Genetic Diversity and Population Structure of Farmed Longfin Batfish (Platax teira) in the South China Sea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Data Collection
2.2. Whole-Genome Resequencing and Variant Calling
2.3. Genetic Diversity and Linkage Disequilibrium
2.4. Genome-Wide Detection of ROH and Estimation of Inbreeding Coefficients
2.5. Population Structure Analysis
3. Results
3.1. Overview of Whole-Genome Resequencing and Variant Detection
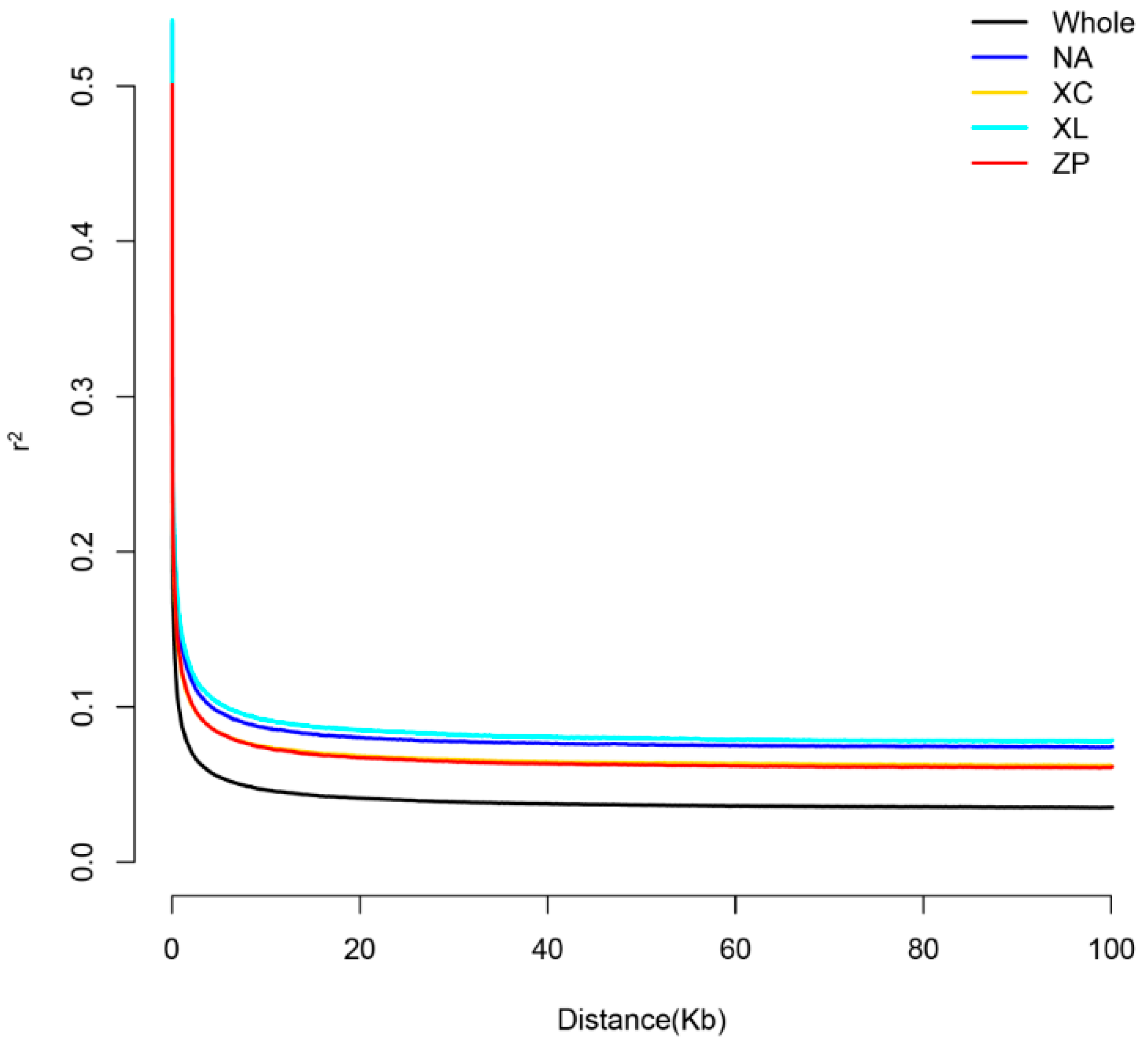
3.2. Genetic Diversity and Linkage Disequilibrium Analysis
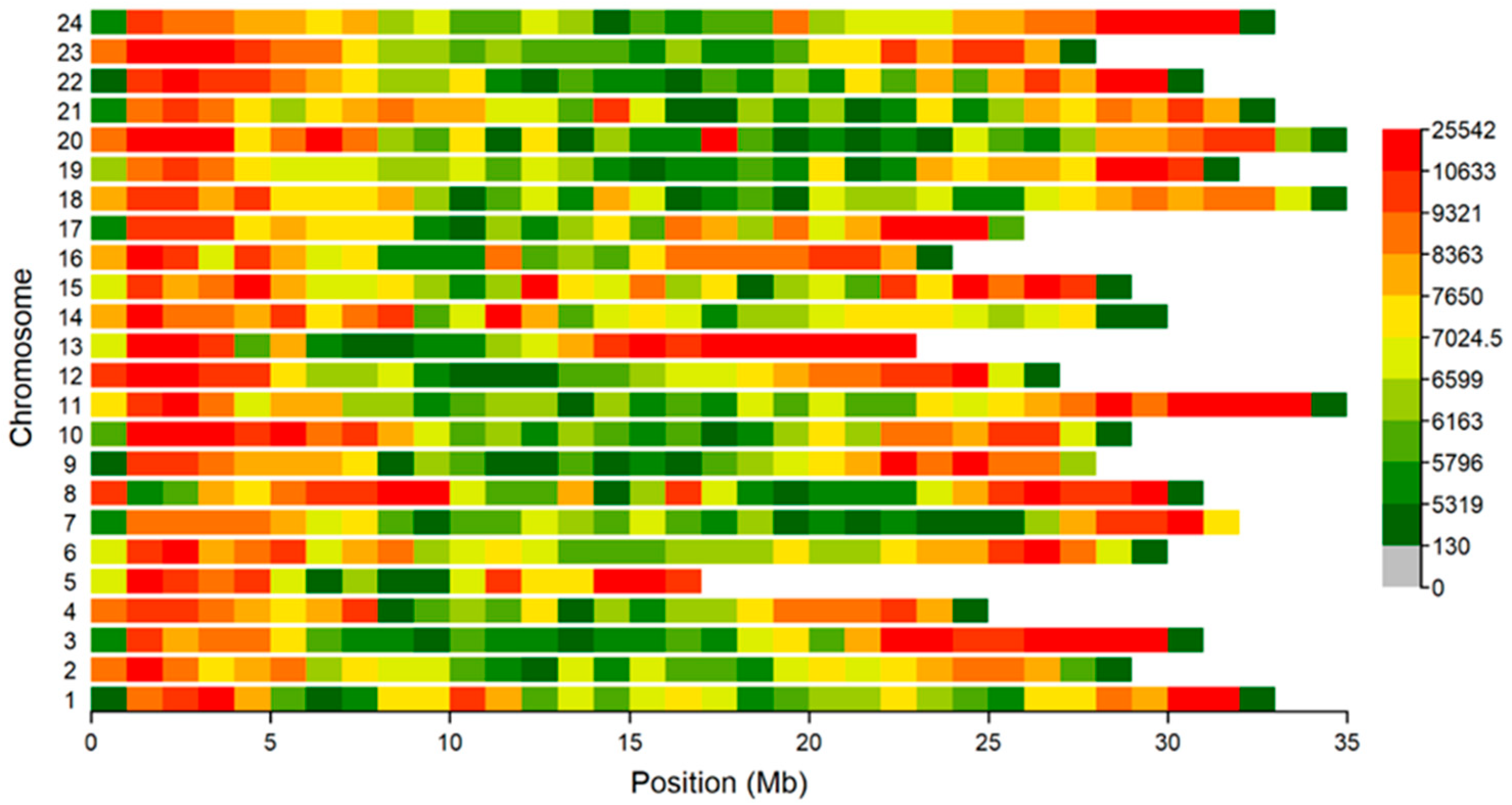
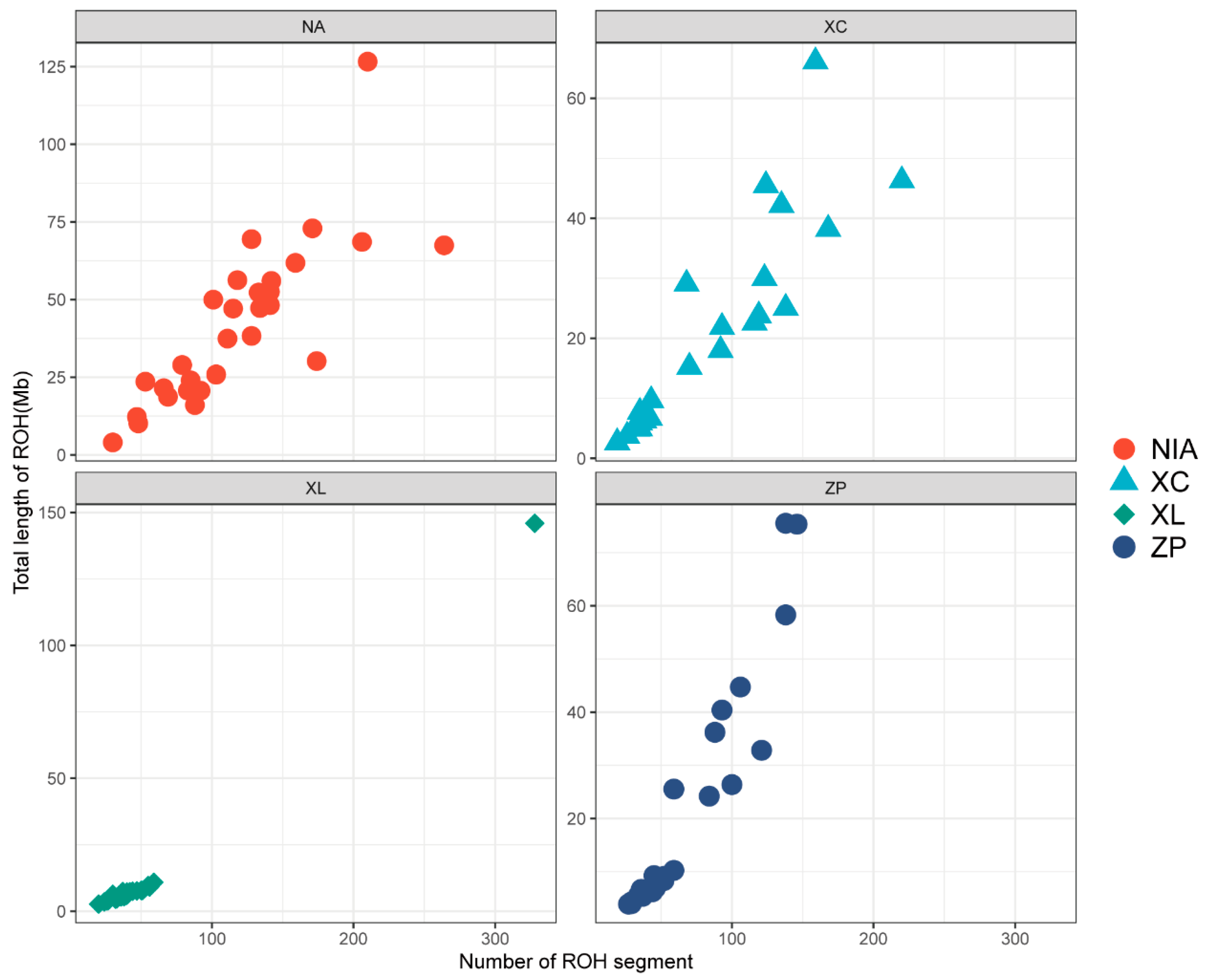
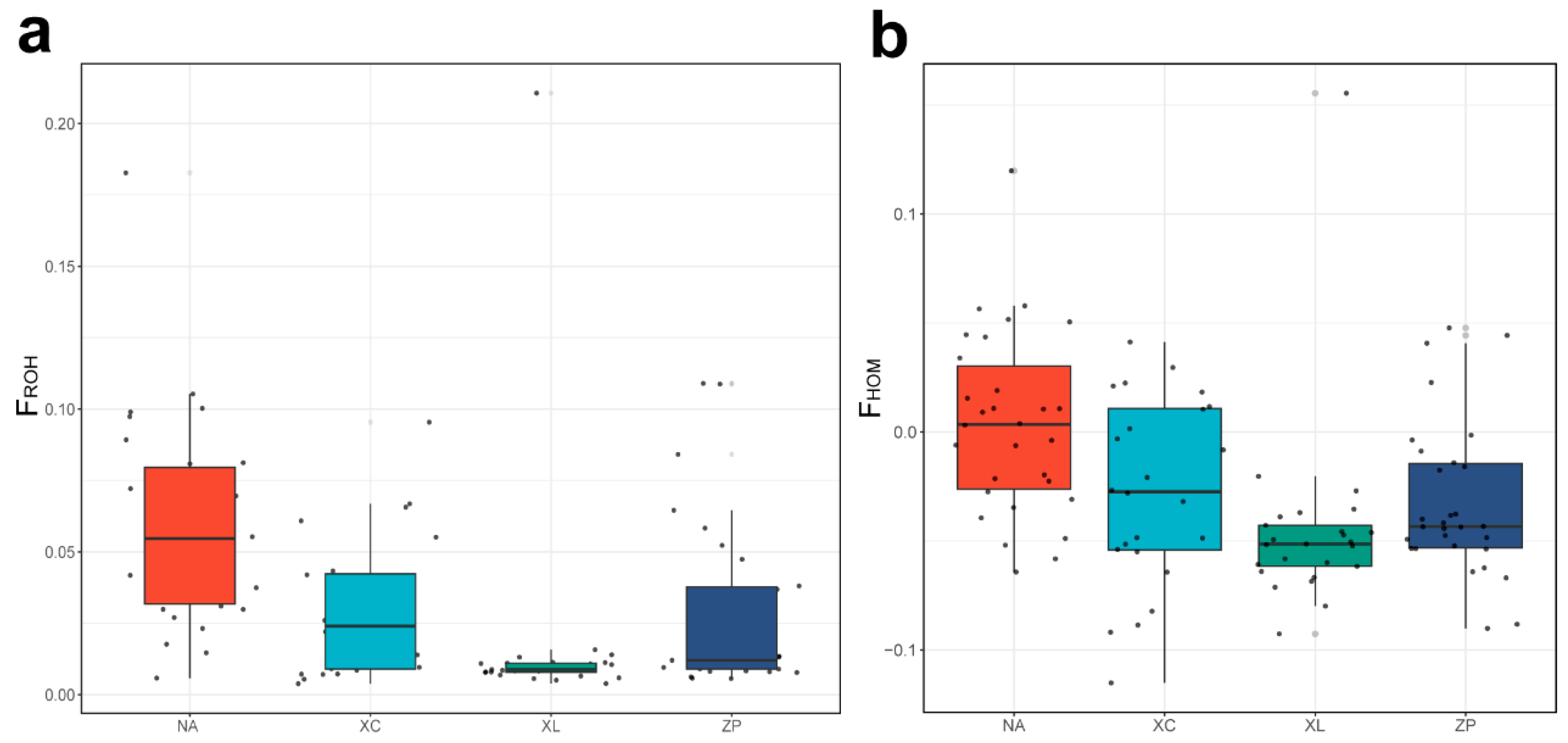
3.3. Genomic Distribution of ROH and Inbreeding Coefficients
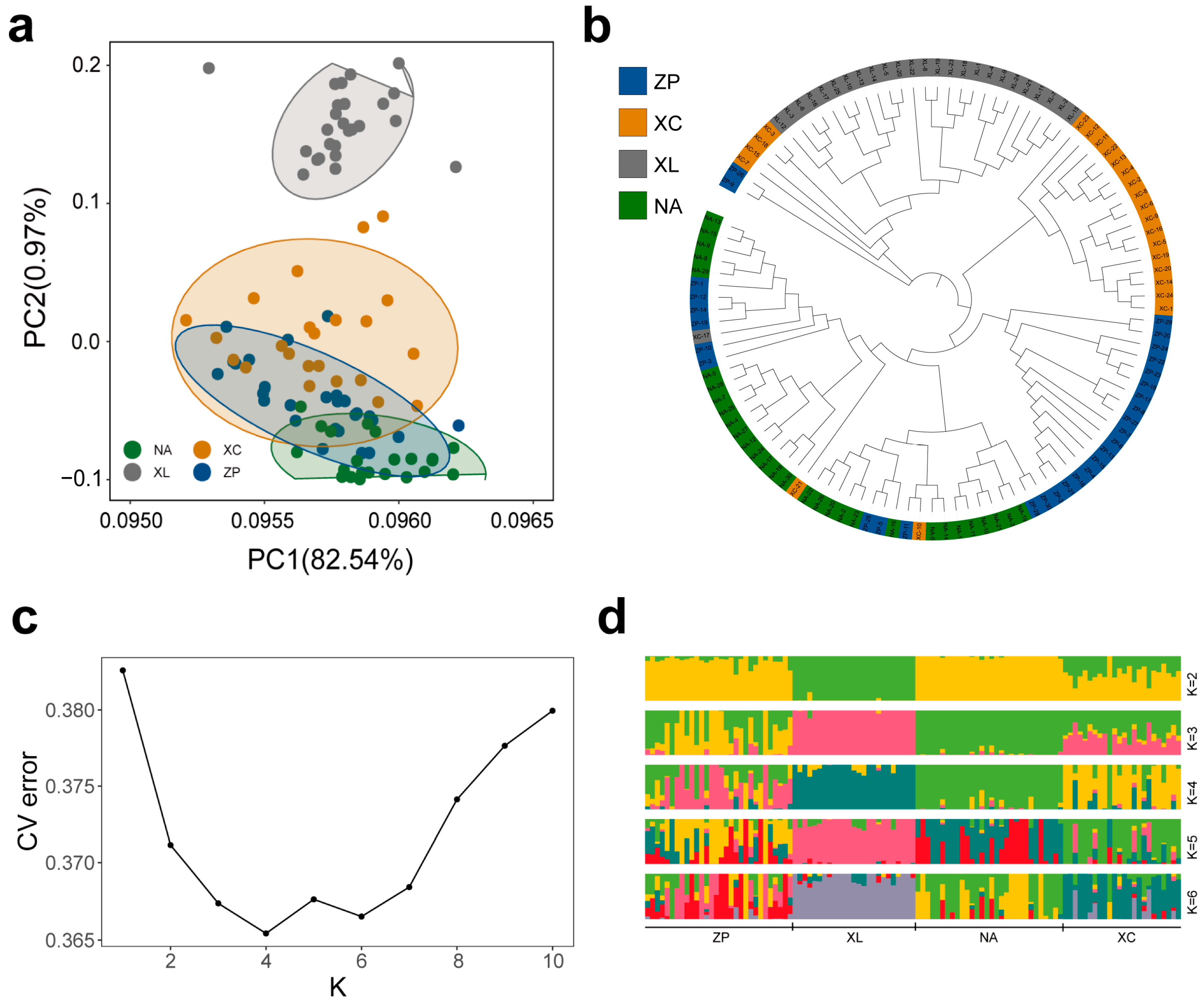
3.4. Population Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, H.; Liu, M.; Gao, J.; Zhu, K.; Liu, B.; Guo, L.; Zhang, N.; Sun, J.; Zeng, C.; Yang, J.; et al. Development of vertebral column and appendicular skeleton in larvae and juveniles of (Platax teira). South China Fish. Sci. 2022, 18, 93–99. [Google Scholar] [CrossRef]
- Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 1997, 145, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Leu, M.-Y.; Tai, K.-Y.; Meng, P.-J.; Tang, C.-H.; Wang, P.-H.; Tew, K.S. Embryonic, larval and juvenile development of the longfin batfish, Platax teira (Forsskål, 1775) under controlled conditions with special regard to mitigate cannibalism for larviculture. Aquaculture 2018, 493, 204–213. [Google Scholar] [CrossRef]
