.; Foidart, J.-M.; Cosson, M.; Nisolle, M. Changes in elastin density in different locations of the vaginal wall in women with pelvic organ prolapse. Int. Urogynecol. J. 2014, 25, 1673–1681. [Google Scholar] [CrossRef]
- Takacs, P.; Nassiri, M.; Candiotti, K.; Yang, J.; Yavagal, S.; Medina, C.A. Differential expression of fibulins in the uterosacral ligaments of women with uterine prolapse. Arch. Gynecol. Obstet. 2010, 282, 389–394. [Google Scholar] [CrossRef]
- Ahn, K.; Kim, T.; Hur, J.; Kim, S.; Lee, K.; Kim, Y. Relationship between the expression of fibulin-3 and anterior vaginal wall prolapse. J. Obstet. Gynaecol. 2012, 32, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ling, O.; Bo, L. Expression and significance of lysyl oxidase-like 1 and fibulin-5 in the cardinal ligament tissue of patients with pelvic floor dysfunction. J. Biomed. Res. 2013, 27, 23. [Google Scholar] [CrossRef]
- Klutke, J.; Ji, Q.; Campeau, J.; Starcher, B.; Carlos Felix, J.; Stanczyk, F.Z.; Klutke, C. Decreased endopelvic fascia elastin content in uterine prolapse. Acta Obstet. Gynecol. Scand. 2008, 87, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Jeon, M.J.; Yim, G.W.; Kim, S.K.; Choi, J.R.; Bai, S.W. Changes in expression of fibulin-5 and lysyl oxidase-like 1 associated with pelvic organ prolapse. Eur. J. Obstet. Gynecol. Reprod. Biol. 2009, 145, 117–122. [Google Scholar] [CrossRef]
- Takacs, P.; Nassiri, M.; Viciana, A.; Candiotti, K.; Fornoni, A.; Medina, C.A. Fibulin-5 expression is decreased in women with anterior vaginal wall prolapse. Int. Urogynecol. J. 2009, 20, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Alarab, M.; Bortolini, M.A.; Drutz, H.; Lye, S.; Shynlova, O. LOX family enzymes expression in vaginal tissue of premenopausal women with severe pelvic organ prolapse. Int. Urogynecol. J. 2010, 21, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Strinic, T.; Vulic, M.; Tomic, S.; Capkun, V.; Stipic, I.; Alujevic, I. Matrix metalloproteinases-1,-2 expression in uterosacral ligaments from women with pelvic organ prolapse. Maturitas 2009, 64, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Vulic, M.; Strinic, T.; Tomic, S.; Capkun, V.; Jakus, I.A.; Ivica, S. Difference in expression of collagen type I and matrix metalloproteinase-1 in uterosacral ligaments of women with and without pelvic organ prolapse. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011, 155, 225–228. [Google Scholar] [CrossRef]
- Dviri, M.; Leron, E.; Dreiher, J.; Mazor, M.; Shaco-Levy, R. Increased matrix metalloproteinases-1,-9 in the uterosacral ligaments and vaginal tissue from women with pelvic organ prolapse. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011, 156, 113–117. [Google Scholar] [CrossRef]
- Usta, A.; Guzin, K.; Kanter, M.; Ozgül, M.; Usta, C.S. Expression of matrix metalloproteinase-1 in round ligament and uterosacral ligament tissue from women with pelvic organ prolapse. J. Mol. Histol. 2014, 45, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Alarab, M.; Kufaishi, H.; Lye, S.; Drutz, H.; Shynlova, O. Expression of extracellular matrix-remodeling proteins is altered in vaginal tissue of premenopausal women with severe pelvic organ prolapse. Reprod. Sci. 2014, 21, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Chen, J.; Guo, X.; Guan, H.; Li, C. Differential expression profiling of matrix metalloproteinases and tissue inhibitors of metalloproteinases in females with or without pelvic organ prolapse. Mol. Med. Rep. 2014, 10, 2004–2008. [Google Scholar] [CrossRef]
- Moon, Y.J.; Choi, J.R.; Jeon, M.J.; Kim, S.K.; Bai, S.W. Alteration of elastin metabolism in women with pelvic organ prolapse. J. Urol. 2011, 185, 1786–1792. [Google Scholar] [CrossRef]
