Spotlight on FAM72B: Pan-Cancer Expression Profiles and Its Potential as a Prognostic and Immunotherapeutic Biomarker
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Processing
2.2. mRNA Expression of FAM72B in Pan-Cancer
2.3. Pan-Cancer Survival Analysis of FAM72B
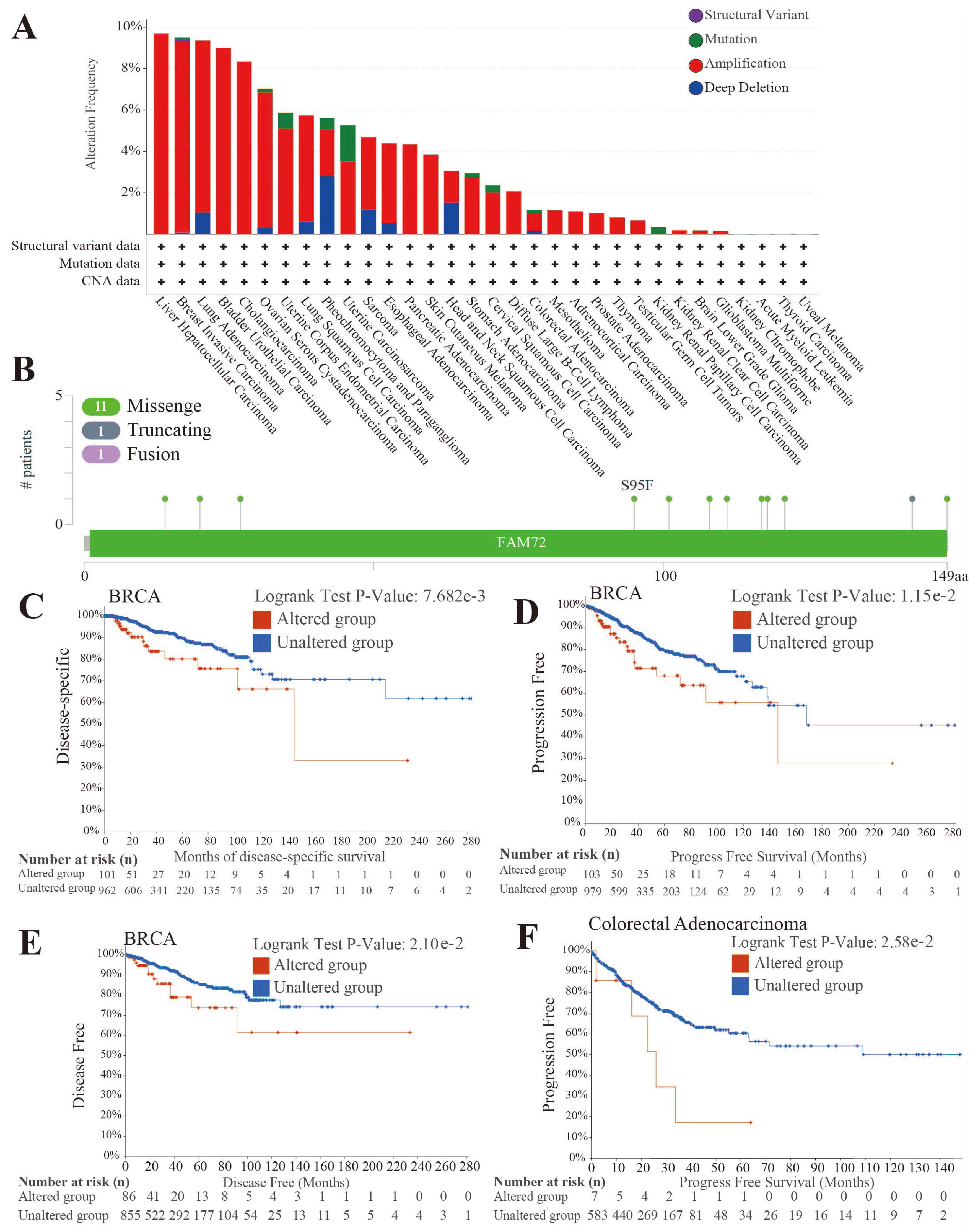
2.4. Analysis of FAM72B Genetic Mutations
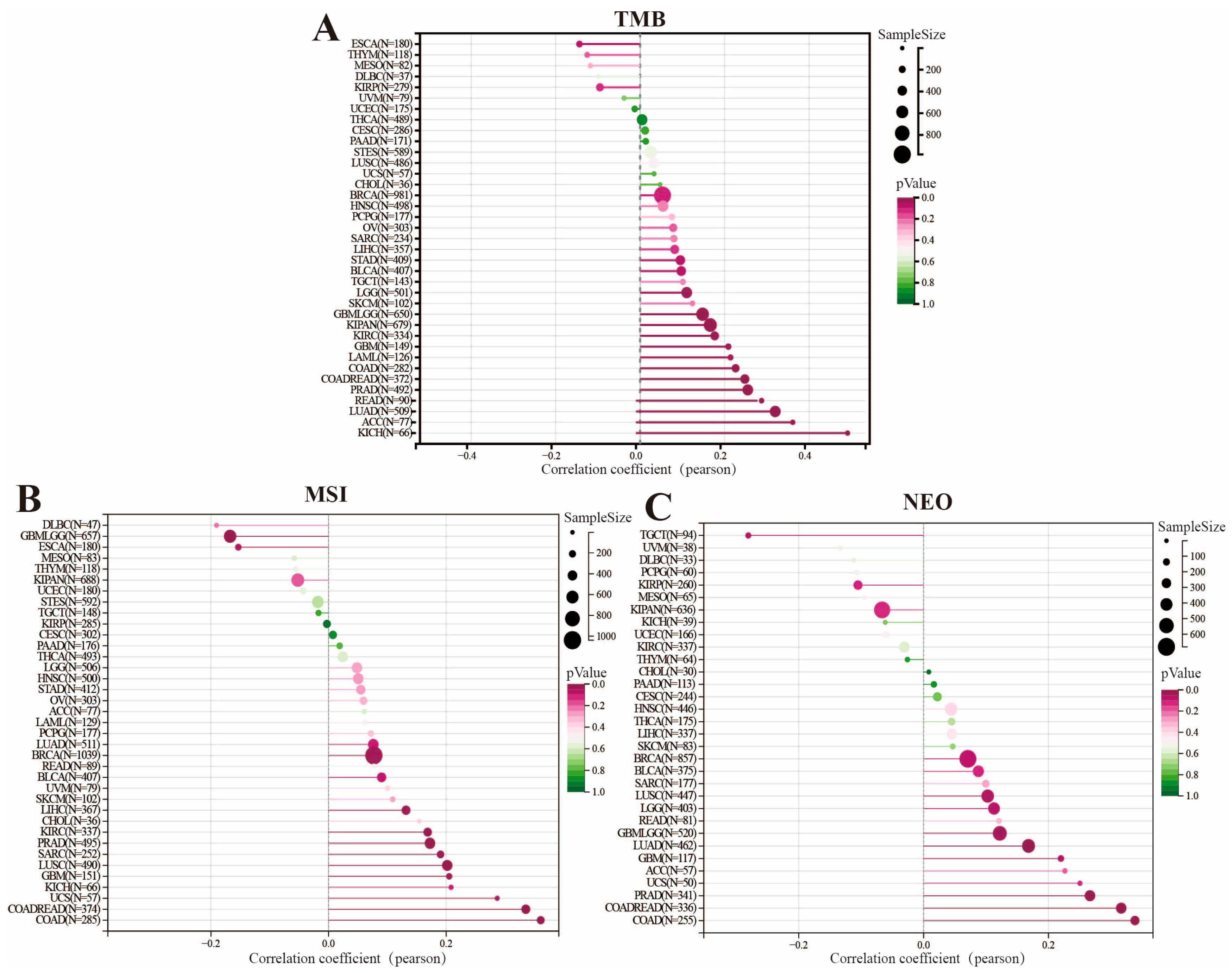
2.5. Tumor Mutational Burden, Microsatellite Instability, and Neoantigen
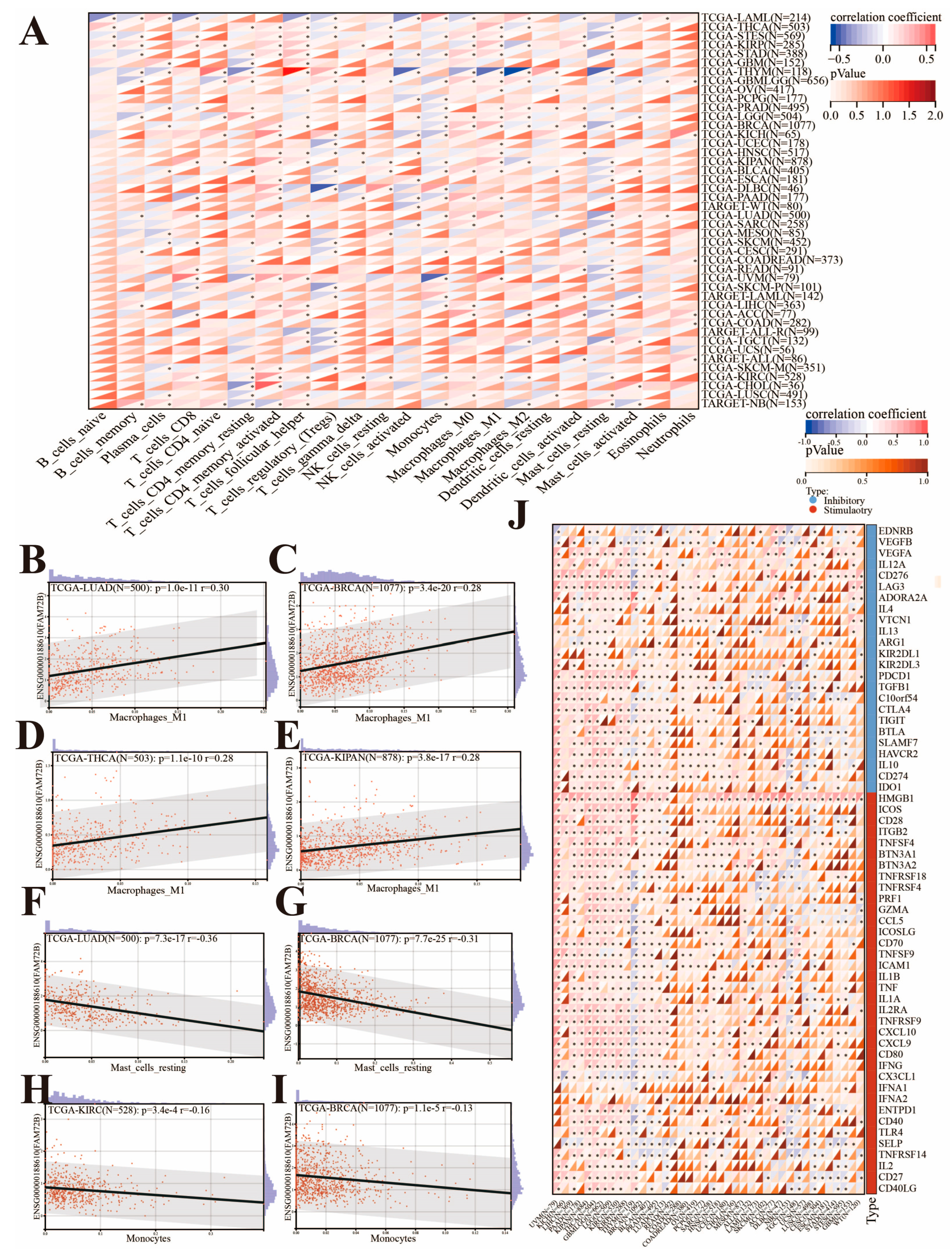
2.6. Analysis of Immune Checkpoint Genes and Immune Cell Infiltration in the Tumor Immune Microenvironment
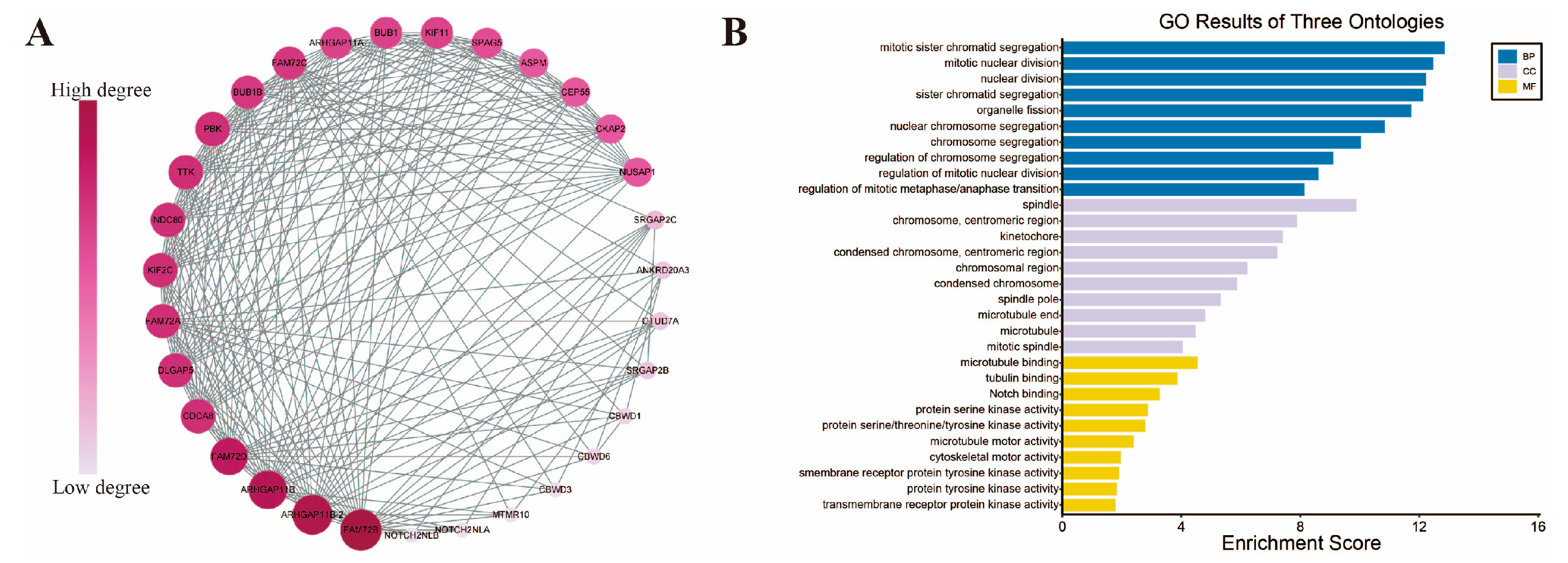
2.7. FAM72B-Related Gene Enrichment Analysis
2.8. Single-Cell Analysis of FAM72B Gene Expression
3. Results
3.1. Analysis of FAM72B mRNA Expression and Its Correlation with Clinicopathological Parameters in Multiple Human Cancers
3.2. Impact of FAM72B mRNA Expression on Prognosis in Multiple Human Cancers
3.3. FAM72B Genetic Mutations in Multiple Human Cancers
3.4. Analysis of the Correlation Between Genomic Heterogeneity and Gene Expression of FAM72B in Multiple Cancers
3.5. Correlation of FAM72B Expression with Immune Checkpoint Genes and Immune Cell Infiltration in Multiple Cancers
3.6. Functional Enrichment Analysis of FAM72B-Related Genes
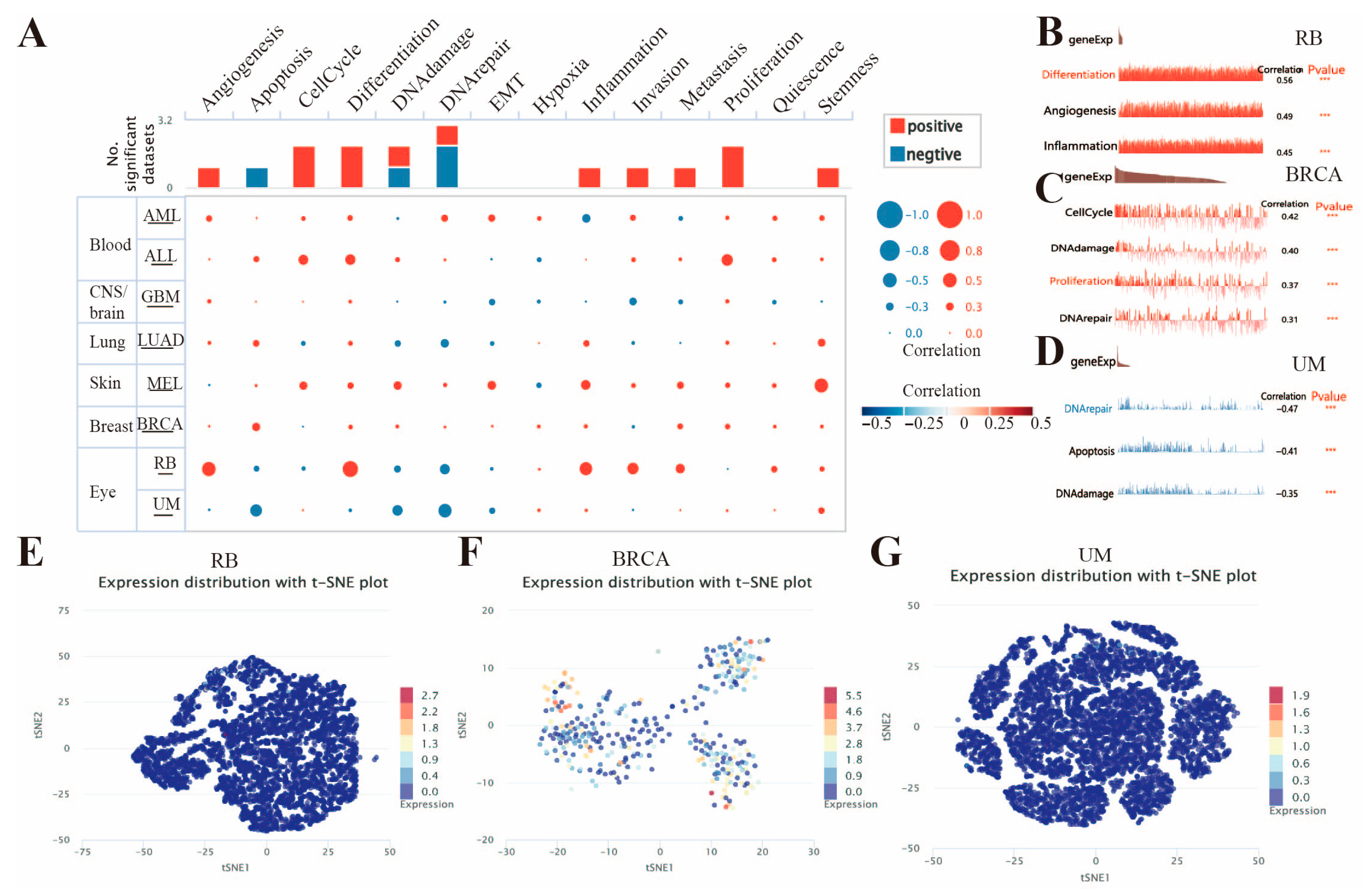
3.7. Expression Patterns of FAM72B at the Single-Cell Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, C.; Zhang, X.; Fink, S.P.; Platzer, P.; Wilson, K.; Willson, J.K.; Wang, Z.; Markowitz, S.D. Ugene, a newly identified protein that is commonly overexpressed in cancer and binds uracil DNA glycosylase. Cancer Res. 2008, 68, 6118–6126. [Google Scholar] [CrossRef]
- Wang, L.T.; Lin, C.S.; Chai, C.Y.; Liu, K.Y.; Chen, J.Y.; Hsu, S.H. Functional interaction of Ugene and EBV infection mediates tumorigenic effects. Oncogene 2011, 30, 2921–2932. [Google Scholar] [CrossRef]
- Nehar, S.; Mishra, M.; Heese, K. Identification and characterisation of the novel amyloid-beta peptide-induced protein p17. FEBS Lett. 2009, 583, 3247–3253. [Google Scholar] [CrossRef]
- Kutzner, A.; Pramanik, S.; Kim, P.S.; Heese, K. All-or-(N)One—An epistemological characterization of the human tumorigenic neuronal paralogous FAM72 gene loci. Genomics 2015, 106, 278–285. [Google Scholar] [CrossRef]
- Fu, Y.; Jia, X.; Yuan, J.; Yang, Y.; Zhang, T.; Yu, Q.; Zhou, J.; Wang, T. Fam72a functions as a cell-cycle-controlled gene during proliferation and antagonizes apoptosis through reprogramming PP2A substrates. Dev. Cell 2023, 58, 398–415.e397. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Li, C.; Stewart, J.A.; Barbulescu, P.; Seija Desivo, N.; Álvarez-Quilón, A.; Pezo, R.C.; Perera, M.L.W.; Chan, K.; Tong, A.H.Y.; et al. FAM72A antagonizes UNG2 to promote mutagenic repair during antibody maturation. Nature 2021, 600, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Rogier, M.; Moritz, J.; Robert, I.; Lescale, C.; Heyer, V.; Abello, A.; Martin, O.; Capitani, K.; Thomas, M.; Thomas-Claudepierre, A.S.; et al. Fam72a enforces error-prone DNA repair during antibody diversification. Nature 2021, 600, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Barbulescu, P.; Chana, C.K.; Wong, M.K.; Ben Makhlouf, I.; Bruce, J.P.; Feng, Y.; Keszei, A.F.A.; Wong, C.; Mohamad-Ramshan, R.; McGary, L.C.; et al. FAM72A degrades UNG2 through the GID/CTLH complex to promote mutagenic repair during antibody maturation. Nat. Commun. 2024, 15, 7541. [Google Scholar] [CrossRef]
- Barbulescu, P.; Wong, M.K.; Baronian, L.; Wang, P.; Aderinto, A.; Kneussel, M.; Martin, A. MKLN1-dependent GID4/CTLH E3 ubiquitin ligase complex assemblies are required to support B-cell antibody diversification. J. Immunol. 2025. [Google Scholar] [CrossRef]