- Liu, M.; Guo, H.; Gao, J.; Zhu, K.; Liu, B.; Guo, L.; Zhang, N.; Yang, J.; Liu, B.; Zhang, D. Embryonic development and morphological characteristics of larvae and juvenile of (Platax teira). South China Fish. Sci. 2022, 18, 103–111. [Google Scholar] [CrossRef]
- Liu, M.-J.; Gao, J.; Guo, H.-Y.; Zhu, K.-C.; Liu, B.-S.; Zhang, N.; Sun, J.-H.; Zhang, D.-C. Transcriptomics Reveal the Effects of Breeding Temperature on Growth and Metabolism in the Early Developmental Stage of Platax teira. Biology 2023, 12, 1161. [Google Scholar] [CrossRef]
- Liu, B.; Guo, H.-Y.; Zhu, K.-C.; Liu, B.-S.; Guo, L.; Zhang, N.; Jiang, S.-G.; Zhang, D.-C. Nutritional compositions in different parts of muscle in the longfin batfish, Platax teira (Forsskål, 1775). J. Appl. Anim. Res. 2019, 47, 403–407. [Google Scholar] [CrossRef]
- Chiu, P.-S.; Chu, Y.-T.; Huang, C.-H.; Ho, S.-W.; Huang, J.-W.; Yeh, S.-L. Effects of stocking density on growth performance, survival and size heterogeneity of juvenile longfin batfish Platax teira. Aquac. Res. 2020, 51, 5269–5272. [Google Scholar] [CrossRef]
- Li, S.; Xie, Z.; Chen, P.; Tang, J.; Tang, L.; Chen, H.; Wang, D.; Zhang, Y.; Lin, H. The complete mitochondrial genome of the Platax teira (Osteichthyes: Ephippidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 796–797. [Google Scholar] [CrossRef]