- Leegant, A.; Zuckerwise, L.C.; Downing, K.; Brouwer-Visser, J.; Zhu, C.; Cossio, M.J.; Strube, F.; Xie, X.; Banks, E.; Huang, G.S. Transforming growth factor β1 and extracellular matrix protease expression in the uterosacral ligaments of patients with and without pelvic organ prolapse. Urogynecology 2015, 21, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-C.; Huang, H.-Y.; Tseng, L.-H.; Chang, S.-D.; Lo, T.-S.; Lee, C.-L. Expression of matrix metalloproteinase-2 and tissue inhibitors of metalloproteinase-1 (TIMP-1, TIMP-2 and TIMP-3) in women with uterine prolapse but without urinary incontinence. Eur. J. Obstet. Gynecol. Reprod. Biol. 2010, 153, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.W.; Chung, D.J.; Yoon, J.M.; Shin, J.S.; Kim, S.K.; Park, K.H. Roles of estrogen receptor, progesterone receptor, p53 and p21 in pathogenesis of pelvic organ prolapse. Int. Urogynecol. J. 2005, 16, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, W.; Elenskaia, K.; Obermayr, E.; Horvat, R.; Mayerhofer, K.; Umek, W.; Zeillinger, R.; Hanzal, E. Relaxin and gonadal steroid receptors in uterosacral ligaments of women with and without pelvic organ prolapse. Int. Urogynecol. J. 2012, 23, 495–500. [Google Scholar] [CrossRef]
- Sun, M.-J.; Cheng, Y.-S.; Sun, R.; Cheng, W.-L. Changes in mitochondrial DNA copy number and extracellular matrix (ECM) proteins in the uterosacral ligaments of premenopausal women with pelvic organ prolapse. Taiwan. J. Obstet. Gynecol. 2016, 55, 9–15. [Google Scholar] [CrossRef]
- Tola, E.N.; Koroglu, N.; Yıldırım, G.Y.; Koca, H.B. The role of ADAMTS-2, collagen type-1, TIMP-3 and papilin levels of uterosacral and cardinal ligaments in the etiopathogenesis of pelvic organ prolapse among women without stress urinary incontinence. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018, 231, 158–163. [Google Scholar] [CrossRef]
- Putra, I.G.M.; Warsita, I.G.N.; Suwiyoga, K.; Manuaba, I.G.F.; Budiana, I.N.G.; Wiradnyana, A.G.P. Low expression of collagen type-1 in sacrouterine ligament as risk factor of stage III–IV uterine prolapse. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 249, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, J.; Hu, C.; Hua, K. Relationship of advanced glycation end products and their receptor to pelvic organ prolapse. Int. J. Clin. Exp. Pathol. 2015, 8, 2288. [Google Scholar]
- Eser, A.; Unlubilgin, E.; Hizli, F.; Acar, M.; Kamalak, Z.; Kosus, A.; Kosus, N.; Hizli, D.; Gunduz, E. Is there a relationship between pelvic organ prolapse and tissue Fibrillin-1 levels? Int. Neurourol. J. 2015, 19, 164. [Google Scholar] [CrossRef] [PubMed]
- Bodner-Adler, B.; Bodner, K.; Kimberger, O.; Halpern, K.; Schneidinger, C.; Haslinger, P.; Schneeberger, C.; Horvat, R.; Umek, W. The role of tenascin-X in the uterosacral ligaments of postmenopausal women with pelvic organ prolapse: An immunohistochemical study. Int. Urogynecol. J. 2020, 31, 101–106. [Google Scholar] [CrossRef]
- Choy, K.; Liu, Y.; Chu, C.; Wang, C.; Lui, W.; Lee, L.; Pang, M.; Rogers, M.; Yip, S. High isoprostane level in cardinal ligament-derived fibroblasts and urine sample of women with uterine prolapse. BJOG Int. J. Obstet. Gynaecol. 2008, 115, 1179–1183. [Google Scholar] [CrossRef]
- Yılmaz, N.; Ozaksit, G.; Terzi, Y.K.; Yılmaz, S.; Budak, B.; Aksakal, O.; Şahin, F.İ. HOXA11 and MMP2 gene expression in uterosacral ligaments of women with pelvic organ prolapse. J. Turk. Ger. Gynecol. Assoc. 2014, 15, 104. [Google Scholar] [CrossRef]
- Connell, K.A.; Guess, M.K.; Tate, A.; Andikyan, V.; Bercik, R.; Taylor, H.S. Diminished vaginal HOXA13 expression in women with pelvic organ prolapse. Menopause 2009, 16, 529–533. [Google Scholar] [CrossRef]