- Ho, N.T.T.; Kutzner, A.; Heese, K. A Novel Divergent Gene Transcription Paradigm-the Decisive, Brain-Specific, Neural |-Srgap2-Fam72a-| Master Gene Paradigm. Mol. Neurobiol. 2019, 56, 5891–5899. [Google Scholar] [CrossRef]
- Zeng, Y.; Xiong, C.; Tang, N.; Wang, S.; Xiong, Z.; Liang, T.; Wang, Q.; Li, M.; Li, J. FAM72A promotes glioma progression by regulating mitophagy through the Pink1/Parkin signaling pathway. J. Cancer 2023, 14, 903–915. [Google Scholar] [CrossRef]
- Rahane, C.S.; Kutzner, A.; Heese, K. A cancer tissue-specific FAM72 expression profile defines a novel glioblastoma multiform (GBM) gene-mutation signature. J. Neuro-Oncol. 2019, 141, 57–70. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, Z.; Zheng, Q.; Li, J. FAM72 serves as a biomarker of poor prognosis in human lung adenocarcinoma. Aging 2021, 13, 8155–8176. [Google Scholar] [CrossRef]
- Zhang, T.; Nie, Y.; Gu, J.; Cai, K.; Chen, X.; Li, H.; Wang, J. Identification of Mitochondrial-Related Prognostic Biomarkers Associated with Primary Bile Acid Biosynthesis and Tumor Microenvironment of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 587479. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, L.; Yang, L.; Zhou, H.; Chen, Y.; Guo, Y. Integrated systemic analysis of FAM72A to identify its clinical relevance, biological function, and relationship to drug sensitivity in hepatocellular carcinoma. Front. Oncol. 2022, 12, 1046473. [Google Scholar] [CrossRef]
- Xu, Y.; Hirachan, S.; Shen, Y.; Huang, Q.; Bhandari, A.; Xia, E. The pan-cancer analysis of the oncogenic role of FAM72A as a BRCA prognostic biomarker and immunotherapeutic target. Environ. Toxicol. 2023, 38, 1100–1117. [Google Scholar] [CrossRef]
- Chi, H.; Gao, X.; Xia, Z.; Yu, W.; Yin, X.; Pan, Y.; Peng, G.; Mao, X.; Teichmann, A.T.; Zhang, J.; et al. FAM family gene prediction model reveals heterogeneity, stemness and immune microenvironment of UCEC. Front. Mol. Biosci. 2023, 10, 1200335. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Cao, K.; Zhang, P.; Ma, J.; Zhu, J. Prognostic and Immunological Implications of FAM72A in Pan-Cancer and Functional Validations. Int. J. Mol. Sci. 2022, 24, 375. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Higgs, R.E.; Hoffman, R.W.; Dow, E.R.; Liu, X.; Petri, M.; Wallace, D.J.; Dörner, T.; Eastwood, B.J.; Miller, B.B.; et al. A Bayesian gene network reveals insight into the JAK-STAT pathway in systemic lupus erythematosus. PLoS ONE 2019, 14, e0225651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wan, S.; Yu, Y.; Ruan, W.; Wang, H.; Xu, L.; Wang, C.; Chen, S.; Cao, T.; Peng, Q.; et al. Identifying potential DNA methylation markers in early-stage colorectal Cancer. Genomics 2020, 112, 3365–3373. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Q.; Xu, L.; Wang, H.; Liu, X.; Li, S.; Hu, T.; Liu, Y.; Peng, Q.; Chen, Z.; et al. Sensitive detection of colorectal cancer in peripheral blood by a novel methylation assay. Clin. Epigenet. 2021, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, Y.; Chen, Y.; Tang, Z.; Huang, M.; Ming, Y.; Wang, M.; Chen, W.; Huang, Z.; Qing, L.; et al. Family with Sequence Similarity 72 (FAM72)—A prospective biomarker for poor prognosis in patients with oral squamous cell carcinoma. Arch. Oral Biol. 2023, 151, 105695. [Google Scholar] [CrossRef]
- Chatonnet, F.; Pignarre, A.; Sérandour, A.A.; Caron, G.; Avner, S.; Robert, N.; Kassambara, A.; Laurent, A.; Bizot, M.; Agirre, X.; et al. The hydroxymethylome of multiple myeloma identifies FAM72D as a 1q21 marker linked to proliferation. Haematologica 2020, 105, 774–783. [Google Scholar] [CrossRef]