- An, H.S.; Byun, S.G.; Kim, Y.C.; Lee, J.W.; Myeong, J.I. Wild and hatchery populations of Korean starry flounder (Platichthys stellatus) compared using microsatellite DNA markers. Int. J. Mol. Sci. 2011, 12, 9189–9202. [Google Scholar] [CrossRef]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Lind, C.E.; Evans, B.S.; Knauer, J.; Taylor, J.J.U.; Jerry, D.R. Decreased genetic diversity and a reduced effective population size in cultured silver-lipped pearl oysters (Pinctada maxima). Aquaculture 2009, 286, 12–19. [Google Scholar] [CrossRef]
- Dong, Y.; Li, Q.; Zhong, X.; Kong, L. Development of Gene-derived SNP Markers and Their Application for the Assessment of Genetic Diversity in Wild and Cultured Populations in Sea Cucumber, Apostichopus japonicus. J. World Aquac. Soc. 2016, 47, 873–888. [Google Scholar] [CrossRef]
- Yu, D.; Wang, H.; Gu, W.; Qin, T.; Zheng, H. Genetic diversity and population structure of popcorn germplasm resources using genome-wide SNPs through genotyping-by-sequencing. Genet. Resour. Crop Evol. 2021, 68, 2379–2389. [Google Scholar] [CrossRef]
- Vandeputte, M.; Gagnaire, P.A.; Allal, F. The European sea bass: A key marine fish model in the wild and in aquaculture. Anim. Genet. 