Research Progress on Gene Regulation of Plant Floral Organogenesis
Abstract
:1. Introduction
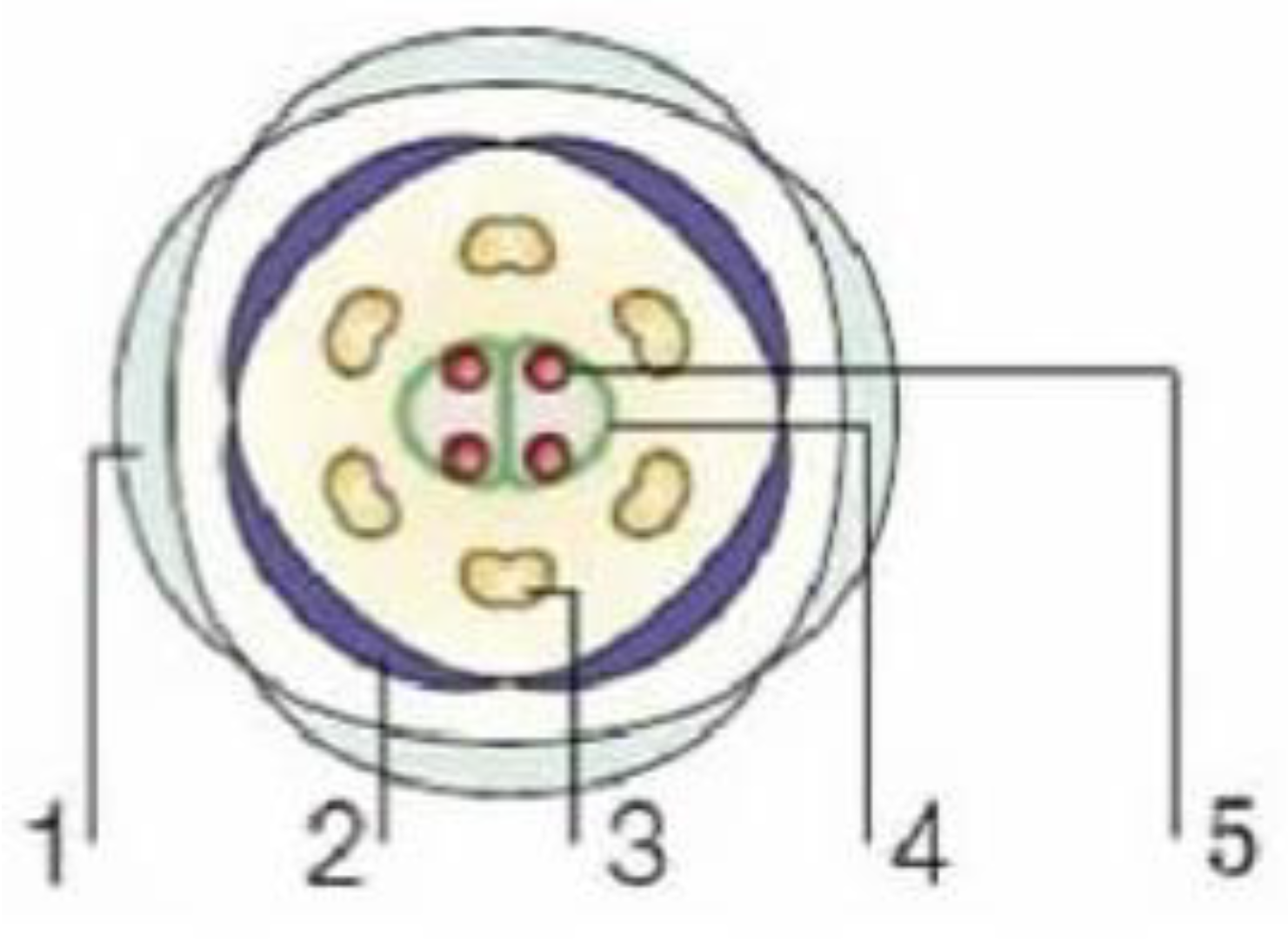
2. Models of Plant Floral Organogenesis
3. The Roles of the AE Floral Organ Model in Flower Development
3.1. The Functional Diversity of AP1 and AP2 Genes in Floral Organ Genesis
3.2. Molecular Regulation of Petals and Stamens by AP3 and PI in Flower Morphogenesis
3.3. Involvement of SEP Genes in Floral Organ Formation and Flowering Regulation
3.4. Contribution of Other Genes in the Modulation of Floral Organogenesis
4. Molecular Regulation of Floral Organogenesis and Its Implications for Ornamental and Economic Crops
4.1. Implications for Ornamental and Economic Crops
4.2. Molecular Regulation of Floral Organogenesis
5. Limitations of Gene Research Methods
5.1. Limitations at the Technical Level
5.2. Limitations of Gene Editing Technologies
6. Limitations at the Sample and Data Levels
6.1. Problems with Sample Representativeness
6.2. Complexity of Data Interpretation
6.3. The Necessity of Conducting Translational Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, F.; Han, T.; Chen, Z. Circadian and photoperiodic regulation of the vegetative to reproductive transition in plants. Commun. Biol. 2024, 7, 579. [Google Scholar] [CrossRef] [PubMed]
- Xu, K. Evolution of flowering time due to variation in the onset of pollen dispersal among individuals. Evolution 2024, 78, 401–412. [Google Scholar] [CrossRef]
- Schiffmann, Y. The generation of the flower by self-organisation. Prog. Biophys. Mol. Biol. 2023, 177, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Schnurbusch, T. The Birth and Death of Floral Organs in Cereal Crops. Annu. Rev. Plant Biol. 2024, 75, 427–458. [Google Scholar] [CrossRef] [PubMed]
- James, A.R.M.; Mayfield, M.M.; Dwyer, J.M. Patterns of frequency and density dependence are highly variable in diverse annual flowering plant communities. Ecology 2023, 104, e4021. [Google Scholar] [CrossRef]
- Judkevich, M.D.; Luaces, P.A.; Gonzalez, A.M. Flower structure, anatomy, and sexuality of Chrysophyllum gonocarpum (Sapotaceae). Protoplasma 2023, 260, 1271–1285. [Google Scholar] [CrossRef]
- Li, R.; Nie, L.; Wang, Q.; Ouyang, D.; Zhang, L.; Yuan, Y.; Hong, Y.; Wang, J.; Hu, X. Phytochemical constituents, chemotaxonomic significance and anti-arthritic effect of Eucommia ulmoides Oliver staminate flowers. Nat. Prod. Res. 2022, 36, 3455–3459. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.X.; Zhang, Y.F.; Xu, K.; Fan, Y.S.; Li, Z.; Li, Y.; Kakishima, M. First Report of the Rust Fungus Melampsora ferrinii on Salix babylonica in China and a New Spermogonial and Aecial Host, Corydalis bungeana. Plant Dis. 2024, 5, 1203–1221. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.; Guo, Z.; Xiao, X.; Peng, C. Effects of tree-herb co-planting on the bacterial community composition and the relationship between specific microorganisms and enzymatic activities in metal (loid)-contaminated soil. Chemosphere 2019, 220, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Xiong, F.; Ren, Q.P.; Wang, X.L. Regulation of flowering transition by alternative splicing: The role of the U2 auxiliary factor. J. Exp. Bot. 2020, 71, 751–758. [Google Scholar] [CrossRef]
- Favero, B.T.; Tan, Y.; Lin, Y.; Hansen, H.B.; Shadmani, N.; Xu, J.; He, J.; Müller, R.; Almeida, A.; Lütken, H. Transgenic Kalanchoe blossfeldiana, Containing Individual rol Genes and Open Reading Frames Under 35S Promoter, Exhibit Compact Habit, Reduced Plant Growth, and Altered Ethylene Tolerance in Flowers. Front. Plant Sci. 2021, 12, 672023. [Google Scholar] [CrossRef] [PubMed]
- Meyerowitz, E.M. Plants and the logic of development. Genetics 1997, 145, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Moyroud, E. Reflections on the ABC model of flower development. Plant Cell 2024, 36, 1334–1357. [Google Scholar] [CrossRef]
- Colombo, L.; Franken, J.; Koetje, E.; van Went, J.; Dons, H.J.; Angenent, G.C.; van Tunen, A.J. The petunia MADS-box gene FBP11 determines ovule identity. Plant Cell 1995, 7, 1859–1868. [Google Scholar]
- Theissen, G.; Becker, A.; Rosa, A.D.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.U.; Saedler, H. A short history of MADSbox genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ye, X.; Sun, Y.; Zhang, M.; Feng, S.; Zhou, A.; Wu, W.; Ma, S.; Liu, S. The underlying molecular conservation and diversification of dioecious flower and leaf buds provide insights into the development, dormancy breaking, flowering, and sex association of willows. Plant Physiol. Biochem. 2021, 167, 651–664. [Google Scholar]