- Fan, C.; Huang, Z.; Xu, H.; Zhang, T.; Wei, H.; Gao, J.; Xu, C.; Fan, C. Machine learning-based identification of co-expressed genes in prostate cancer and CRPC and construction of prognostic models. Sci. Rep. 2025, 15, 5679. [Google Scholar] [CrossRef]
- Hu, F.; Zeng, W.; Liu, X. A Gene Signature of Survival Prediction for Kidney Renal Cell Carcinoma by Multi-Omic Data Analysis. Int. J. Mol. Sci. 2019, 20, 5720. [Google Scholar] [CrossRef]
- Yao, Z.; Jia, C.; Tai, Y.; Liang, H.; Zhong, Z.; Xiong, Z.; Deng, M.; Zhang, Q. Serum exosomal long noncoding RNAs lnc-FAM72D-3 and lnc-EPC1-4 as diagnostic biomarkers for hepatocellular carcinoma. Aging 2020, 12, 11843–11863. [Google Scholar] [CrossRef]
- Zhou, Z.; Fan, B.; Qiu, Q.; Cheng, H.; Wang, L.; Wu, Y.; Xie, J.; Ni, C.; Li, N. Pan-cancer analysis and experimental validation reveal FAM72D as a potential novel biomarker and therapeutic target in lung adenocarcinoma. Gene 2024, 928, 148764. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Li, Y.; Su, X.; Tang, Y.; Ren, Y.; Luo, J.; Yuan, F.; Yang, G.; He, Z.; Shi, Z.; et al. Exosome-derived lnc-FAM72D-3 promotes lenvatinib resistance by remodeling hepatocellular carcinoma cytoskeleton via MBNL1/FAK axis. Drug Resist. Updates 2025, 82, 101271. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, X.; Yuan, Y.; Zhai, R.; Cho, W.C.; Jiang, X. Identifying Immune Cell Infiltration and Effective Diagnostic Biomarkers in Lung Adenocarcinoma by Comprehensive Bioinformatics Analysis and In Vitro Study. Front. Oncol. 2022, 12, 916947. [Google Scholar] [CrossRef]
- Balraj, A.S.; Muthamilselvan, S.; Raja, R.; Palaniappan, A. PRADclass: Hybrid Gleason Grade-Informed Computational Strategy Identifies Consensus Biomarker Features Predictive of Aggressive Prostate Adenocarcinoma. Technol. Cancer Res. Treat. 2024, 23, 15330338231222389. [Google Scholar] [CrossRef] [PubMed]
- Rajan, P.; Stockley, J.; Sudbery, I.M.; Fleming, J.T.; Hedley, A.; Kalna, G.; Sims, D.; Ponting, C.P.; Heger, A.; Robson, C.N.; et al. Identification of a candidate prognostic gene signature by transcriptome analysis of matched pre- and post-treatment prostatic biopsies from patients with advanced prostate cancer. BMC Cancer 2014, 14, 977. [Google Scholar] [CrossRef]
- Gao, W.; Sun, L.; Gai, J.; Cao, Y.; Zhang, S. Exploring the resistance mechanism of triple-negative breast cancer to paclitaxel through the scRNA-seq analysis. PLoS ONE 2024, 19, e0297260. [Google Scholar] [CrossRef]
- Ramesh, J.; Gopalakrishnan, R.M.; Nguyen, T.H.A.; Lai, S.K.; Li, H.Y.; Kim, P.S.; Kutzner, A.; Inoue, N.; Heese, K. Deciphering the molecular landscape of the FAM72 gene family: Implications for stem cell biology and cancer. Neurochem. Int. 2024, 180, 105853. [Google Scholar] [CrossRef]
- Schou, K.B.; Mandacaru, S.; Tahir, M.; Tom, N.; Nilsson, A.S.; Andersen, J.S.; Tiberti, M.; Papaleo, E.; Bartek, J. Exploring the structural landscape of DNA maintenance proteins. Nat. Commun. 2024, 15, 7748. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Zheng, R.S.; Chen, R.; Han, B.F.; Wang, S.M.; Li, L.; Sun, K.X.; Zeng, H.M.; Wei, W.W.; He, J. Cancer incidence and mortality in China, 2022. Zhonghua Zhong Liu Za Zhi 2024, 46, 221–231. [Google Scholar]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef]