2019, 50, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Meng, Z.; Liu, X.; Zhang, Y.; Lin, H. Genetic Diversity an A program for annotating ad Differentiation of the Orange-Spotted Grouper (Epinephelus coioides) Between and Within Cultured Stocks and Wild Populations Inferred from Microsatellite DNA Analysis. Int. J. Mol. Sci. 2011, 12, 4378–4394. [Google Scholar] [CrossRef]
- Cádiz, M.I.; López, M.E.; Díaz-Domínguez, D.; Cáceres, G.; Yoshida, G.M.; Gomez-Uchida, D.; Yáñez, J.M. Whole genome re-sequencing reveals recent signatures of selection in three strains of farmed Nile tilapia (Oreochromis niloticus). Sci. Rep. 2020, 10, 11514. [Google Scholar] [CrossRef]
- Longo, A.; Kurta, K.; Vanhala, T.; Jeuthe, H.; de Koning, D.-J.; Palaiokostas, C. Genetic diversity patterns in farmed rainbow trout (Oncorhynchus mykiss) populations using genome-wide SNP and haplotype data. Anim. Genet. 2024, 55, 87–98. [Google Scholar] [CrossRef]
- Tiangen Biotech. TIANamp Marine Animals DNA Kit Handbook; Tiangen Biotech (Beijing) Co., Ltd.: Beijing, China; Available online: https://www.tiangen.com/product/ (accessed on 15 October 2025).
- Ouyang, Y.; Pan, J.; Xian, L.; Liu, B.; Guo, H.; Zhu, T.; Zhang, N.; Zhu, K.; Zhang, D. Chromosome-level genome and characteristic analysis of (Platax teira). South China Fish. Sci. 2024, 20, 31–42. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, F.C.; Hazelhurst, S.; Ramsay, M. Assessing runs of Homozygosity: A comparison of SNP Array and whole genome sequence low coverage data. BMC Genom. 