- Patil, R.V.; Pawar, K.D. Comparative de novo flower transcriptome analysis of polygamodioecious tree Garcinia indica. 3 Biotech 2019, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Tang, A.; Wan, H.; Zhang, T.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. An APETALA2 homolog, RcAP2, regulates the number of rose petals derived from stamens and response to temperature fluctuations. Front. Plant Sci. 2018, 9, 481. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Tang, A.; Yu, J.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. RcAP1, a Homolog of APETALA1, is Associated with Flower Bud Differentiation and Floral Organ Morphogenesis in Rosa chinensis. Int. J. Mol. Sci. 2019, 20, 3557. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.L.; Li, X.L.; Zhang, Y.; Wu, S.; Song, Z.X.; Yin, H.F.; Liu, W.X.; Fan, Z.Q.; Li, J.Y. Floral organ transcriptome in Camellia sasanqua provided insight into stamen petaloid. BMC Plant Biol. 2022, 22, 474. [Google Scholar] [CrossRef] [PubMed]
- Ferrario, S.; Shchennikova, A.V.; Franken, J.; Richard, G.; Immink, H.; Angenent, G.C. Control of floral meristem determinacy in petunia by MADS-box transcription factors. Plant Physiol. 2006, 140, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.F.; Huang, C.H.; Chou, L.H.; Yang, C.H. Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Huang, F.; Liu, H.; Yang, S.; Yu, D. An APETALA1-like gene of soybean regulates flowering time and specifies floral organs. J. Plant Physiol. 2011, 168, 2251–2259. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Ge, H.S.; Ahmad, N.; Li, J.; Wang, Y.J.; Liu, X.Y.; Liu, W.C.; Li, X.W.; Wang, N.; Wang, F.; et al. Genome-Wide Identification of MADS-Box Family Genes in Safflower (Carthamus tinctorius L.) and Functional Analysis of CtMADS24 during Flowering. Int. J. Mol. Sci. 2023, 24, 1026. [Google Scholar] [CrossRef]
- Chen, M.K.; Lin, I.C.; Yang, C.H. Functional analysis of three lily (Lilium longiflorum) APETALA1-like MADS box genes in regulating floral transition and formation. Plant Cell Physiol. 2008, 49, 704–717. [Google Scholar] [CrossRef]
- Huang, H.J.; Chen, S.; Li, H.Y.; Jiang, J. Next-generation transcriptome analysis in transgenic birch overexpressing and suppressing APETALA1 sheds lights in reproduction development and diterpenoid biosynthesis. Plant Cell Rep. 2015, 34, 1663–1680. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, T.; Qiu, L.; Guo, X.; Li, P.; Yong, X.; Li, L.; Ahmad, S.; Wang, J.; Cheng, T.; et al. A 49-bp deletion of PmAG results in a double flower phenotype in Prunus mume. Hortic. Res. 2023, 11, uhad278. [Google Scholar] [CrossRef] [PubMed]
- Yoshihiro, N.; Yohei, H.; Yuichi, Y.; Tamotsu, H. Environmental responses of the FT/TFL1 gene family and their involvement in flower induction in Fragaria × ananassa. J. Plant Physiol. 2015, 177, 60–66. [Google Scholar]
- Gao, M.; Jiang, W.; Lin, Z.; Lin, Q.; Ye, Q.; Wang, W.; Xie, Q.; He, X.; Luo, C.; Chen, Q. SMRT and illumina RNA-Seq identifies potential candidate genes related to the double flower phenotype and unveils SsAP2 as a key regulator of the doubleflower trait in Sagittaria sagittifolia. Int. J. Mol. Sci. 2022, 23, 2240. [Google Scholar] [CrossRef] [PubMed]
- Maes, T.; Van de Steene, N.; Zethof, J.; Karimi, M.; D′Hauw, M.; Mares, G.; Van Montagu, M.; Gerats, T. Petunia Ap2-like genes and their role in flower and seed development. Plant Cell 2001, 13, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Irish, V.F. Evolution of petal identity. J. Exp. Bot. 2009, 60, 2517–2527. [Google Scholar] [CrossRef] [PubMed]
- Jing, D.; Xia, Y.; Chen, F.; Wang, Z.; Zhang, S.; Wang, J. Ectopic expression of a Catalpa bungei (Bignoniaceae) PISTILLATA homologue rescues the petal and stamen identities in Arabidopsis pi-1 mutant. Plant Sci. 2015, 231, 40–51. [Google Scholar] [CrossRef]
- Liu, W.; Shen, X.; Liang, H.; Wang, Y.; He, Z.; Zhang, D.; Chen, F. Isolation and functional analysis of PISTILLATA homolog from Magnolia wufengensis. Front. Plant Sci. 2018, 9, 1743. [Google Scholar] [CrossRef]
- Fei, Y.; Liu, Z.X. Isolation and characterization of the PISTILLATA ortholog gene from Cymbidium faberi Rolfe. Agronomy 2019, 9, 425. [Google Scholar] [CrossRef]
- Chen, M.K.; Hsieh, W.P.; Yang, C.H. Functional analysis reveals the possible role of the C-terminal sequences and PI motif in the function of lily (Lilium longiflorum) PISTILLATA (PI) orthologues. J. Exp. Bot. 2012, 63, 941–961. [Google Scholar] [CrossRef]
- Tsai, W.C.; Lee, P.F.; Chen, H.I.; Hsiao, Y.Y.; Wei, W.J.; Pan, Z.J.; Chuang, M.H.; Kuoh, C.S.; Chen, W.H.; Chen, H.H. PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol. 2005, 46, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.L.; Xu, J.; Tomes, S.; Cui, W.; Luo, Z.; Deng, C.; Ireland, H.S.; Schaffer, R.J.; Gleave, A.P. Ectopic expression of the PISTILLATA homologous MdPI inhibits fruit tissue growth and changes fruit shape in apple. Plant Direct 2018, 2, 15–19. [Google Scholar] [CrossRef]