- Rodriguez-Martin, B.; Alvarez, E.G.; Baez-Ortega, A.; Zamora, J.; Supek, F.; Demeulemeester, J.; Santamarina, M.; Ju, Y.S.; Temes, J.; Garcia-Souto, D.; et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat. Genet. 2020, 52, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Hu, A.; Sun, L.; Lin, H.; Liao, Y.; Yang, H.; Mao, Y. Harnessing innate immune pathways for therapeutic advancement in cancer. Signal Transduct. Target. Ther. 2024, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Han, J.; Lin, C.; Zhou, Y.; Wang, W. Beyond the tumor microenvironment: Orchestrating systemic T-cell response for next-generation cancer immunotherapy (Review). Int. J. Oncol. 2025, 67, 56. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, J.; Wang, J.; Yang, S.; Wang, R.; Zhang, G.; Li, Z.; Shi, R.; Wang, Z.; Lu, Q. Deciphering the tumor immune microenvironment from a multidimensional omics perspective: Insight into next-generation CAR-T cell immunotherapy and beyond. Mol. Cancer 2024, 23, 131. [Google Scholar] [CrossRef]
- Qin, S.; Xie, B.; Wang, Q.; Yang, R.; Sun, J.; Hu, C.; Liu, S.; Tao, Y.; Xiao, D. New insights into immune cells in cancer immunotherapy: From epigenetic modification, metabolic modulation to cell communication. MedComm 2024, 5, e551. [Google Scholar] [CrossRef]
- Lv, B.; Wang, Y.; Ma, D.; Cheng, W.; Liu, J.; Yong, T.; Chen, H.; Wang, C. Immunotherapy: Reshape the Tumor Immune Microenvironment. Front. Immunol. 2022, 13, 844142. [Google Scholar] [CrossRef]
- Fu, T.; Dai, L.J.; Wu, S.Y.; Xiao, Y.; Ma, D.; Jiang, Y.Z.; Shao, Z.M. Spatial architecture of the immune microenvironment orchestrates tumor immunity and therapeutic response. J. Hematol. Oncol. 2021, 14, 98. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017, 1, 1–15. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef]
- Huang, R.J.; Huang, Y.S.; An, N.; Hu, J.J.; Wu, C.Y.; Chen, Y.X.; Chen, J.Y.; Zhao, Q.; Xu, R.H.; Yuan, S.Q.; et al. Pan-cancer analysis of heterogeneity of tumor mutational burden and genomic mutation under treatment pressure. ESMO Open 2024, 9, 103494. [Google Scholar] [CrossRef]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef]
- Wang, P.; Chen, Y.; Wang, C. Beyond Tumor Mutation Burden: Tumor Neoantigen Burden as a Biomarker for Immunotherapy and Other Types of Therapy. Front. Oncol. 2021, 11, 672677. [Google Scholar] [CrossRef]
- Gou, H.; Chen, P.; Wu, W. FAM72 family proteins as poor prognostic markers in clear cell renal carcinoma. Biochem. Biophys. Rep. 2023, 35, 101506. [Google Scholar] [CrossRef] [PubMed]
- Ho, N.T.T.; Rahane, C.S.; Pramanik, S.; Kim, P.S.; Kutzner, A.; Heese, K. FAM72, Glioblastoma Multiforme (GBM) and Beyond. Cancers 2021, 13, 1025. [Google Scholar] [CrossRef]
- Gorka, J.; Marona, P.; Kwapisz, O.; Waligórska, A.; Pospiech, E.; Dobrucki, J.W.; Rys, J.; Jura, J.; Miekus, K. MCPIP1 inhibits Wnt/β-catenin signaling pathway activity and modulates epithelial-mesenchymal transition during clear cell renal cell carcinoma progression by targeting miRNAs. Oncogene 2021, 40, 6720–6735. [Google Scholar] [CrossRef]
- Bo, J.; Mao, S.; Yang, J.; Wang, L.; Zheng, J.; Zhang, C.; Song, M.; Chen, S.; Liu, C. Rhodiolin inhibits the PI3K/AKT/mTOR signaling pathway via the glycolytic enzyme GPI in human papillary thyroid cancer. Phytomedicine 2024, 132, 155804. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, M.; Sa, R.; Fu, H.; Cheng, L.; Chen, L. Mouse models of thyroid cancer: Bridging pathogenesis and novel therapeutics. Cancer Lett. 2020, 469, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Tsimberidou, A.M.; Hong, D.S.; Fu, S.; Karp, D.D.; Piha-Paul, S.; Kies, M.S.; Ravi, V.; Subbiah, V.; Patel, S.M.; Tu, S.M.; et al. Precision medicine: Preliminary results from the Initiative for Molecular Profiling and Advanced Cancer Therapy 2 (IMPACT2) study. npj Precis. Oncol. 2021, 5, 21. [Google Scholar] [CrossRef]
- Cifaldi, C.; Rivalta, B.; Amodio, D.; Mattia, A.; Pacillo, L.; Di Cesare, S.; Chiriaco, M.; Ursu, G.M.; Cotugno, N.; Giancotta, C.; et al. Clinical, Immunological, and Molecular Variability of RAG Deficiency: A Retrospective Analysis of 22 RAG Patients. J. Clin. Immunol. 2022, 42, 130–145. [Google Scholar] [CrossRef]