2018, 19, 106. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-Efficient, Visualization-Enhanced, and Parallel-Accelerated Tool for Genome-Wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Sun, X.; Guo, J.; Li, R.; Zhang, H.; Zhang, Y.; Liu, G.E.; Emu, Q.; Zhang, H. Whole-Genome Resequencing Reveals Genetic Diversity and Wool Trait-Related Genes in Liangshan Semi-Fine-Wool Sheep. Animals 2024, 14, 444. [Google Scholar] [CrossRef]
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. detectRUNS: An R Package to Detect Runs of Homozygosity and Heterozygosity in Diploid Genomes; R foundation: Vienna, Austria, 2019. [Google Scholar]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Rousset, F. genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Xu, L.; Wang, J.X.; Yue, Z.; Jing, Y.; Tai, S.; Yang, J.; Fang, X. VCF2PCACluster: A simple, fast and memory-efficient tool forprincipal component analysis of tens of millions of SNPs. BMC Bioinform. 2024, 25, 173. [Google Scholar] [CrossRef]
- Behr, A.A.; Liu, K.Z.; Liu-Fang, G.; Nakka, P.; Ramachandran, S. pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 2016, 32, 2817–2823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, Z.; Luo, L.; Gu, S.; Hu, Z.; Wan, S.; Gao, Z. Genetic Diversity and Population Structure of Coilia nasus Revealed by 2b-RAD Sequencing. Biology 2023, 12, 600. [Google Scholar] [CrossRef]