- Fang, Z.W.; Qi, R.; Li, X.F.; Liu, Z.X. Ectopic expression of FaesAP3, a Fagopyrum esculentum (Polygonaceae) AP3 orthologous gene rescues stamen development in an Arabidopsis ap3 mutant. Gene 2014, 55, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Roque, E.; Serwatowska, J.; Rochina, M.C.; Wen, J.; Mysore, K.S.; Yenush, L.; Beltrán, J.P.; Cañas, L.A. Functional specialization of duplicated AP3-like genes in Medicago truncatula. Plant J. 2013, 73, 663–675. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Zhang, W.; Yu, F.; Tian, J.; Li, D.; Guo, A. Functional analysis of the two Brassica AP3 genes involved in apetalous and stamen carpelloid phenotypes. PLoS ONE 2011, 6, e20930. [Google Scholar] [CrossRef]
- Jing, D.; Chen, W.; Shi, M.; Wang, D.; Xia, Y.; He, Q.; Dang, J.; Guo, Q.; Liang, G. Ectopic expression of an Eriobotrya japonica APETALA3 ortholog rescues the petal and stamen identities in Arabidopsis ap3-3 mutant. Biochem. Biophys. Res. Commun. 2020, 523, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Pelayo, M.A.; Yamaguchi, N.; Ito, T. One factor, many systems: The floral homeotic protein AGAMOUS and its epigenetic regulatory mechanisms. Curr. Opin. Plant Biol. 2021, 61, 102009. [Google Scholar] [CrossRef] [PubMed]
- Sage-Ono, K.; Ozeki, Y.; Hiyama, S. Induction of double flowers in Pharbitis nil using a class-C MADS-box transcription factor with Chimeric REpressor gene-Silencing Technology. Plant Biotechnol. 2011, 28, 153–165. [Google Scholar] [CrossRef]
- Ma, N.; Chen, W.; Fan, T.; Tian, Y.; Zhang, S.; Zeng, D.; Li, Y. Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biol. 2015, 15, 237. [Google Scholar] [CrossRef]
- Ma, X.; Fan, L.; Ye, S.; Chen, Y.; Huang, Y.; Wu, L.; Zhao, L.; Yi, B.; Ma, C.; Tu, J.; et al. Identification of candidate genes associated with double flowers via integrating BSA-seq and RNA-seq in Brassica napus. BMC Genom. 2024, 25, 799. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Oshima, Y.; Yamamura, T.; Sugiyama, M.; Mitsuda, N.; Ohtsubo, N.; Ohme-Takagi, M.; Terakawa, T. Multi-petal cyclamen flowers produced by AGAMOUS chimeric repressor expression. Sci. Rep. 2013, 3, 2641. [Google Scholar] [CrossRef] [PubMed]
- Rudaya, E.S.; Kozyulina, P.Y.; Pavlova, O.A.; Dolgikh, A.V.; Ivanova, A.N.; Dolgikh, E.A. Regulation of the Later Stages of Nodulation Stimulated by IPD3/CYCLOPS Transcription Factor and Cytokinin in Pea Pisum sativum L. Plants 2021, 11, 56. [Google Scholar] [CrossRef]
- Sasaki, K.; Yoshioka, S.; Aida, R.; Ohtsubo, N. Production of petaloid phenotype in the reproductive organs of compound flowerheads by the co-suppression of class-C genes in hexaploid Chrysanthemum morifolium. Planta 2021, 253, 100. [Google Scholar] [CrossRef]
- Rodríguez-Cazorla, E.; Ortuño-Miquel, S.; Candela, H.; BaileySteinitz, L.J.; Yanofsky, M.F.; Martínez-Laborda, A.; Ripoll, J.J.; Vera, A. Ovule identity mediated by pre-mRNA processing in Arabidopsis. PLoS Genet. 2018, 14, e1007182. [Google Scholar] [CrossRef]
- Tani, E.; Polidoros, A.N.; Flemetakis, E.; Stedel, C.; Kalloniati, C.; Demetriou, K.; Katinakis, P.; Tsaftaris, A.S. Characterization and expression analysis of AGAMOUS-like, SEEDSTICK-like, and SEPALLATA-like MADS-box genes in peach (Prunus persica) fruit. Plant Physiol. Biochem. 2009, 47, 690–700. [Google Scholar] [CrossRef] [PubMed]
- Acri-Nunes-Miranda, R.; Mondragón-Palomino, M. Expression of paralogous SEP-, FUL-, AG- and STK-like MADS-box genes in wild-type and peloric Phalaenopsis flowers. Front. Plant Sci. 2014, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Dirks-Mulder, A.; Butôt, R.; Schaik, P.V.; Wijnands, J.W.; Berg, P.D.; Krol, L.; Doebar, S.; Kooperen, K.V.; Boer, H.D.; Kramer, E.M.; et al. Exploring the evolutionary origin of floral organs of Erycina pusilla, an emerging orchid model system. BMC Evol. Biol. 2017, 17, 89. [Google Scholar] [CrossRef]
- Yang, F.; Zhu, G.; Wei, Y.; Gao, J.; Liang, G.; Peng, L.; Lu, C.; Jin, J. Low-temperature-induced changes in the transcriptome reveal a major role of CgSVP genes in regulating flowering of Cymbidium goeringii. BMC Genom. 2019, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.T.; Wang, Q.; Park, W.; Feng, Y.; Kumar, D.; Meeley, R.; Dooner, H.K. Competitive Ability of Maize Pollen Grains Requires Paralogous Serine Threonine Protein Kinases STK1 and STK2. Genetics 2017, 207, 1361–1370. [Google Scholar] [CrossRef]