- Fang, G.; Hong, L.; Liu, C.; Yang, Q.; Zhang, Q.; Li, Y.; Li, B.; Wu, D.; Wu, W.; Shi, H. Oxidative status of cardinal ligament in pelvic organ prolapse. Exp. Ther. Med. 2018, 16, 3293–3302. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Chung, N.; Park, S.H.; Lee, K.-H.; Kim, S.W.; Kim, J.Y.; Bai, S.W.; Jeon, M.J. Involvement of oxidative stress and mitochondrial apoptosis in the pathogenesis of pelvic organ prolapse. J. Urol. 2013, 189, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Jakus, I.A.; Jakus, D.; Aračić, N.; Stipić, I.; Vilović, K. Immunohistochemical expression of hypoxia-inducible factor-1α in stromal cells of vaginal tissue in post-menopausal women with pelvic organ prolapse. Indian J. Med. Res. 2017, 146, S63–S67. [Google Scholar] [CrossRef]
- Sun, M.-J.; Cheng, Y.-S.; Liu, C.-S.; Sun, R. Changes in the PGC-1α and mtDNA copy number may play a role in the development of pelvic organ prolapse in pre-menopausal patients. Taiwan. J. Obstet. Gynecol. 2019, 58, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Huang, Y.; Tan, Y.; Zhao, W.; Tian, Q. Single-cell RNA sequencing in hematological diseases. Proteomics 2020, 20, 1900228. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.-Y.; Sun, B.-F.; Ma, Y.; Zhang, Y.; Wang, M.; Ma, C.; Shi, H.; Sun, Z.; Chen, J. Single-cell transcriptome profiling of the vaginal wall in women with severe anterior vaginal prolapse. Nat. Commun. 2021, 12, 87. [Google Scholar] [CrossRef]
- Fan, W.; Wu, D.; Zhang, L.; Ye, J.; Guan, J.; Yang, Y.; Mei, X.; Chen, R. Single-cell transcriptomic data reveal the increase in extracellular matrix organization and antigen presentation abilities of fibroblasts and smooth muscle cells in patients with pelvic organ prolapse. Int. Urogynecol. J. 2023, 34, 2529–2537. [Google Scholar] [CrossRef]
- Miao, Y.; Wen, J.; Wang, L.; Wen, Q.; Cheng, J.; Zhao, Z.; Wu, J. scRNA-seq reveals aging-related immune cell types and regulators in vaginal wall from elderly women with pelvic organ prolapse. Front. Immunol. 2023, 14, 1084516. [Google Scholar] [CrossRef]
- Liu, X.; Su, M.; Wei, L.; Zhang, J.; Wang, W.; Hao, Q.; Lin, X.; Wang, L. Single-cell analysis of uterosacral ligament revealed cellular heterogeneity in women with pelvic organ prolapse. Commun. Biol. 2024, 7, 159. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Mak, T.S.-H.; O’Reilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef] [PubMed]
- Eichler, E.E.; Flint, J.; Gibson, G.; Kong, A.; Leal, S.M.; Moore, J.H.; Nadeau, J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010, 11, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.P. A global reference for human genetic variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef] [PubMed]
- Zarrei, M.; MacDonald, J.R.; Merico, D.; Scherer, S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015, 16, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zheng, P.; Duan, A.; Hao, Y.; Lu, C.; Lu, D. Genome-wide DNA methylation analysis of uterosacral ligaments in women with pelvic organ prolapse. Mol. Med. Rep. 2019, 19, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Qin, M.; Luan, M.; Xia, Z. miR-19-3p Promotes Autophagy and Apoptosis in Pelvic Organ Prolapse Through the AKT/mTOR/p70S6K Pathway: Function of miR-19-3p on Vaginal Fibroblasts by Targeting IGF-1. Urogynecology 2021, 27, e630–e638. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Niu, G.; Gao, J.; Liu, J.X.; Qu, H. MicroRNA-92 expression may be associated with reduced estrogen receptor β1 mRNA levels in cervical portion of uterosacral ligaments in women with pelvic organ prolapse. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 198, 94–99. [Google Scholar] [CrossRef]
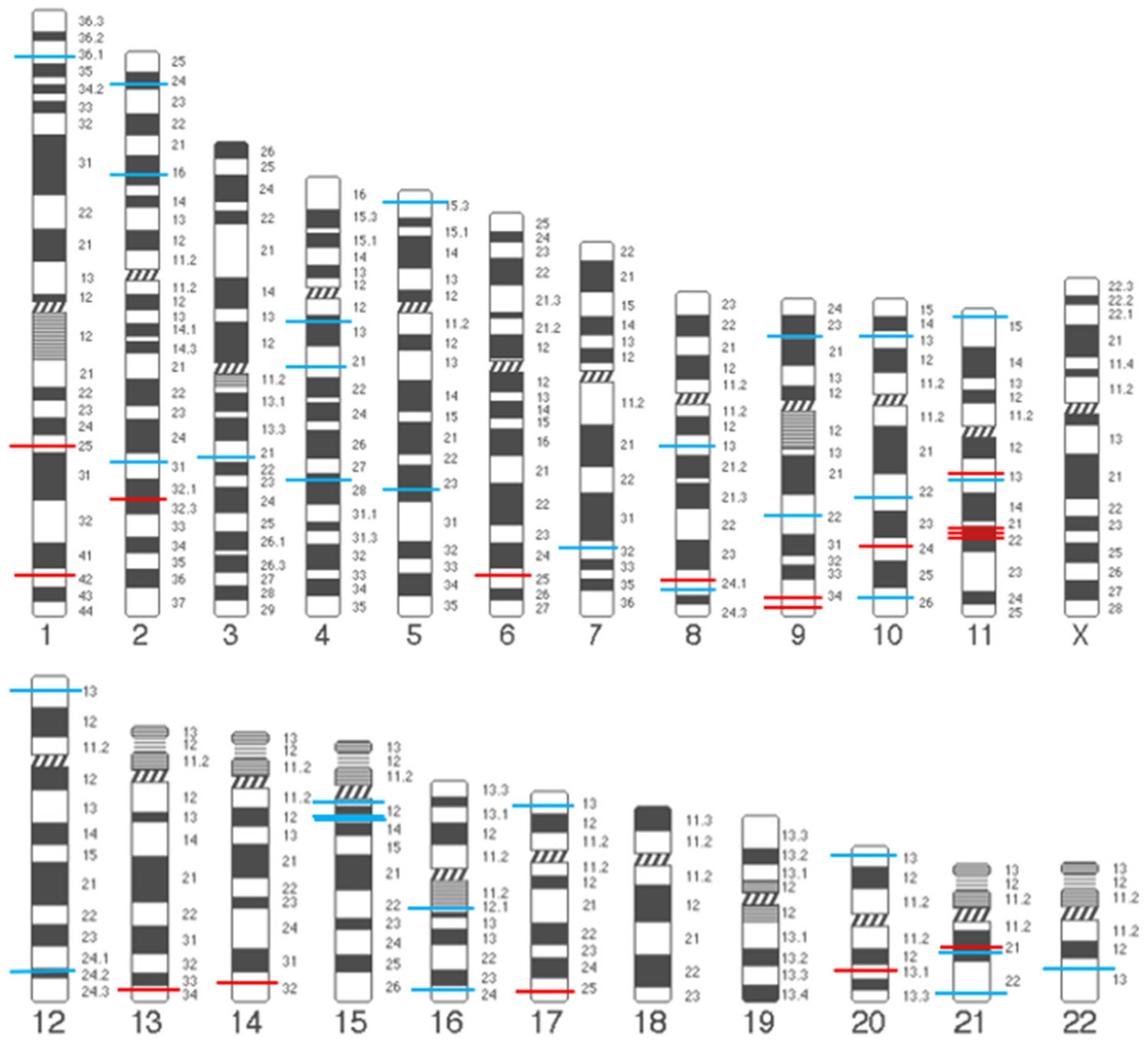
| Candidate Gene Association Studies | |||||||||||