- Villanueva, B.; Fernández, A.; Peiró-Pastor, R.; Peñaloza, C.; Houston, R.D.; Sonesson, A.K.; Tsigenopoulos, C.S.; Bargelloni, L.; Gamsız, K.; Karahan, B.; et al. Population structure and genetic variability in wild and farmed Mediterranean populations of gilthead seabream and European seabass inferred from a 60K combined species SNP array. Aquac. Rep. 2022, 24, 101145. [Google Scholar] [CrossRef]
- Caballero, A. Quantitative Genetics; Cambridge University Press: Cambridge, UK, 2020. [Google Scholar]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.A.; Carim, K.J.; Kovach, R.; Eby, L.A.; Barfoot, C.; Painter, S.; Lodmell, A.; Amish, S.J.; Smith, S.; Rosenthal, L.; et al. Genomic Insights Into Inbreeding and Adaptive Divergence of Trout Populations to Inform Genetic Rescue. Evol. Appl. 2025, 18, e70090. [Google Scholar] [CrossRef]
- Kajungiro, R.A.; Palaiokostas, C.; Pinto, F.A.L.; Mmochi, A.J.; Mtolera, M.; Houston, R.D.; de Koning, D.J. Population Structure and Genetic Diversity of Nile Tilapia (Oreochromis niloticus) Strains Cultured in Tanzania. Front. Genet. 2019, 10, 1269. [Google Scholar] [CrossRef]
- Houston, R.D.; Bean, T.P.; Macqueen, D.J.; Gundappa, M.K.; Jin, Y.H.; Jenkins, T.L.; Selly, S.L.C.; Martin, S.A.M.; Stevens, J.R.; Santos, E.M.; et al. Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 2020, 21, 389–409. [Google Scholar] [CrossRef] [PubMed]