- Monniaux, M.; Vandenbussche, M. Flower Development in the Solanaceae. Methods Mol. Biol. 2023, 2686, 39–58. [Google Scholar] [PubMed]
- Zhao, X.Y.; Cheng, Z.J.; Zhang, X.S. Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 2006, 223, 698–707. [Google Scholar] [CrossRef]
- Kaufmann, K.; Muiño, J.M.; Jauregui, R.; Airoldi, C.A.; Smaczniak, C.; Krajewski, P.; Angenent, G.C.; Weigel, D. Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLOS Biol. 2009, 7, e1000090. [Google Scholar] [CrossRef]
- Wang, J.Y.; Jiu, S.T.; Xu, Y.; Ali Sabir, I.; Wang, L.; Ma, C.; Xu, W.P.; Wang, S.P.; Zhang, C.X. SVP-like gene PavSVP potentially suppressing flowering with PavSEP, PavAP1, and PavJONITLESS in sweet cherries (Prunus avium L.). Plant Physiol. Biochem. 2021, 159, 277–284. [Google Scholar] [CrossRef]
- Cheng, Z.H.; Zhuo, S.B.; Liu, X.F.; Che, G.; Wang, Z.Y.; Gu, R.; Shen, J.J.; Song, W.Y.; Zhou, Z.Y.; Han, D.G.; et al. The MADS-box gene CsSHP participates in fruit maturation and floral organ development in cucumber. Front. Plant Sci. 2019, 10, 1781. [Google Scholar] [CrossRef]
- Pu, Z.Q.; Ma, Y.Y.; Lu, M.X.; Ma, Y.Q.; Xu, Z.Q. Cloning of a SEPALLATA4-like gene (IiSEP4) in Isatis indigotica Fortune and characterization of its function in Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 154, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Ampomah-Dwamena, C.; Morris, B.A.; Sutherland, P.; Veit, B.; Yao, J.L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002, 130, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.W.; Yang, L.; Wu, D.; Meng, Q.C.; Deng, X.; Huang, G.Q.; Zhang, J.; Chen, X.F.; Ferrándiz, C.; Liang, W.Q.; et al. Rice SEPALLATA genes OsMADS5 and OsMADS34 cooperate to limit inflorescence branching by repressing the TERMINAL FLOWER1-like gene RCN4. New Phytol. 2022, 233, 1682–1700. [Google Scholar] [CrossRef]
- Zhou, Y.Z.; Xu, Z.D.; Yong, X.; Ahmad, S.; Yang, W.R.; Cheng, T.R.; Wang, J.; Zhang, Q.X. SEP-class genes in Prunus mume and their likely role in floral organ development. BMC Plant Biol. 2017, 17, 10. [Google Scholar] [CrossRef]
- Yu, X.; Duan, X.; Zhang, R.; Fu, X.; Ye, L.; Kong, H.; Xu, G.; Shan, H. Prevalent exon-intron structural changes in the APETALA1/FRUITFULL, SEPALLATA, AGAMOUS-LIKE6, and FLOWERING LOCUS C MADS-box gene subfamilies provide new insights into their evolution. Front. Plant Sci. 2016, 7, 598. [Google Scholar] [CrossRef]
- Ma, J.; Deng, S.; Chen, L.; Jia, Z.; Sang, Z.; Zhu, Z.; Ma, L.; Chen, F. Gene duplication led to divergence of expression patterns, protein-protein interaction patterns and floral development functions of AGL6-like genes in the basal angiosperm Magnolia wufengensis (Magnoliaceae). Tree Physiol. 2019, 39, 861–876. [Google Scholar] [CrossRef]
- Yu, X.; Chen, G.; Guo, X.; Lu, Y.; Zhang, J.; Hu, J.; Tian, S.; Hu, Z. Silencing SlAGL6, a tomato AGAMOUS-LIKE6 lineage gene, generates fused sepal and green petal. Plant Cell Rep. 2017, 36, 959–969. [Google Scholar] [CrossRef]
- Hsu, H.F.; Chen, W.H.; Shen, Y.H.; Hsu, W.H.; Mao, W.T.; Yang, C.H. Multifunctional evolution of B and AGL6 MADS box genes in orchids. Nat. Commun. 2021, 12, 902. [Google Scholar] [CrossRef] [PubMed]
- Rijpkema, A.S.; Zethof, J.; Gerats, T.; Vandenbussche, M. The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 2009, 60, 1–9. [Google Scholar] [CrossRef]
- Li, B.J.; Zheng, B.Q.; Wang, J.Y.; Tsai, W.C.; Lu, H.C.; Zou, L.H.; Wan, X.; Zhang, D.Y.; Qiao, H.J.; Liu, Z.J.; et al. New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun. Biol. 2020, 3, 89. [Google Scholar] [CrossRef]
- Vandenbussche, M.; Zethof, J.; Souer, E.; Koes, R.; Tornielli, G.B.; Pezzotti, M.; Ferrario, S.; Angenent, G.C.; Gerats, T. Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 2003, 15, 2680–2693. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Lee, J.H.; Kim, W.; Jung, H.S.; Huijser, P.; Ahn, J.H. The micro RNA156-SQUAMOSA PROMOTER BINDING PROTEINLIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012, 159, 461–478. [Google Scholar] [CrossRef]
- Usami, T.; Horiguchi, G.; Yano, S.; Tsukaya, H. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 2009, 136, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, Z.; Yang, Y.; Chen, X.; Chen, G. Function annotation of an SBP-box gene in Arabidopsis based on Journal of Molecular Sciences. Int. J. Mol. Sci. 2009, 10, 116–132. [Google Scholar] [CrossRef]
- Zhang, X.; Dou, L.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Wang, C.; Yu, S. Genomic organization, differential expression, and functional analysis of the SPL gene family in Gossypium hirsutum. Mol. Genet. Genom. 2015, 290, 115–126. [Google Scholar] [CrossRef]