| Component | SNP ID | Annotation | Change in Nucleotide | Candidate Gene | Effect Direction | Significant Association with POP * | Race/Ethnicity | Sample Size (POP/Control) | Reference | ||
| Collagen | rs1800255 | Exonic | G>A | COL3A1 | Risk | genotype AA, OR 5.05 | East Asian (Chinese) | 84 vs. 147 | [29] | ||
| Risk | genotype AA, OR 5.00 | European (Dutch) | 202 vs. 102 | [30] | |||||||
| rs445348 | Exonic | A>G | COL4A2 | Risk | allele G, OR 2.15 | East Asian (Chinese) | 48 vs. 48 | [31] | |||
| rs76425569 | Exonic | G>A | COL4A2 | Risk | allele A, OR 2.02 | East Asian (Chinese) | 48 vs. 48 | [31] | |||
| rs388222 | Intronic | C>T | COL4A2 | Protective | allele T, OR 0.50 | East Asian (Chinese) | 48 vs. 48 | [31] | |||
| rs2281968 | Intronic | G>A | COL4A2 | Risk | allele A, OR 2.02 | East Asian (Chinese) | 48 vs. 48 | [31] | |||
| rs3827852 | Intronic | A>G | COL5A1 | Protective | allele G, OR 0.40 | East Asian (Chinese) | 48 vs. 48 | [31] | |||
| rs4870723 | Exonic | A>C | COL14A1 | Protective | allele C, OR 0.46 | East Asian (Chinese) | 48 vs. 48 | [31] | |||
| rs2305600 | Exonic | T>C | COL14A1 | Protective | allele C, OR 0.48 | East Asian (Chinese) | 48 vs. 48 | [31] | |||
| rs2305598 | Exonic | T>C | COL14A1 | Protective | allele C, OR 0.50 | East Asian (Chinese) | 48 vs. 48 | [31] | |||
| Elastic | rs2018736 | Intronic | C>A | FBLN5 | Protective | allele A, OR 0.73 | Russian | 210 vs. 292 | [32] | ||
| fibers | rs12589592 | Intronic | G>A | FBLN5 | Protective | allele A, OR 0.42 | Russian | 210 vs. 292 | [32] | ||
| Protective | genotype AA, OR 0.11, allele A, OR 0.48 | East Asian (minority/non-minority Chinese) | 88 vs. 108 | [33] | |||||||
| Lysyl oxidase | rs2862296 | Intergenic | A>G | LOXL4 | Risk | genotype AG, OR 3.80; genotype GG, OR 4.50 | East Asian (Japanese) | 52 vs. 28 | [34] | ||
| Laminin | rs10911241 | Intronic | A>G | LAMC1 | Risk | allele G, OR 1.71 | East Asian (Chinese) | 161 vs. 235 | [35] | ||
| Proteases | - | Upstream Variant | G→GG | MMP1 | Risk | genotype GG/GG (OR not analyzed) | European (Italian) | 137 vs. 96 | [36] | ||
| rs17576 | Exonic | A>G | MMP9 | Risk | genotype AG, OR 5.41; genotype GG, OR 5.77 | East Asian (Chinese) | 92 vs. 152 | [37] | |||
| rs3918253 | Intronic | C>T | MMP9 | Risk | allele T, OR 1.56 | Non-Hispanic White | 239 vs. 197 | [38] | |||
| rs3918256 | Intronic | G>A | MMP9 | Risk | allele A, OR 1.56 | Non-Hispanic White | 239 vs. 197 | [38] | |||
| rs17435959 | Exonic | G>C | MMP10 | Risk | genotype GC, OR 9.59; genotype CC, OR 4.30 | East Asian (Chinese) | 91 vs. 172 | [39] | |||
| rs370850 | Intronic | C>T | ADAMTS1 | Risk | allele T, OR 3.71 | East Asian (Chinese) | 48 vs. 48 | [40] | |||
| rs422803 | Intronic | C>A | ADAMTS1 | Risk | allele A, OR 3.71 | East Asian (Chinese) | 48 vs. 48 | [40] | |||
| rs402007 | 5’UTR | C>G | ADAMTS1 | Risk | allele G, OR 2.18 | East Asian (Chinese) | 48 vs. 48 | [40] | |||
| rs428785 | Exonic | C>G | ADAMTS1 | Risk | allele G, OR 2.18 | East Asian (Chinese) | 48 vs. 48 | [40] | |||
| rs434857 | Exonic | T>G | ADAMTS1 | Risk | allele G, OR 2.18 | East Asian (Chinese) | 48 vs. 48 | [40] | |||
| rs445784 | Exonic | G>T | ADAMTS1 | Risk | allele T, OR 2.18 | East Asian (Chinese) | 48 vs. 48 | [40] | |||
| rs149586801 | Intronic | C>T | ADAMTS13 | Protective | allele T, OR 0.18 | East Asian (Chinese) | 48 vs. 48 | [40] | |||
| rs2277698 | Exonic | C>T | TIMP2 | Protective | allele T, OR 0.37 | East Asian (Chinese) | 48 vs. 48 | [40] | |||
| Genome-Wide Association Study | |||||||||||
| SNP ID | Band Region | Annotation | Effect Allele | Other Allele | EAF | Effect Direction | OR (95% CI) | p Value | Mapped Gene(s) ^ | Reference | |
| rs9306894 | 2p24.1 | 3’UTR | G | A | 0.16 | Risk | 1.10 (1.08–1.12) | 5.61 × 10−24 | GDF7 | [41,42] | |