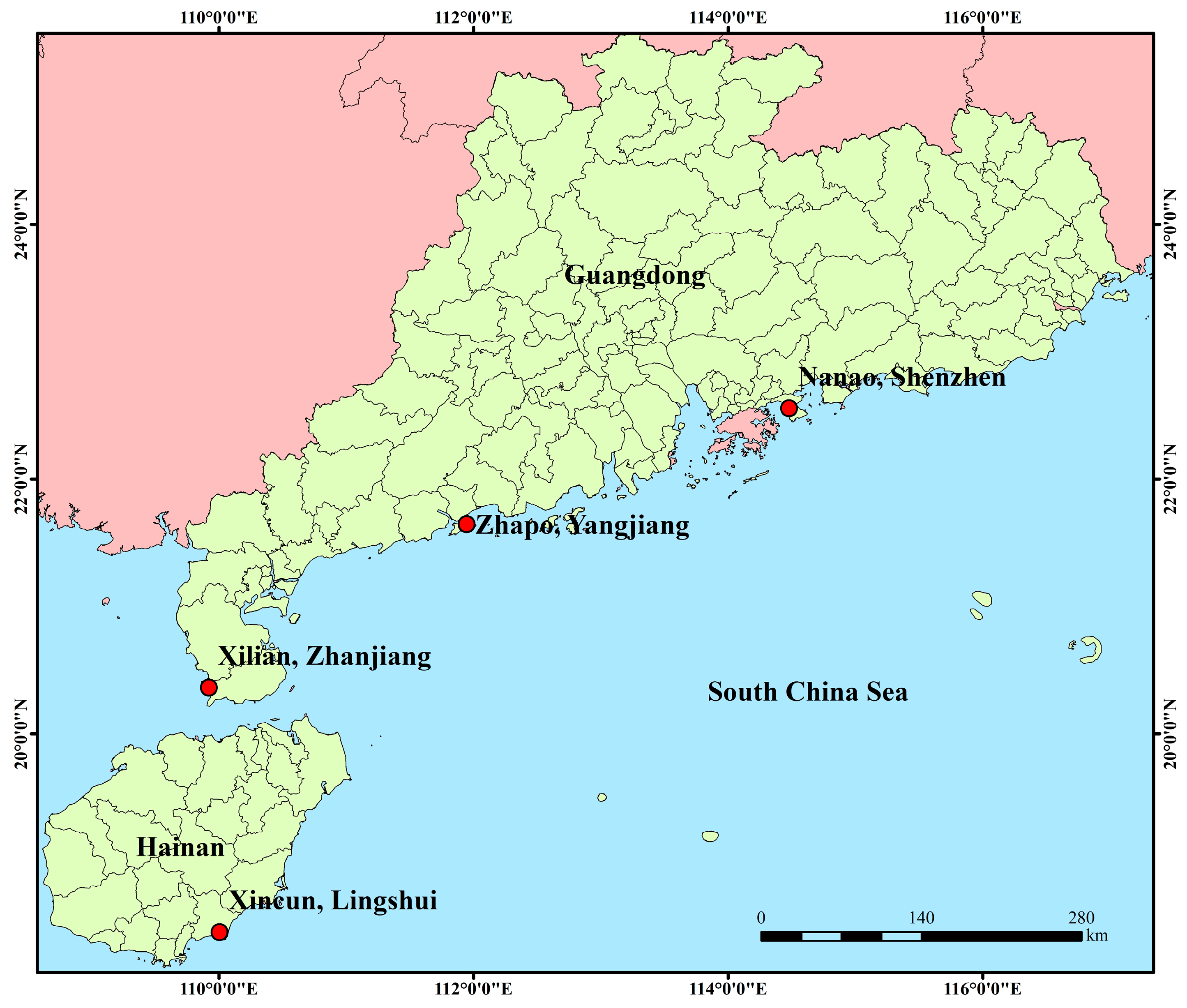
| Location | Abbreviation | Coordinate | Sample Size |
|---|---|---|---|
| Nanao, Shenzhen | NA | 114.31° E, 22.33° N | 30 |
| Zhapo, Yangjiang | ZP | 111.80° E, 21.57° N | 30 |
| Xilian, Zhanjiang | XL | 109.90° E, 20.42° N | 25 |
| Xincun, Lingshui | XC | 109.97° E, 18.40° N | 24 |
| Genomic Region | Count | Percentage (%) |
|---|---|---|
| Intergenic | 1,655,036 | 30.74 |
| Upstream | 762,891 | 14.17 |
| Intro | 2,189,178 | 40.66 |
| Exon | 207,637 | 3.86 |
| Splice | 19,968 | 0.37 |
| Downstream | 549,319 | 10.20 |
| Total | 5,384,029 | 100 |
| Population | SNP (Num) | Ho | He | PIC | Hardy–Weinberg (HWP) | π | FROH | FHOM |
|---|---|---|---|---|---|---|---|---|
| NA | 4,225,699 | 0.269 | 0.257 | 0.164 | 0.7744 | 0.00158 | 0.059 | 0.003 |
| ZP | 4,667,155 | 0.253 | 0.242 | 0.172 | 0.7876 | 0.00164 | 0.028 | −0.030 |
| XL | 4,224,080 | 0.282 | 0.252 | 0.160 | 0.7709 | 0.00155 | 0.017 | −0.034 |
| XC | 4,625,229 | 0.254 | 0.245 | 0.171 | 0.8025 | 0.00165 | 0.029 | −0.052 |
| Count | Proportion (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Insert Size (MB) | NA | XC | XL | ZP | NA | XC | XL | ZP |
| 0.1–0.3 MB | 2406 | 1615 | 1065 | 1473 | 68.3 | 80.8 | 84.3 | 77.5 |
| 0.3–0.6 MB | 620 | 263 | 128 | 226 | 17.6 | 13.2 | 10.1 | 11.9 |
| 0.6–1 MB | 254 | 83 | 40 | 88 | 7.2 | 4.2 | 3.2 | 4.6 |
| 1–2 MB | 195 | 35 | 24 | 88 | 5.5 | 1.8 | 1.9 | 4.6 |