- Shikata, M.; Koyama, T.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis SBP-Box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009, 50, 2133–2145. [Google Scholar] [CrossRef]
- Murmu, J.; Bush, M.J.; DeLong, C.; Li, S.; Xu, M.; Khan, M.; Malcolmson, C.; Fobert, P.R.; Zachgo, S.; Hepworth, S.R. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for another development. Plant Physiol. 2010, 154, 1492–1504. [Google Scholar] [CrossRef]
- Thurow, C.; Schiermeyer, A.; Krawczyk, S.; Butterbrodt, T.; Nickolov, K.; Gatz, C. Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 2005, 44, 100–113. [Google Scholar] [CrossRef]
- Deveaux, Y.; Toffano-Nioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef]
- Minh-Thu, P.T.; Kim, J.S.; Chae, S.; Jun, K.M.; Lee, G.S.; Kim, D.E.; Cheng, J.J.; Song, S.I.; Nahm, B.H.; Kim, Y.K. A WUSCHEL homeobox transcription factor, OsWOX13, enhances drought tolerance and triggers early flowering in rice. Mol. Cells 2018, 41, 781–798. [Google Scholar]
- Zhang, C.; Wang, J.; Wang, X.; Li, C.; Ye, Z.; Zhang, J. UF, a WOX gene, regulates a novel phenotype of un-fused flower in tomato. Plant Sci. 2020, 297, 110523. [Google Scholar] [CrossRef]
- Li, Z.; Liu, D.; Xia, Y.; Li, Z.; Jing, D.; Du, J.; Niu, N.; Ma, S.; Wang, J.; Song, Y.; et al. Identification of the WUSCHEL-related homeobox (WOX) gene family, and interaction and functional analysis of TaWOX9 and TaWUS in wheat. Int. J. Mol. Sci. 2020, 21, 1581. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ning, K.; Zhang, W.; Chen, H.; Lu, X.; Zhang, D.; El-Kassaby, Y.A.; Bian, J. Phenotypic variation of floral organs in flowering crabapples and its taxonomic significance. BMC Plant Biol. 2021, 21, 503. [Google Scholar] [CrossRef] [PubMed]
- Maio, K.A.; Moubayidin, L. 'Organ'ising Floral Organ Development. Plants 2024, 13, 1595. [Google Scholar] [CrossRef]
- Ahmad, S.; Lu, C.; Gao, J.; Wei, Y.; Xie, Q.; Jin, J.; Zhu, G.; Yang, F. Integrated proteomic, transcriptomic, and metabolomic profiling reveals that the gibberellin-abscisic acid hub runs flower development in the Chinese orchid Cymbidium sinense. Hortic. Res. 2024, 11, uhae073. [Google Scholar] [CrossRef]
- Zhou, G.; Yin, H.; Chen, F.; Wang, Y.; Gao, Q.; Yang, F.; He, C.; Zhang, L.; Wan, Y. The genome of Areca catechu provides insights into sex determination of monoecious plants. New Phytol. 2022, 236, 2327–2343. [Google Scholar] [CrossRef]
- Li, C.; Chen, L.; Fan, X.; Qi, W.; Ma, J.; Tian, T.; Zhou, T.; Ma, L.; Chen, F. MawuAP1 promotes flowering and fruit development in the basal angiosperm Magnolia wufengensis (Magnoliaceae). Tree Physiol. 2020, 40, 1247–1259. [Google Scholar] [CrossRef]
- Ramage, E.; Soza, V.L.; Yi, J.; Deal, H.; Chudgar, V.; Hall, B.D.; Di Stilio, V.S. Gene Duplication and Differential Expression of Flower Symmetry Genes in Rhododendron (Ericaceae). Plants 2021, 10, 1994. [Google Scholar] [CrossRef]
- Madrigal, Y.; Alzate, J.F.; Pabón-Mora, N. Evolution of major flowering pathway integrators in Orchidaceae. Plant Reprod. 2024, 37, 85–109. [Google Scholar] [CrossRef] [PubMed]
- Kurokura, T.; Mimida, N.; Battey, N.H.; Hytönen, T. The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 2013, 64, 4131–4141. [Google Scholar] [CrossRef]
- Wu, T.; Liu, Z.; Yu, T.; Zhou, R.; Yang, Q.; Cao, R.; Nie, F.; Ma, X.; Bai, Y.; Song, X. Flowering genes identification, network analysis, and database construction for 837 plants. Hortic. Res. 2024, 11, uhae013. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Mishina, K.; Wang, Q.; Zhu, H.; Tagiri, A.; Kikuchi, S.; Sassa, H.; Oono, Y.; Komatsuda, T. Organ-enriched gene expression during floral morphogenesis in wild barley. Plant J. 2023, 116, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.L.; Kang, C.; Gu, C.; Gleave, A.P. The Roles of Floral Organ Genes in Regulating Rosaceae Fruit Development. Front. Plant Sci. 2022, 12, 644424. [Google Scholar] [CrossRef] [PubMed]
- Freytes, S.N.; Canelo, M.; Cerdán, P.D. Regulation of Flowering Time: When and Where? Curr. Opin. Plant Biol. 2021, 63, 102049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Cheng, G.; Shu, X.; Wang, N.; Wang, Z. Transcriptome Analysis of Lycoris chinensis Bulbs Reveals Flowering in the Age-Mediated Pathway. Biomolecules 2022, 12, 899. [Google Scholar] [CrossRef] [PubMed]
- Winter, C.M.; Yamaguchi, N.; Wu, M.F.; Wagner, D. Transcriptional programs regulated by both LEAFY and APETALA1 at the time of flower formation. Physiol. Plant 2015, 155, 55–73. [Google Scholar] [CrossRef]
- Zhang, Y.; Kan, L.; Hu, S.; Liu, Z.; Kang, C. Roles and evolution of four LEAFY homologs in floral patterning and leaf development in woodland strawberry. Plant Physiol. 2023, 192, 240–255. [Google Scholar] [CrossRef]