| rs1430191 | 2p16.1 | Intergenic | T | C | 0.48 | Risk | 1.09 (1.06–1.12) | 1.00 × 10−9 | EFEMP1 | [41] | |
| rs11899888 | 2p16.1 | Intron | G | A | 0.11 | Risk | 1.11 (1.09–1.14) | 4.01 × 10−16 | EFEMP1 | [42] | |
| rs3791675 | 2p16.1 | Intron | T | C | 0.25 | Protective | 0.92 (0.90–0.94) | 1.23 × 10−13 | EFEMP1 | [41,42] | |
| rs77648136 | 2q31.1 | Intergenic | T | G | 0.16 | Protective | 0.94 (0.92–0.96) | 4.81 × 10−8 | HOXD13 | [42] | |
| rs42400 | 5p15.32 | Intergenic | G | C | 0.36 | Protective | 0.94 (0.92–0.96) | 1.65 × 10−10 | ADAMTS16 | [42] | |
| rs10810888 | 9p22.3 | Intron | G | A | 0.65 | Risk | 1.05 (1.03–1.07) | 4.00 × 10−8 | ADAMTSL1 | [42] | |
| rs7072877 | 10q26.13 | Intergenic | C | T | 0.80 | Risk | 1.06 (1.04–1.08) | 4.11 × 10−8 | FGFR2 | [43] | |
| rs35166569 | 11p13 | Intergenic | C | T | 0.09 | Protective | 0.89 (0.86–0.93) | 2.54 × 10−8 | WT1 | [42] | |
| rs11031796 | 11p13 | Intron | A | G | 0.31 | Protective | 0.93 (0.91–0.94) | 2.47 × 10−15 | WT1-AS | [42] | |
| rs10742277 | 11p13 | Intron | C | G | 0.33 | Risk | 1.48 (1.29–1.68) | 6.72 × 10−9 | WT1 | [43] | |
| rs4886778 | 15q24.1 | Intron | A | C | 0.47 | Risk | 1.05 (1.03–1.07) | 4.12 × 10−8 | LOXL1 | [42] | |
| rs235929 | 21q21.3 | Intron | C | G | 0.39 | Protective | 0.93 (0.92–0.95) | 2.01 × 10−12 | ADAMTS5, ADAMTS1 | [42] | |
| rs2236479 | 21q22.3 | Intron | A | G | 0.59 | Risk | 2.23 | 2.80 × 10−7 | COL18A1 | [44] | |
| Candidate Gene Association Studies | |||||||||||
| Component | SNP ID | Annotation | Change in Nucleotide | Candidate Gene | Effect Direction | Significant Association with POP * | Race/Ethnicity | Sample Size (POP/Control) | Reference | ||
| Estrogen receptor | rs17847075/rs2077647 | Exonic | T>C | ESR1 | Risk | genotype TC, OR 2.7 | East Asian (minority/non-minority Chinese) | 88 vs. 108 | [33] | ||
| rs2234693 | Intronic | T>C | ESR1 | Risk | genotype TC, OR 2.99 | East Asian (minority/non-minority Chinese) | 88 vs. 108 | [33] | |||
| rs2228480 | Exonic | G>A | ESR1 | Risk | genotype GA, OR 2.05 | East Asian (Chinese) | 88 vs. 153 | [75] | |||
| Risk | genotype AA, OR 39.70 genotype GA, OR 19.20 | Ashkenazi-Jewish origin | 33 vs. 33 | [65] | |||||||
| Progestogen receptor | rs484389 | Exonic | T>C | PGR | Risk | genotype TC, OR 4.77 | East Asian (Chinese) | 87 vs. 150 | [76] | ||
| Genome-Wide Association Studies | |||||||||||
| SNP ID | Band Region | Annotation | Effect Allele | Other Allele | EAF | Effect Direction | OR (95% CI) | p Value | Mapped Gene(s) ^ | Reference | |
| rs3820282 | 1p36.12 | Intron | T | C | 0.17 | Protective | 0.85 (0.82–0.88) | 3.30 × 10−21 | WNT4 | [41,42] | |
| rs72839768 | 17p13.1 | Exon | A | G | 0.02 | Risk | 1.19 (1.12–1.26) | 4.66 × 10−9 | DVL2 | [42] | |
| Candidate Gene Association Studies | |||||||||||
| Component | SNP ID | Annotation | Change in Nucleotide | Candidate Gene | Effect Direction | Significant Association with POP * | Race/Ethnicity | Sample Size (POP/Control) | Reference | ||
| OS | rs1695 | Exonic | A>G | GSTP1 | Protective | genotype AG+GG, OR 0.63 allele G, OR 0.60 | East Asian (Korean) | 189 vs. 156 | [89] | ||
| OS | rs1136410 | Exonic | T>C | PARP1 | Protective | genotype CC, OR 0.46 allele C, OR 0.72 | East Asian (Korean) | 185 vs. 155 | [90] | ||
| Genome-Wide Association Studies | |||||||||||
| SNP ID | Band Region | Annotation | Effect Allele | Other Allele | EAF | Effect Direction | OR (95% CI) | p Value | Mapped Gene(s) ^ | Reference | |
| OS-related | |||||||||||
| rs1036819 | 8q24.22 | Intron | C | A | 0.31 | Risk | 4.03 | 3.57 × 10−21 | ZFAT | [44] | |
| rs1810636 | 20p13 | Intron | C | A | 0.57 | Risk | 2.32 | 6.06 × 10−8 | IDH3B | [44] | |
| rs2267372 | 22q13.1 | Intron | G | A | 0.61 | Protective | 0.93 (0.91–0.95) | 1.07 × 10−13 | MAFF | [42] | |