- Schnuck, J.O.; Chauhan, S.S.B.; Sham, J.G. Surrogate endpoints in clinical trials: When is good…good enough? Hepatobil. Surg. Nutr. 2024, 13, 1062–1064. [Google Scholar] [CrossRef] [PubMed]
- Marcadis, A.R.; Marti, J.L.; Ehdaie, B.; Hakimi, A.A.; Davies, L.; Morris, L.G.T. Characterizing Relative and Disease-Specific Survival in Early-Stage Cancers. JAMA Intern. Med. 2020, 180, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.G.; Booth, C.M.; Eisenhauer, E.A. Disease-free survival as an end-point in the treatment of solid tumours--perspectives from clinical trials and clinical practice. Eur. J. Cancer 2014, 50, 2298–2302. [Google Scholar] [CrossRef]
- Courtinard, C.; Gourgou, S.; Jacot, W.; Carton, M.; Guérin, O.; Vacher, L.; Bertaut, A.; Le Deley, M.C.; Pérol, D.; Marino, P.; et al. Association between progression-free survival and overall survival in women receiving first-line treatment for metastatic breast cancer: Evidence from the ESME real-world database. BMC Med. 2023, 21, 87. [Google Scholar] [CrossRef]
- Wang, J.; Su, W.; Zhang, T.; Zhang, S.; Lei, H.; Ma, F.; Shi, M.; Shi, W.; Xie, X.; Di, C. Aberrant Cyclin D1 splicing in cancer: From molecular mechanism to therapeutic modulation. Cell Death Dis. 2023, 14, 244. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, K.; Zhang, Z.; Zeng, X.; Huang, Z.; Qin, P.; Xie, Z.; Cai, X.; Ashrafizadeh, M.; Tian, Y.; et al. PI3K/AKT/mTOR Axis in Cancer: From Pathogenesis to Treatment. MedComm 2025, 6, e70295. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, X.; Huang, S.; Liu, W. Identification of a Cancer Stem Cell-Related Gene Signature in Hepatocellular Carcinoma Based on Single-Cell RNA-Seq and Bulk RNA-Seq Analysis. Int. J. Mol. Sci. 2025, 26, 2933. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, Y.; Zhang, S.; Wang, X.; Dou, H.; Yu, X.; Zhang, Z.; Yang, S.; Xiao, M. Extracellular matrix remodeling in tumor progression and immune escape: From mechanisms to treatments. Mol. Cancer 2023, 22, 48. [Google Scholar] [CrossRef]
- Mejía-García, A.; Orozco, C.A.; Herzog, J.; Alarcón-Betancourth, O.; Meneses-Torres, A.; Ramírez, M.; González, J.; Zambrano, Y.; Combita, A.L.; Bonilla, D.A.; et al. Development and Validation of an Extracellular Matrix Gene Expression Signature for Prognostic Prediction in Patients with Uveal Melanoma. Int. J. Mol. Sci. 2025, 26, 4317. [Google Scholar] [CrossRef]
- Overacre-Delgoffe, A.E.; Bumgarner, H.J.; Cillo, A.R.; Burr, A.H.P.; Tometich, J.T.; Bhattacharjee, A.; Bruno, T.C.; Vignali, D.A.A.; Hand, T.W. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity 2021, 54, 2812–2824.e2814. [Google Scholar] [CrossRef]
- Gutiérrez-Melo, N.; Baumjohann, D. T follicular helper cells in cancer. Trends Cancer 2023, 9, 309–325. [Google Scholar] [CrossRef]
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef]
- Subbiah, V.; Solit, D.B.; Chan, T.A.; Kurzrock, R. The FDA approval of pembrolizumab for adult and pediatric patients with tumor mutational burden (TMB) ≥10: A decision centered on empowering patients and their physicians. Ann. Oncol. 2020, 31, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients with Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Germano, G.; Amirouchene-Angelozzi, N.; Rospo, G.; Bardelli, A. The Clinical Impact of the Genomic Landscape of Mismatch Repair-Deficient Cancers. Cancer Discov. 2018, 8, 1518–1528. [Google Scholar] [CrossRef]
- Ludford, K.; Ho, W.J.; Thomas, J.V.; Raghav, K.P.S.; Murphy, M.B.; Fleming, N.D.; Lee, M.S.; Smaglo, B.G.; You, Y.N.; Tillman, M.M.; et al. Neoadjuvant Pembrolizumab in Localized Microsatellite Instability High/Deficient Mismatch Repair Solid Tumors. J. Clin. Oncol. 2023, 41, 2181–2190. [Google Scholar] [CrossRef]