- Zhu, Y.; Klasfeld, S.; Jeong, C.W.; Jin, R.; Goto, K.; Yamaguchi, N.; Wagner, D. TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 2020, 11, 5118. [Google Scholar] [CrossRef]
- Paull, R.E.; Ksouri, N.; Kantar, M.; Zerpa-Catanho, D.; Chen, N.J.; Uruu, G.; Yue, J.; Guo, S.; Zheng, Y.; Wai, C.M.J.; et al. Differential gene expression during floral transition in pineapple. Plant Direct 2023, 14, e541. [Google Scholar] [CrossRef]
- Lv, T.; Wang, L.; Zhang, C.; Liu, S.; Wang, J.; Lu, S.; Fang, C.; Kong, L.; Li, Y.; Li, Y.; et al. Identification of two quantitative genes controlling soybean flowering using bulked-segregant analysis and genetic mapping. Front. Plant Sci. 2022, 13, 987073. [Google Scholar] [CrossRef] [PubMed]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Cheng, X.; Fan, S.; Wen, C.; Du, X. CRISPR/Cas9 for cancer treatment: Technology, clinical applications and challenges. Brief Funct Genom. 2020, 19, 209–214. [Google Scholar] [CrossRef]
- Kosiyaporn, H.; Chanvatik, S.; Issaramalai, T.; Kaewkhankhaeng, W.; Kulthanmanusorn, A.; Saengruang, N.; Witthayapipopsakul, W.; Viriyathorn, S.; Kirivan, S.; Kunpeuk, W.; et al. Surveys of knowledge and awareness of antibiotic use and antimicrobial resistance in general population: A systematic review. PLoS ONE 2020, 15, e0227973. [Google Scholar] [CrossRef]
- Kofler, J.; Milyaev, A.; Würtz, B.; Pfannstiel, J.; Flachowsky, H.; Wünsche, J.N. Proteomic differences in apple spur buds from high and non-cropping trees during floral initiation. J. Proteomics 2022, 253, 104459. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.B.; Kar, D.; Datta, S.; Vijay, N. Genomic and Transcriptomic Analyses Illuminates Unique Traits of Elusive Night Flowering Jasmine Parijat (Nyctanthes arbor-tristis). Physiol. Plant 2023, 175, e14119. [Google Scholar] [CrossRef] [PubMed]
- Olmedo, P.; Núñez-Lillo, G.; Vidal, J.; Leiva, C.; Rojas, B.; Sagredo, K.; Arriagada, C.; Defilippi, B.G.; Pérez-Donoso, A.G.; Meneses, C.; et al. Proteomic and metabolomic integration reveals the effects of pre-flowering cytokinin applications on central carbon metabolism in table grape berries. Food Chem. 2023, 411, 135498. [Google Scholar] [CrossRef]
- Khan, T.; Farley, C.M.; Wilson, J.J.; Chang, C.H.; Chaussabel, D. Tackling the Complexity of Spatial Transcriptomics Data Interpretation with Large Language Models. bioRxiv 2024, 28, 625773. [Google Scholar]
- Salse, J. Translational research from models to crops: Comparative genomics for plant breeding. Comptes Rendus Biol. 2023, 345, 111–128. [Google Scholar] [CrossRef]

| Main Features | Mutant Phenotype |
|---|---|
| Determine the formation of floral meristems and the initiation of sepal and petal floral organ primordia. | Strong mutants: sepals undeveloped or transformed into leaf-like structures. |
| Together with AP1, determine the initiation of the sepal and petal two-whorl floral organ primordia. | A four-whorl floral organ structure with carpels, stamens, pistils, and carpels from the outside in. |
| AP3 and PI jointly regulate the development of petals and stamens. | When the petals and stamens are, respectively, transformed into sepals and carpels, the strong AP3 mutant will present an enlarged central stamen and no petal structure. |
| Regulate the development of stamens and carpels. | The flower turns into a five-whorl structure of sepal, petal, petal, sepal, and sepal, and in some plants, the ovule mutates into a small flower with only petals. |
| Regulate ovule development. | The flower transforms into a structure with a sepal, petal, stamen, and pistil, and some ovules degenerate into leaf-like or carpel-like forms. |
| Flower organ development co-factors, participating in the development of five-whorl flower organs. | The sep1sep2sep3sep4 quadruple mutant causes all five-whorl structures of the flower to become leaf-like structures, while the sep1sep2sep3 triple mutant results in the transformation of the five-whorl structures into sepals. |
| Species | Function Description | Reference |
|---|---|---|
| Rosa chinensis | The RcAP1 gene enables the transition from inflorescence meristem to floral meristem, making it flower earlier, and regulates the formation of sepals during floral organ development. | [20] |
| Camellia japonica L. | CjAP2 is involved in the sepal petalization process, resulting in the double-petal phenomenon in camellia. | [21] |
| Petunia hybrida (Hook.) E. Vilm. | The transgenic petunia R0 generation plants exhibit the characteristics of early and continuous flowering. | [22] |
| Malus pumila Mill., Cymbidium ssp. | When the apple MdMADS2 and orchid OMADS1 genes are transferred into tobacco, both cause the phenomena of early flowering and changes in floral organs in tobacco. | [23] |