| The variants overlapped with other metabolic and cardiovascular health | |||||||||||
| rs10762631 | 10q22.1 | Intron | A | G | 0.10 | Protective | 0.92 (0.90–0.95) | 3.76 × 10−8 | ADK | [42] | |
| rs12314243 | 12p13.2 | Intron | T | C | 0.54 | Protective | 0.91 (0.90–0.93) | 3.66 × 10−9 | DUSP16 | [42] | |
| rs73197353 | 12q24.21 | Intergenic | C | T | 0.08 | Risk | 1.12 (1.08–1.17) | 1.63 × 10−8 | TBX5 | [42] | |
| rs1247943 | 12q24.21 | Intergenic | A | G | 0.12 | Risk | 1.09 (1.06–1.12) | 1.68 × 10−21 | TBX5 | [41,42] | |
| rs4779517 | 15q13.2 | Intron | G | C | 0.49 | Risk | 1.07 (1.05–1.09) | 1.10 × 10−11 | KLF13 | [42] | |
| Others | |||||||||||
| rs58170120 | 3q21.3 | Intergenic | A | T | 0.18 | Risk | 1.08 (1.06–1.11) | 1.17 × 10−10 | SEC61A1 | [42] | |
| rs201194999 | 4q13.2 | Intergenic | T | C | 0.30 | Protective | 0.89 (0.86–0.93) | 2.42 × 10−8 | EPHA5 | [42] | |
| rs1455311 | 4q21.21 | Intron | G | A | 0.34 | Risk | 2.58 | 7.65 × 10−12 | PAQR3, BMP2K, ANTXR2 | [44] | |
| rs28403275 | 4q28.1 | Intergenic | C | G | 0.18 | Risk | 1.12 (1.10–1.15) | 1.58 × 10−22 | FAT4 | [42] | |
| rs7682992 | 4q28.1 | Intergenic | T | A | 0.21 | Risk | 1.13 (1.10–1.16) | 4.50 × 10−16 | FAT4 | [41] | |
| rs10013769 | 4q28.1 | Intergenic | G | A | 0.65 | Risk | 1.07 (1.05–1.09) | 1.26 × 10−10 | FAT4 | [42] | |
| rs251217 | 5q23.3 | Intron | G | A | 0.61 | Risk | 1.06 (1.05–1.08) | 4.22 × 10−11 | SLC12A2, FBN2 | [42] | |
| rs72624976 | 7q32.1 | 3’UTR | T | C | 0.01 | Protective | 0.84 (0.79–0.89) | 1.14 × 10−9 | IMPDH1 | [41,42] | |
| rs1493202 | 8q13.2 | Intergenic | G | T | 0.52 | Risk | 1.05 (1.03–1.07) | 3.56 × 10−8 | LACTB2 | [42] | |
| rs430794 | 9q22.2 | Intron | T | G | 0.13 | Protective | 0.35 | 6.74 × 10−5 | AUH, NFIL3 | [44] | |
| rs6484161 | 11p15.4 | Intron | T | G | 0.31 | Risk | 1.06 (1.04–1.08) | 5.89 × 10−9 | SBF2, ADM | [42] | |
| rs4944936 | 11q13.4 | Intergenic | C | T | 0.72 | Protective | 0.93 (0.91–0.95) | 7.13 × 10−12 | CHRDL2 | [42] | |
| rs8027714 | 15q11.2 | Intergenic | A | G | 0.26 | Risk | 9.04 | 5.65 × 10−43 | NPAP1 | [44] | |
| rs12915554 | 15q13.1 | 3’UTR | A | C | 0.32 | Protective | 0.95 (0.93–0.96) | 1.06 × 10−8 | GREM1 | [42] | |
| rs12325192 | 16q21.1 | Intergenic | T | C | 0.18 | Protective | 0.89 (0.87–0.91) | 1.14 × 10−21 | SALL1 | [41,42] | |
| rs1874008 | 16q24.1 | 3’UTR | C | T | 0.77 | Protective | 0.94 (0.92–0.96) | 5.77 × 10−9 | CRISPLD2 | [42] | |
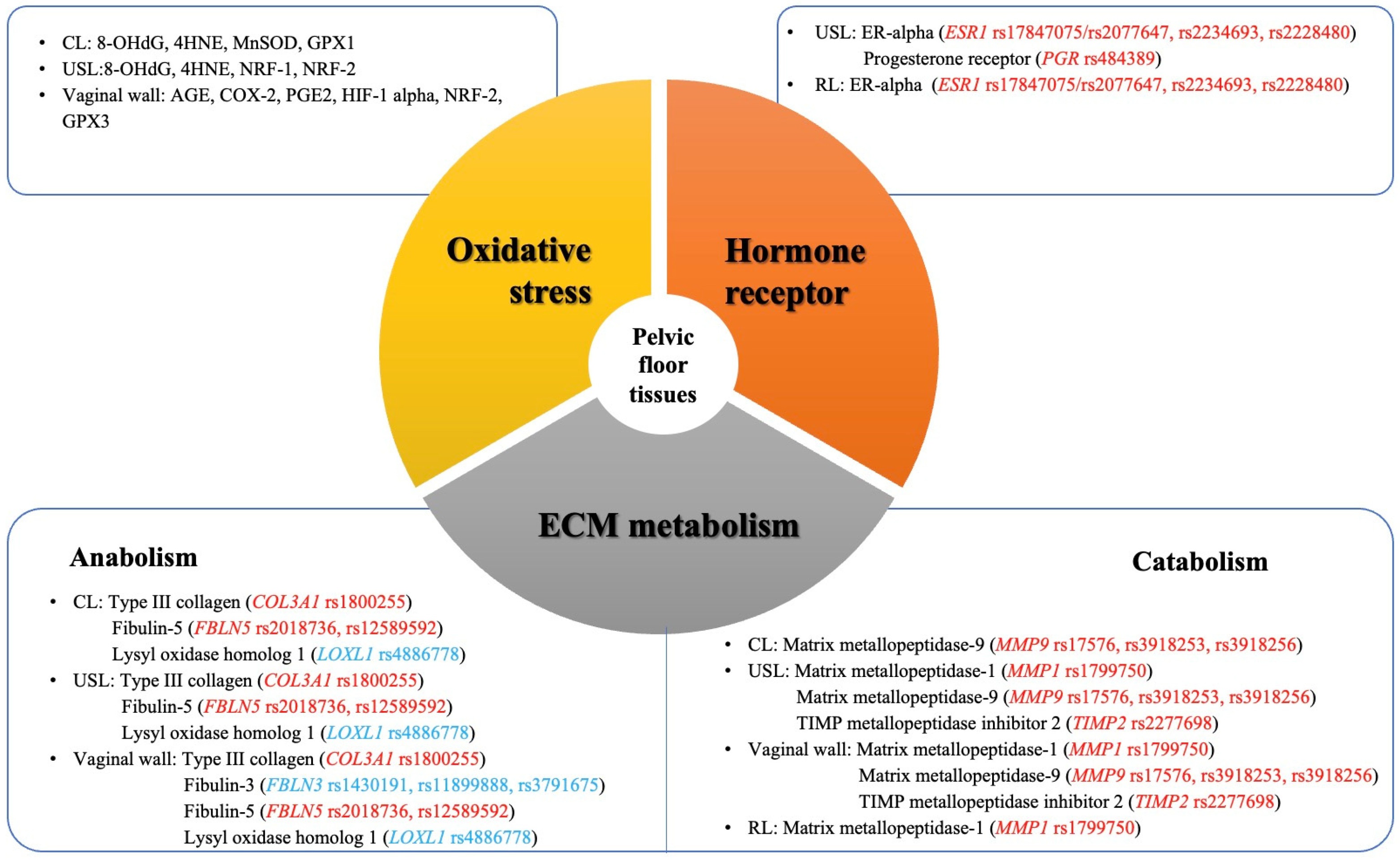
| Components | Tissues | Gene | RNA | Protein | Race/Ethnicity | Sample Size (POP/Control) | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| & Methods | # POP vs. Control | Name | & Methods | # POP vs. Control | ||||||
| ECM-related | ||||||||||
| Collagen | ||||||||||
| Cardinal ligament | - | - | - | Type III collagen | IHC | ↑ | Caucasian | 33 vs. 25 | [26] | |
| - | - | - | Type III collagen | IHC, WB | ↓ | East Asian (Chinese) | 30 vs. 30 | [113] | ||
| Uterosacral ligament | - | - | - | Type III collagen | IHC | ↑ | European (German) | 25 vs. 16 | [45] | |
| - | - | - | Type III collagen | IHC | ↑ | Turk | 22 vs. 23 | [114] | ||
| COL3A1 | qRT-PCR | ↑ | Type III collagen | IHC | ↑ | East Asian (Chinese) | 22 vs. 34 | |||
| COL3A1 | qRT-PCR | ND | - | - | - | Turk | 32 vs. 8 | [115] | ||
| COL3A1 | qRT-PCR | ND | Type III collagen | IHC | ND | East Asian (Chinese) | 35 vs. 20 | [116] | ||
| COL3A1 | qRT-PCR | ↓ | Type III collagen | IHC | ↓ | East Asian (Chinese) | 30 vs. 30 | [117] | ||
| Round ligament | COL3A1 | qRT-PCR | ND | - | - | - | Turk | 32 vs. 8 | [115] | |
| Para-urethral tissues | COL3A1 | qRT-PCR | ND | Type III collagen | IHC | ND | European (Sweden) | 15 vs. 14 | [118] | |