- Ott, P.A.; Hu-Lieskovan, S.; Chmielowski, B.; Govindan, R.; Naing, A.; Bhardwaj, N.; Margolin, K.; Awad, M.M.; Hellmann, M.D.; Lin, J.J.; et al. A Phase Ib Trial of Personalized Neoantigen Therapy Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-small Cell Lung Cancer, or Bladder Cancer. Cell 2020, 183, 347–362.e324. [Google Scholar] [CrossRef]
- Awad, M.M.; Govindan, R.; Balogh, K.N.; Spigel, D.R.; Garon, E.B.; Bushway, M.E.; Poran, A.; Sheen, J.H.; Kohler, V.; Esaulova, E.; et al. Personalized neoantigen vaccine NEO-PV-01 with chemotherapy and anti-PD-1 as first-line treatment for non-squamous non-small cell lung cancer. Cancer Cell 2022, 40, 1010–1026.e1011. [Google Scholar] [CrossRef]
- Lopez, J.; Powles, T.; Braiteh, F.; Siu, L.L.; LoRusso, P.; Friedman, C.F.; Balmanoukian, A.S.; Gordon, M.; Yachnin, J.; Rottey, S.; et al. Autogene cevumeran with or without atezolizumab in advanced solid tumors: A phase 1 trial. Nat. Med. 2025, 31, 152–164. [Google Scholar] [CrossRef]
- Dai, E.; Zhu, Z.; Wahed, S.; Qu, Z.; Storkus, W.J.; Guo, Z.S. Epigenetic modulation of antitumor immunity for improved cancer immunotherapy. Mol. Cancer 2021, 20, 171. [Google Scholar] [CrossRef] [PubMed]
- House, I.G.; Savas, P.; Lai, J.; Chen, A.X.Y.; Oliver, A.J.; Teo, Z.L.; Todd, K.L.; Henderson, M.A.; Giuffrida, L.; Petley, E.V.; et al. Macrophage-Derived CXCL9 and CXCL10 Are Required for Antitumor Immune Responses Following Immune Checkpoint Blockade. Clin. Cancer Res. 2020, 26, 487–504. [Google Scholar] [CrossRef]
- Voron, T.; Colussi, O.; Marcheteau, E.; Pernot, S.; Nizard, M.; Pointet, A.L.; Latreche, S.; Bergaya, S.; Benhamouda, N.; Tanchot, C.; et al. VEGF-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J. Exp. Med. 2015, 212, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, Y. HMGB1 in inflammation and cancer. J. Hematol. Oncol. 2020, 13, 116. [Google Scholar] [CrossRef]
- Han, Y.; Sheng, W.; Liu, X.; Liu, H.; Jia, X.; Li, H.; Wang, C.; Wang, B.; Hu, T.; Ma, Y. Glycyrrhizin ameliorates colorectal cancer progression by regulating NHEJ pathway through inhibiting HMGB1-induced DNA damage response. Sci. Rep. 2024, 14, 24948. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Lyu, Q.; Yi, R.; Sun, Y.; Zhang, W.; Wei, T.; Zhang, Y.; Shi, J.; Zhang, J. HMGB1 promotes chemoresistance in small cell lung cancer by inducing PARP1-related nucleophagy. J. Adv. Res. 2024, 66, 165–180. [Google Scholar] [CrossRef]
- Yuan, S.; Liu, Z.; Xu, Z.; Liu, J.; Zhang, J. High mobility group box 1 (HMGB1): A pivotal regulator of hematopoietic malignancies. J. Hematol. Oncol. 2020, 13, 91. [Google Scholar] [CrossRef]
- Tan, G.; Huang, C.; Chen, J.; Zhi, F. HMGB1 released from GSDME-mediated pyroptotic epithelial cells participates in the tumorigenesis of colitis-associated colorectal cancer through the ERK1/2 pathway. J. Hematol. Oncol. 2020, 13, 149. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, L.; Jiang, S.; Yang, T.; Lan, J.; Lei, Y.; Tan, H.; Pan, K. HMGB1 mediates lipopolysaccharide-induced inflammation via interacting with GPX4 in colon cancer cells. Cancer Cell Int. 2020, 20, 205. [Google Scholar] [CrossRef]
- Hao, W.; Yan, A.; Guo, X.; Chen, Z.; Chen, N.; Li, H.; Wu, F.; Sun, P.; Zhao, Y.; Zhao, G.; et al. Targeting HMGB1 modulates cancer-associated fibroblasts and enhances radiotherapy in lung adenocarcinoma. J. Control. Release 2025, 387, 114199. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Zhao, Y.; Wang, X.; Chen, N.; Mao, F.; Teng, Y.; Wang, T.; Peng, L.; Zhang, J.; Cheng, P.; et al. Increased intratumoral mast cells foster immune suppression and gastric cancer progression through TNF-α-PD-L1 pathway. J. Immunother. Cancer 2019, 7, 54. [Google Scholar] [CrossRef]