| >2 MB | 47 | 4 | 7 | 25 | 1.3 | 0.2 | 0.6 | 1.3 |
| Total | 3522 | 2000 | 1264 | 1900 | ||||
| Population | NA | ZP | XL | XC |
|---|---|---|---|---|
| NA | - | 0.026 | 0.067 | 0.029 |
| ZP | 0.026 | - | 0.052 | 0.021 |
| XL | 0.065 | 0.051 | - | 0.040 |
| XC | 0.029 | 0.021 | 0.039 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Liu, B.; Guo, H.; Zhu, K.; Xian, L.; Zhang, N.; Zhu, T.; Zhang, D. Genetic Diversity and Population Structure of Farmed Longfin Batfish (Platax teira) in the South China Sea. Genes 2025, 16, 1254. https://doi.org/10.3390/genes16111254
Gao Y, Liu B, Guo H, Zhu K, Xian L, Zhang N, Zhu T, Zhang D. Genetic Diversity and Population Structure of Farmed Longfin Batfish (Platax teira) in the South China Sea. Genes. 2025; 16(11):1254. https://doi.org/10.3390/genes16111254
Chicago/Turabian StyleGao, Yayang, Baosuo Liu, Huayang Guo, Kecheng Zhu, Lin Xian, Nan Zhang, Tengfei Zhu, and Dianchang Zhang. 2025. "Genetic Diversity and Population Structure of Farmed Longfin Batfish (Platax teira) in the South China Sea" Genes 16, no. 11: 1254. https://doi.org/10.3390/genes16111254
APA StyleGao, Y., Liu, B., Guo, H., Zhu, K., Xian, L., Zhang, N., Zhu, T., & Zhang, D. (2025). Genetic Diversity and Population Structure of Farmed Longfin Batfish (Platax teira) in the South China Sea. Genes, 16(11), 1254. https://doi.org/10.3390/genes16111254