| Vaginal wall | - | - | - | Type III collagen | IF | ↑ | American | 62 vs. 15 | [119] | |
| - | - | - | Type III collagen | IHC | ↓ | East Asian (Chinese) | 23 vs. 15 | [120] | ||
| COL3A1 | qRT-PCR | ↑ | - | - | - | American | 47 vs. 7 | [121] | ||
| - | - | - | Type III collagen | WB | ↓ | American | 17 vs. 5 | [122] | ||
| - | - | - | Type III collagen | IHC | ND | Caucasian | 13 vs. 13 | [123] | ||
| - | - | - | Type III collagen | IHC, IF | ↑ | European (Italian) | 14 vs. 10 | [124] | ||
| COL3A1 | qRT-PCR | ↓ | Type III collagen | IHC | ↓ | East Asian (Chinese) | 60 vs. 35 | [125] | ||
| - | - | - | Type III collagen | IHC, WB | ↑ | European (Italian) | 20 vs. 10 | [126] | ||
| - | - | - | Type III collagen | IHC, WB | ↓ | East Asian (Chinese) | 35 vs. 35 | [127] | ||
| - | - | - | Type IV collagen | IHC | ND | East Asian (Chinese) | 23 vs. 15 | [120] | ||
| - | - | - | Type V collagen | IF | ND | American | 62 vs. 15 | [119] | ||
| - | - | - | Type V collagen | IHC | ND | East Asian (Chinese) | 23 vs. 15 | [120] | ||
| Elastic fibers | ||||||||||
| Cardinal ligament | - | - | - | Elastin | IHC | ↓ | Caucasian | 33 vs. 25 | [26] | |
| Uterosacral ligament | - | - | - | Elastin | IHC, IF | ↓ | European (German) | 59 vs. 30 | [128] | |
| - | - | - | Elastin | IHC | ND | East Asian (Chinese) | 30 vs. 30 | [58] | ||
| Vaginal wall | - | - | - | Elastin | IHC | ↓ | European (Belgian) | 15 vs. 0 | [129] | |
| - | - | - | Elastin | IHC | ND | East Asian (Chinese) | 23 vs. 15 | [120] | ||
| ELN | qRT-PCR | ND | - | - | - | American | 47 vs. 7 | [121] | ||
| - | - | - | Elastin | IHC | ND | Caucasian | 13 vs. 13 | [123] | ||
| Uterosacral ligament | EFEMP1 | qRT-PCR | ND | Fibulin-3 | IHC | ND | American | 8 vs. 8 | [130] | |
| Vaginal wall | EFEMP1 | qRT-PCR | ND | Fibulin-3 | IHC | ND | East Asian (Korean) | 12 vs. 12 | [131] | |
| Cardinal ligament | - | - | - | Fibulin-5 | IHC | ↓ | East Asian (Chinese) | 53 vs. 25 | [132] | |
| Uterosacral ligament | FBLN5 | qRT-PCR | ↑ | - | - | - | American | 31 vs. 29 | [133] | |
| FBLN5 | qRT-PCR | ↓ | Fibulin-5 | WB | ↓ | East Asian (Korean) | 30 vs. 30 | [134] | ||
| FBLN5 | qRT-PCR | ↓ | Fibulin-5 | IHC | ↓ | American | 8 vs. 8 | [130] | ||
| FBLN5 | - | - | Fibulin-5 | IHC | ↓ | East Asian (Chinese) | 30 vs. 30 | [58] | ||
| Para-urethral tissues | FBLN5 | qRT-PCR | ↓ | Fibulin-5 | IHC | ND | European (Sweden) | 15 vs. 14 | [118] | |
| Vaginal wall | FBLN5 | qRT-PCR | ↓ | Fibulin-5 | IHC | ↓ | American | 12 vs. 10 | [135] | |
| FBLN5 | qRT-PCR | ND | - | - | - | Caucasian | 15 vs. 11 | [136] | ||
| Lysyl oxidase | ||||||||||
| Cardinal ligament | - | - | - | Lysyl oxidase homolog 1 | IHC | ↓ | East Asian (Chinese) | 53 vs. 25 | [132] | |
| Uterosacral ligament | LOXL1 | qRT-PCR | ↑ | Lysyl oxidase homolog 1 | WB | ↑ | East Asian (Korean) | 30 vs. 30 | [134] | |
| - | - | - | Lysyl oxidase homolog 1 | IHC | ↓ | East Asian (Chinese) | 30 vs. 30 | [58] | ||
| Vaginal wall | LOXL1 | qRT-PCR | ↓ | Lysyl oxidase homolog 1 | IHC, WB | ND | Caucasian | 15 vs. 11 | [136] | |
| Vaginal wall | LOXL4 | qRT-PCR | ND | - | - | - | Caucasian | 15 vs. 11 | [136] | |
| Glycoprotein | ||||||||||
| Vaginal wall | - | - | - | Laminin | IHC | ND | East Asian (Chinese) | 23 vs. 15 | [120] | |
| Extracellular proteases | ||||||||||
| Uterosacral ligament | - | - | - | Interstitial collagenase | IHC | ↑ | European (Croatian) | 40 vs. 40 | [137] | |
| - | - | - | Interstitial collagenase | IHC | ↑ | European (Croatian) | 46 vs. 49 | [138] | ||
| - | - | - | Interstitial collagenase | IHC | ↑ | Israeli | 20 vs. 20 | [139] | ||
| - | - | - | Interstitial collagenase | IHC | ↑ | Turk | 42 vs. 49 | [140] | ||
| MMP1 | qRT-PCR | ND | Interstitial collagenase | IHC | ND | East Asian (Chinese) | 35 vs. 20 | [116] | ||