- Somasundaram, R.; Connelly, T.; Choi, R.; Choi, H.; Samarkina, A.; Li, L.; Gregorio, E.; Chen, Y.; Thakur, R.; Abdel-Mohsen, M.; et al. Tumor-infiltrating mast cells are associated with resistance to anti-PD-1 therapy. Nat. Commun. 2021, 12, 346. [Google Scholar] [CrossRef]
- Terada, T.; Matsunaga, Y. Increased mast cells in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J. Hepatol. 2000, 33, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, L.; Hu, X. Mast Cells Resting-Related Prognostic Signature in Hepatocellular Carcinoma. J. Oncol. 2021, 2021, 4614257. [Google Scholar] [CrossRef] [PubMed]
- van Elsas, M.J.; Middelburg, J.; Labrie, C.; Roelands, J.; Schaap, G.; Sluijter, M.; Tonea, R.; Ovcinnikovs, V.; Lloyd, K.; Schuurman, J.; et al. Immunotherapy-activated T cells recruit and skew late-stage activated M1-like macrophages that are critical for therapeutic efficacy. Cancer Cell 2024, 42, 1032–1050.e1010. [Google Scholar] [CrossRef]
- Han, D.; Ma, Q.; Ballar, P.; Zhang, C.; Dai, M.; Luo, X.; Gu, J.; Wei, C.; Guo, P.; Zeng, L.; et al. Reprogramming tumor-associated macrophages and inhibiting tumor neovascularization by targeting MANF-HSF1-HSP70-1 pathway: An effective treatment for hepatocellular carcinoma. Acta Pharm. Sin. B 2024, 14, 4396–4412. [Google Scholar] [CrossRef]
- Shi, Q.; Shen, Q.; Liu, Y.; Shi, Y.; Huang, W.; Wang, X.; Li, Z.; Chai, Y.; Wang, H.; Hu, X.; et al. Increased glucose metabolism in TAMs fuels O-GlcNAcylation of lysosomal Cathepsin B to promote cancer metastasis and chemoresistance. Cancer Cell 2022, 40, 1207–1222.e1210. [Google Scholar] [CrossRef]
- Wu, J.M.; Chen, C.T.; Coumar, M.S.; Lin, W.H.; Chen, Z.J.; Hsu, J.T.; Peng, Y.H.; Shiao, H.Y.; Lin, W.H.; Chu, C.Y.; et al. Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules. Proc. Natl. Acad. Sci. USA 2013, 110, E1779–E1787. [Google Scholar] [CrossRef]
- Friese, A.; Faesen, A.C.; Huis in ‘t Veld, P.J.; Fischböck, J.; Prumbaum, D.; Petrovic, A.; Raunser, S.; Herzog, F.; Musacchio, A. Molecular requirements for the inter-subunit interaction and kinetochore recruitment of SKAP and Astrin. Nat. Commun. 2016, 7, 11407. [Google Scholar] [CrossRef]
- Sun, M.; Wang, Y.; Xin, G.; Yang, B.; Jiang, Q.; Zhang, C. NuSAP regulates microtubule flux and Kif2A localization to ensure accurate chromosome congression. J. Cell Biol. 2024, 223, e202108070. [Google Scholar] [CrossRef]
- Chen, M.; Cen, K.; Song, Y.; Zhang, X.; Liou, Y.C.; Liu, P.; Huang, J.; Ruan, J.; He, J.; Ye, W.; et al. NUSAP1-LDHA-Glycolysis-Lactate feedforward loop promotes Warburg effect and metastasis in pancreatic ductal adenocarcinoma. Cancer Lett. 2023, 567, 216285. [Google Scholar] [CrossRef] [PubMed]
- Canu, V.; Donzelli, S.; Sacconi, A.; Lo Sardo, F.; Pulito, C.; Bossel, N.; Di Benedetto, A.; Muti, P.; Botti, C.; Domany, E.; et al. Aberrant transcriptional and post-transcriptional regulation of SPAG5, a YAP-TAZ-TEAD downstream effector, fuels breast cancer cell proliferation. Cell Death Differ. 2021, 28, 1493–1511. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Deng, Z.; Shen, D.; Lu, M.; Li, M.; Yu, J.; Xiao, Y.; Wang, G.; Qian, K.; Ju, L.; et al. DLGAP5 triggers proliferation and metastasis of bladder cancer by stabilizing E2F1 via USP11. Oncogene 2024, 43, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Zhou, F.; Li, M.; Jin, W.; Yu, J.; Wang, G.; Qian, K.; Ju, L.; Zhang, Y.; Xiao, Y.; et al. DLGAP5 enhances bladder cancer chemoresistance by regulating glycolysis through MYC stabilization. Theranostics 2025, 15, 2375–2392. [Google Scholar] [CrossRef]