| Glycine max (L.) Merr. | The overexpression of the soybean GmAP1 gene in tobacco can promote early flowering in tobacco, cause the specialization of floral organs, and affect the formation of floral meristems. | [24] |
| Arabidopsis thaliana | The overexpression of CtMADS24 leads to the upregulation of some flower development characteristic genes, thereby shortening the flowering time, while the flowering time of the silenced lines is significantly delayed. | [25] |
| Lilium longiflorum Thumb. | The overexpression of the three genes, LMADS5, LMADS6, and LMADS7, causes the plants to flower earlier. Homeotic transformation produces carpelloid sepals and stamenoid petals. | [26] |
| Betula platyphylla Suk | The overexpression of the BpAP1 gene will cause early flowering in Betula platyphylla, and also affect the expression of many flowering-related genes and the synthesis of diterpenoid compounds. The Betula platyphylla offspring inheriting the BpAP1 gene still show early flowering and fruiting, and transgenic BpAP1 tobacco also shows the early flowering trait. | [27] |
| Prunus mume | After the PmAG overexpression vector was transferred into wild-type Arabidopsis thaliana, the petals and stamens of the transgenic plants degenerated, and the inflorescences and pods showed abortion phenomena. | [28] |
| Fragaria × ananassa Duch. | The transgenic lines show obvious early flowering, abnormal floral organs and are unable to form seeds. Meanwhile, vegetative growth is inhibited, resulting in dwarf plants and a reduced number of rosette leaves. | [29] |
| Sagittaria sagittifolia L. | The overexpression of SsAP2 delays the flowering time and increases the number of petals in Sagittaria sagittifolia. | [30] |
| Picea abies | The overexpression of PaAPETALA2-LIKE2 (AP2L2) leads to an increase in the number of pistils and stamens in Arabidopsis thaliana and postpones the flowering time. It can determine petal characteristics in the ap1 mutant of Arabidopsis thaliana. In petunia plants, the expression signal intensity gradually decreases with the maturation of organs in the outer layers of organs such as bracts, sepals, petals, and ovary walls, showing a spatiotemporal pattern. | [31] |
| Species | Function Description | Reference |
|---|---|---|
| Arabidopsis thaliana | When the Arabidopsis thaliana PI gene AtPI is transferred to tobacco, the floral organs of tobacco obviously show phenomena such as a smaller corolla, shorter stamens, and abnormal fruits and ovaries. | [32] |
| Catalpa bungei C. A. Mey | When the PI gene CabuPI was transferred into Arabidopsis thaliana, the Arabidopsis thaliana with the 35S:CabuPI gene produced normal petals and different numbers of stamens. | [33] |
| Magnolia wufengensis L. Y. Ma et L. R. Wang | The MAwuPI gene is only expressed in tepals and stamens and is involved in stamen development in Yulania wufengensis. The ectopic expression of this gene in the Arabidopsis pi-1 mutant can cause the third whorl floral organs to present a filamentous form. | [34] |
| Cymbidium faberi Rolfe | HoPI is widely expressed in all floral organs and can restore the stamen and petal development of the Arabidopsis pi-1 mutant, but it cannot restore the development of anthers on the stamens. | [35] |
| Lilium longiflorum Thumb. | The ectopic expression of LMADS8/9 can rescue the development of the second-round petals in the Arabidopsis pi-1 mutant and transform some sepals into petal-like structures. | [36] |
| Phalaenopsis aphrodite Rchb. f. | The overexpression of the PI-like gene PeMADS6 in Arabidopsis thaliana will lead to the transformation of sepals into petals in Arabidopsis thaliana. | [37] |
| Malus pumila Mill. | The overexpression of MdPI also shows the phenomenon of sepals transforming into petals. In wild-type apples, the length of anthers is similar to that of stigmas, while transgenic apples show that the length of anthers is half of the length of stigmas. | [38] |
| Fagopyrum esculentum Moench | When the buckwheat AP3 gene FaesAP3 is overexpressed in Arabidopsis thaliana, the outer whorl short stamens of the plant become petal-like, and the inner whorl long stamens become filament-like. | [39] |
| Medicago truncatula | Reducing the expression level of MtNMH7 (RNAi-MtNMH7) will lead to slight petal shape defects and stamen carpel-like phenomena in the plant. A decrease in the expression level of MtTomato MADS6 (TM6) will cause some stamens to differentiate into anthers and filaments, but no pollen grains will be produced. When MtTM6 completely loses its expression, all anthers are completely transformed into carpels. | [40] |