| Round ligament | - | - | - | Interstitial collagenase | IHC | ↑ | Turk | 42 vs. 49 | [140] | |
| Vaginal wall | - | - | - | Interstitial collagenase | IHC | ↑ | Israeli | 20 vs. 20 | [139] | |
| MMP1 | qRT-PCR | ND | Interstitial collagenase | IHC, IB | ND | Caucasian | 17 vs. 19 | [141] | ||
| MMP1 | qRT-PCR | ↑ | Interstitial collagenase | IHC | ↑ | East Asian (Chinese) | 72 vs. 72 | [142] | ||
| MMP1 | qRT-PCR | ↑ | Interstitial collagenase | IHC | ↑ | East Asian (Chinese) | 60 vs. 35 | [125] | ||
| Cardinal ligament | - | - | - | Matrix metalloproteinase-9 | IHC, WB | ↑ | East Asian (Chinese) | 30 vs. 30 | [113] | |
| Uterosacral ligament | - | - | - | Matrix metalloproteinase-9 | IHC | ↑ | Israeli | 20 vs. 20 | [139] | |
| MMP9 | qRT-PCR | ↑ | Matrix metalloproteinase-9 | ELISA | ↑ | East Asian (Korean) | 35 vs. 39 | [143] | ||
| - | - | - | Matrix metalloproteinase-9 | IHC | ND | American | 21 vs. 19 | [144] | ||
| MMP9 | qRT-PCR | ↑ | Matrix metalloproteinase-9 | IHC | ↑ | East Asian (Chinese) | 35 vs. 20 | [116] | ||
| Vaginal wall | - | - | - | Matrix metalloproteinase-9 | IF | ↑ | American | 62 vs. 15 | [119] | |
| - | - | - | Matrix metalloproteinase-9 | IHC | ↑ | Israeli | 20 vs. 20 | [139] | ||
| MMP9 | qRT-PCR | ND | Matrix metalloproteinase-9 | IHC, IB | ND | Caucasian | 17 vs. 19 | [141] | ||
| Vaginal wall | MMP10 | qRT-PCR | ND | - | - | - | American | 47 vs. 7 | [121] | |
| Uterosacral ligament | TIMP2 | qRT-PCR | ↓ | Metalloproteinase inhibitor 2 | IHC | ↓ | East Asian (Chinese) | 19 vs. 9 | [145] | |
| TIMP2 | qRT-PCR | ↓ | Metalloproteinase inhibitor 2 | IHC | ND | East Asian (Chinese) | 35 vs. 20 | [116] | ||
| Vaginal wall | TIMP2 | qRT-PCR | ↓ | Metalloproteinase inhibitor 2 | IHC, IB | ND | Caucasian | 17 vs. 19 | [141] | |
| Cervix tissue | TIMP2 | qRT-PCR | ND | Metalloproteinase inhibitor 2 | IHC | ND | East Asian (Chinese) | 19 vs. 9 | [145] | |
| Hormone metabolism-related | ||||||||||
| Estrogen receptor | ||||||||||
| Uterosacral ligament | - | - | - | ER-α | WB | ↓ | East Asian (Korean) | 20 vs. 24 | [146] | |
| ESR1 | qRT-PCR | ↑ | ER-α | IHC | ↑ | Caucasian | 13 vs. 13 | [147] | ||
| ESR1 | qRT-PCR | ↓ | - | - | - | Turk | 32 vs. 8 | [115] | ||
| ESR1 | qRT-PCR | ↓ | ER-α | IHC | ↓ | East Asian (Chinese) | 35 vs. 20 | [116] | ||
| Round ligament | ESR1 | qRT-PCR | ↓ | - | - | - | Turk | 32 vs. 8 | [115] | |
| Progesterone receptor | ||||||||||
| Uterosacral ligament | - | - | - | Progesterone receptor | WB | ↓ | East Asian (Korean) | 20 vs. 24 | [146] | |
| PGR | qRT-PCR | ND | Progesterone receptor | IHC | ND | Caucasian | 13 vs. 13 | [147] | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, W.; Cheung, R.Y.K.; Chung, C.Y.; Chan, S.S.C.; Choy, K.W. Genetic Etiology in Pelvic Organ Prolapse: Role of Connective Tissue Homeostasis, Hormone Metabolism, and Oxidative Stress. Genes 2025, 16, 5. https://doi.org/10.3390/genes16010005
Jiang W, Cheung RYK, Chung CY, Chan SSC, Choy KW. Genetic Etiology in Pelvic Organ Prolapse: Role of Connective Tissue Homeostasis, Hormone Metabolism, and Oxidative Stress. Genes. 2025; 16(1):5. https://doi.org/10.3390/genes16010005
Chicago/Turabian StyleJiang, Wenxuan, Rachel Yau Kar Cheung, Cheuk Yan Chung, Symphorosa Shing Chee Chan, and Kwong Wai Choy. 2025. "Genetic Etiology in Pelvic Organ Prolapse: Role of Connective Tissue Homeostasis, Hormone Metabolism, and Oxidative Stress" Genes 16, no. 1: 5. https://doi.org/10.3390/genes16010005
APA StyleJiang, W., Cheung, R. Y. K., Chung, C. Y., Chan, S. S. C., & Choy, K. W. (2025). Genetic Etiology in Pelvic Organ Prolapse: Role of Connective Tissue Homeostasis, Hormone Metabolism, and Oxidative Stress. Genes, 16(1), 5. https://doi.org/10.3390/genes16010005