- Xing, X.; Zhong, J.; Biermann, J.; Duan, H.; Zhang, X.; Shi, Y.; Gao, Y.; He, K.; Zhai, D.; Luo, F.; et al. Pan-cancer human brain metastases atlas at single-cell resolution. Cancer Cell 2025, 43, 1242–1260.e1249. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, X.; Pan, J.; Ning, H.; Zhang, Y.; Bo, Y.; Ren, X.; Li, J.; Qin, S.; Wang, D.; et al. Pan-cancer single-cell dissection reveals phenotypically distinct B cell subtypes. Cell 2024, 187, 4790–4811.e4722. [Google Scholar] [CrossRef]
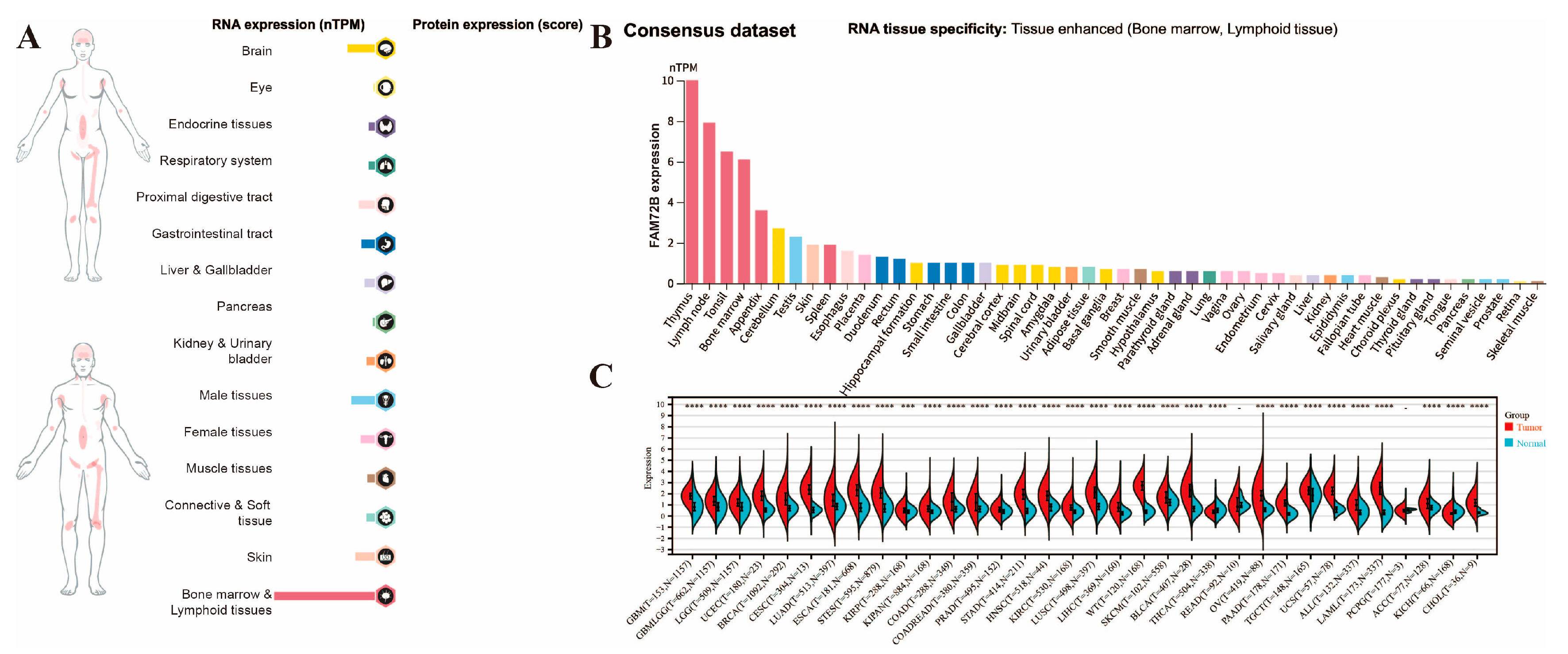
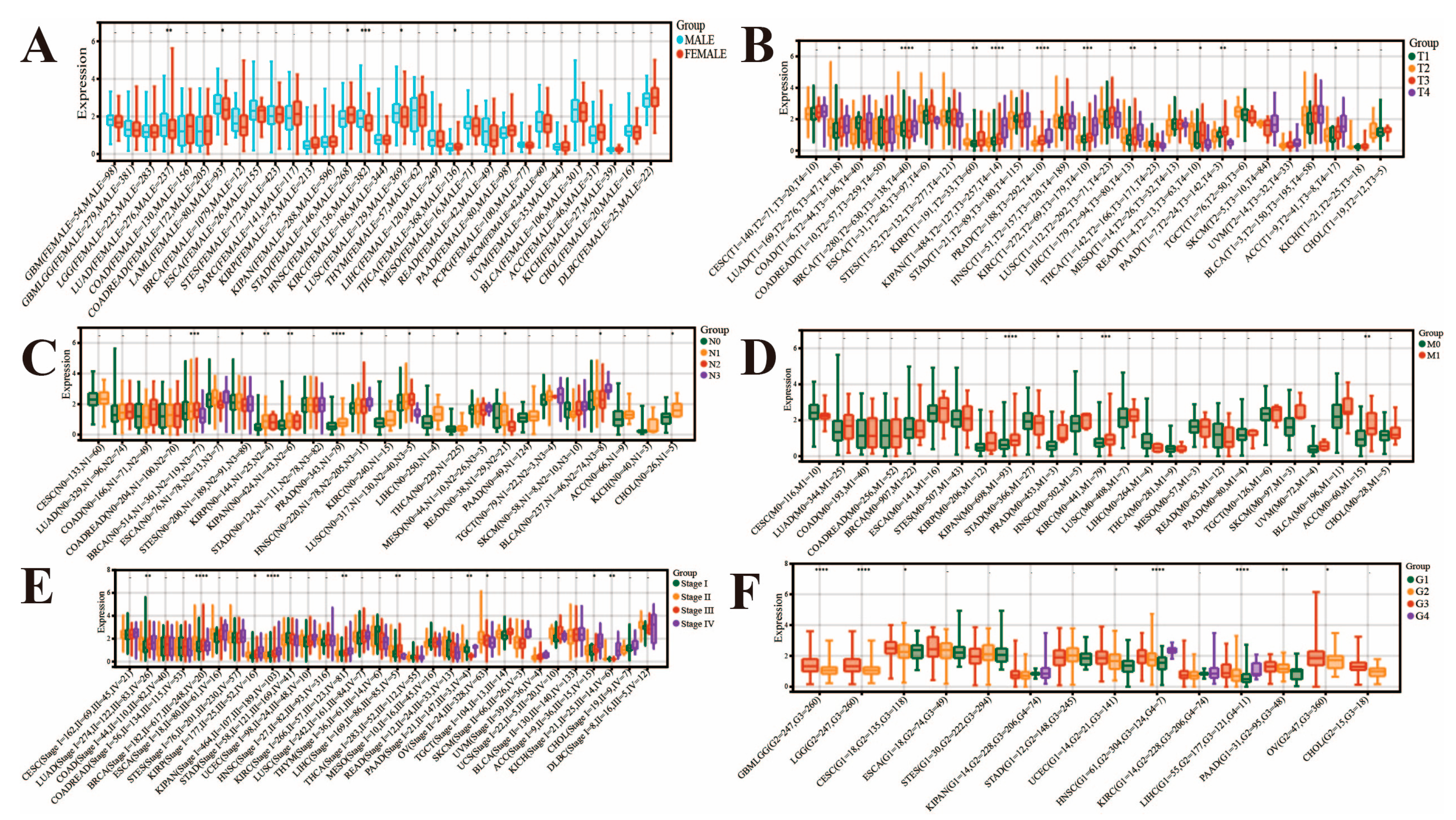
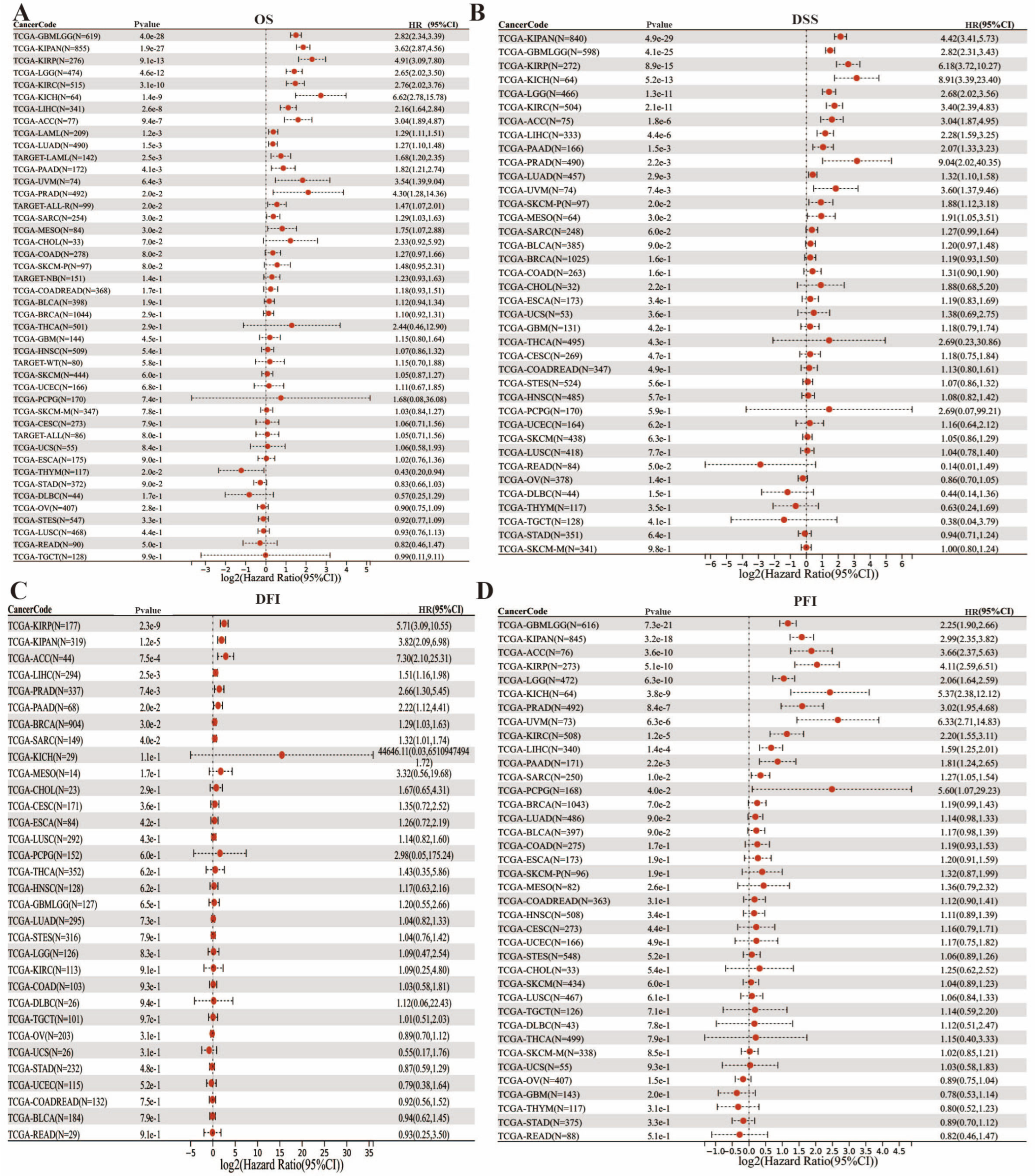

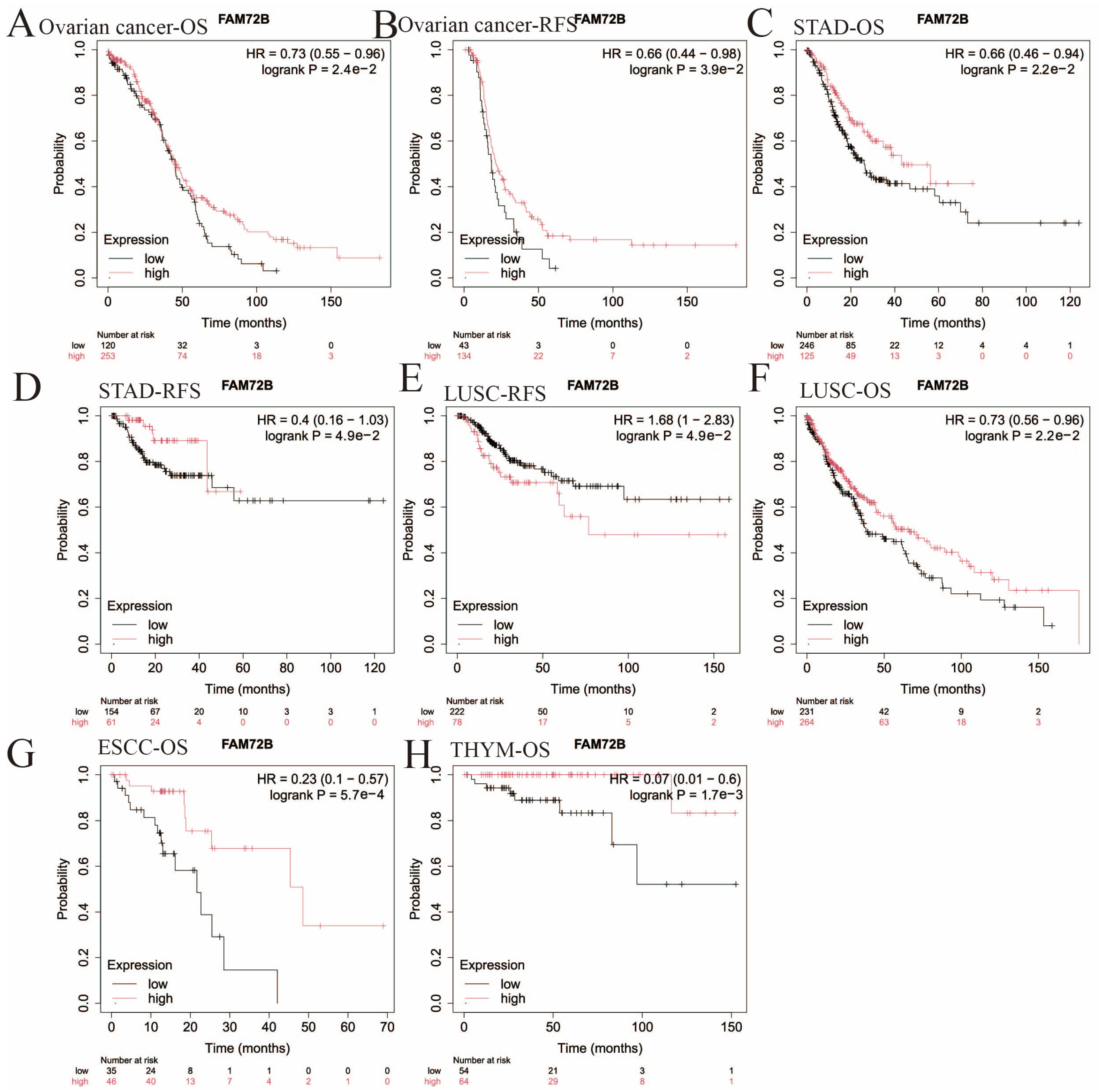
| Abbreviation | Full Name |
|---|---|
| ACC | Adrenocortical carcinoma |
| ALL | Acute lymphoblastic leukemia |
| BLCA | Bladder urothelial carcinoma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon adenocarcinoma |
| COADREAD | Colon adenocarcinoma/rectum adenocarcinoma |
| ESCA | Esophageal carcinoma |
| GBM | Glioblastoma multiforme |
| GBMLGG | Low-grade glioma and glioblastoma |
| HNSC | Head and neck squamous cell carcinoma |
| KICH | Kidney chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIPAN | Pan-kidney cohort (KICH + KIRC + KIRP) |
| KIRP | Kidney renal papillary cell carcinoma |
| LAML | Acute myeloid leukemia |
| LGG | Brain lower-grade glioma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PCPG | Pheochromocytoma and paraganglioma |
| PRAD | Prostate adenocarcinoma |
| READ | Rectum adenocarcinoma |
| SKCM | Skin cutaneous melanoma |
| STAD | Stomach adenocarcinoma |
| STES | Stomach and esophageal carcinoma |
| TGCT | Testicular germ cell tumors |
| THCA | Thyroid carcinoma |
| UCEC | Uterine corpus endometrial carcinoma |
| UCS | Uterine carcinosarcoma |
| WT | High-risk Wilms tumor |
| Cancer Type | Overall Survival | Recurrence-Free Survival | ||
|---|---|---|---|---|
| Hazard Ratio | p-Value | Hazard Ratio | p-Value | |
| KIRP | 5.46 (2.99–9.98) | 6.4e−10 | 5.74 (2.59–12.69) | 1.2e−6 |
| LIHC | 1.9 (1.34–2.68) | 2.2e−4 | 1.69 (1.19–2.4) | 2.7e−3 |
| LUAD | 2.11 (1.54–2.89) | 2e−6 | 1.66 (1.03–2.68) | 3.4e−2 |
| UCEC | 2.03 (1.34–3.08) | 6.8e−4 | 2.44 (1.37–4.35) | 1.8e−3 |
| SARC | 2 (1.29–3.09) | 1.5e−3 | 1.91 (1.07–3.42) | 2.6e−2 |
| CSCC | 1.7 (1.05–2.76) | 2.8e−2 | 1.65 (0.75–3.64) | 2.1e−1 |
| KIRC | 2.12 (1.56–2.9) | 1.2e−6 | 2.4 (0.76–7.57) | 1.2e−1 |
| THCA | 2.09 (0.78–5.63) | 1.3e−1 | 2.52 (1.16–5.49) | 1.6e−2 |
| Breast Cancer | 1.32 (0.95–1.84) | 9.8e−2 | 1.96 (1.06–3.62) | 2.8e−2 |
| Ovarian Cancer | 0.73 (0.55–0.96) | 2.4e−2 | 0.66 (0.44–0.98) | 3.9e−2 |
| STAD | 0.66 (0.46–0.94) | 2.2e−2 | 0.4 (0.16–1.03) | 4.9e−2 |
| LUSC | 0.73 (0.56–0.96) | 2.2e−2 | 1.68 (1–2.83) | 4.9e−2 |
| ESCC | 0.23 (0.1–0.57) | 5.7e−4 | 0.43 (0.17–1.14) | 8.1e−2 |
| THYM | 0.07 (0.01–0.6) | 1.7e−3 | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, A.; Wang, Y. Spotlight on FAM72B: Pan-Cancer Expression Profiles and Its Potential as a Prognostic and Immunotherapeutic Biomarker. Genes 2025, 16, 1140. https://doi.org/10.3390/genes16101140
Chu A, Wang Y. Spotlight on FAM72B: Pan-Cancer Expression Profiles and Its Potential as a Prognostic and Immunotherapeutic Biomarker. Genes. 2025; 16(10):1140. https://doi.org/10.3390/genes16101140
Chicago/Turabian StyleChu, Anran, and Yuchan Wang. 2025. "Spotlight on FAM72B: Pan-Cancer Expression Profiles and Its Potential as a Prognostic and Immunotherapeutic Biomarker" Genes 16, no. 10: 1140. https://doi.org/10.3390/genes16101140
APA StyleChu, A., & Wang, Y. (2025). Spotlight on FAM72B: Pan-Cancer Expression Profiles and Its Potential as a Prognostic and Immunotherapeutic Biomarker. Genes, 16(10), 1140. https://doi.org/10.3390/genes16101140