| Brassica L. Plants | The loss of function of the AP3 gene also shows a trend of stamen-to-carpel transformation. | [41] |
| Eriobotrya japonica (Thunb.) Lindl. | When the EjAP3 mutant was introduced into Arabidopsis thaliana, the transgenic plants showed abnormal traits such as narrower petals and greener stamens. | [42] |
| Species | Function Description | Reference |
|---|---|---|
| Rosa ssp. | Silencing the AG homologous gene RhAG in roses and low temperatures can both increase the number of petals. Restricting the expression of RhAG in double-petaled roses also results in double-petaled roses. | [45] |
| Rosa chinensis | In double-petaled flowers, the expression level of RhAG is lower than that in single-petaled flowers, and the expression domain of RhAG in double-petaled flowers shrinks, thereby resulting in a decrease in the number of stamens and an increase in the number of petals in the flower. | [46] |
| Cyclamen persicum Mill. | When the expression of the AG gene CpAG1 in cyclamen is inhibited, semi-double-petaled flowers with 10 petals appear. When the expressions of CpAG1/2 are inhibited simultaneously, double-petaled flowers with 40 petals appear. | [47] |
| Pisum sativum L. | After silencing the PsAGs genes in peas, the flowers of peas show phenotypes with features such as petalization of stamens and dehiscence of carpels, and an incomplete small flower is also endogenously generated. | [48] |
| Chrysanthemum morifolium Ramat. | By knocking out the CAG1s and CAG2s genes, it was found that chrysanthemums showed a multi-petal phenotype, and the reproductive organs of both tubular and ligulate flowers were transformed into tubular or ligulate petals. | [49] |
| Prunus mume | PmAG in Prunus mume is involved in the growth and development processes of multiple vegetative organs. When the PmAG gene of Prunus mume is overexpressed in Arabidopsis thaliana, the petals of the transgenic Arabidopsis thaliana plants become smaller, and the stamens and pistils are obviously enlarged. | [28] |
| Species | Function Description | Reference |
|---|---|---|
| Triticum aestivum L. | When the SEP-like gene TaMADS1 of wheat was transferred into Arabidopsis thaliana, Arabidopsis thaliana showed the phenomenon of early flowering and also changed the development of floral organs, such as sepals turning into leaves and a reduction in the number of petals and stigmas. | [57] |
| Arabidopsis thaliana | In the Arabidopsis sep1sep2sep3sep4 quadruple mutant, all four types of floral organs are mutated into leaf-like structures. The sep1sep2sep3 triple mutant is mutated into sepal-like structures. Except that the expression of AtSEP3 occurs in the later stage of flower development, AtSEP1/2/4 are all expressed in the early stage. The SEP3 mutation will lead to a significant reduction in the number of stamens in the flower, and the stamens are transformed into filamentous carpel structures or fused with carpels. | [58] |
| Prunus avium (L.) | The interaction between PavSEP and the Pav Short Vegetative Phase (SVP) can promote the floral transition. | [59] |
| Cucumis sativus L. | In cucumbers, it has also been found that SEP and SHP genes interact to regulate the formation of floral organs. | [60] |
| Isatis indigotica Fortune | In Isatis indigotica, IiSEP4 can interact with IiSVP, IiSHP2, and IiFruitfull (FUL) to regulate flowering time and the development of stigmas and fruits. | [61] |
| Solanum lycopersicum L. | The inhibition of the SEP1 homologous gene Tomato MADS29 (TM29) causes the partial transformation of tomato stamens and petals into sepals. | [62] |
| Oryza sativa L. | The SEP gene OsMADS5/34 can regulate the branching state of rice inflorescences. | [63] |
| Prunus mume | PmSEP2 and PmSEP3 are involved in the formation of stamens and pistils in Prunus mume, while PmSEP4 and PmSEP1/2 interact pairwise and are involved in the formation of sepals. | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Iqbal, A.; Yang, M.; Yang, Y. Research Progress on Gene Regulation of Plant Floral Organogenesis. Genes 2025, 16, 79. https://doi.org/10.3390/genes16010079
Zhou L, Iqbal A, Yang M, Yang Y. Research Progress on Gene Regulation of Plant Floral Organogenesis. Genes. 2025; 16(1):79. https://doi.org/10.3390/genes16010079
Chicago/Turabian StyleZhou, Lixia, Amjad Iqbal, Mengdi Yang, and Yaodong Yang. 2025. "Research Progress on Gene Regulation of Plant Floral Organogenesis" Genes 16, no. 1: 79. https://doi.org/10.3390/genes16010079
APA StyleZhou, L., Iqbal, A., Yang, M., & Yang, Y. (2025). Research Progress on Gene Regulation of Plant Floral Organogenesis. Genes, 16(1), 79. https://doi.org/10.3390/genes16010079






