Examining the Concordance of Detection of Hereditary Cancer Gene Variants Between Blood, Tumour, and Normal Tissue in Patients with High-Grade Serous Ovarian Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Preparation and Testing
2.3. Hereditary Cancer Panel
2.4. Data Analysis
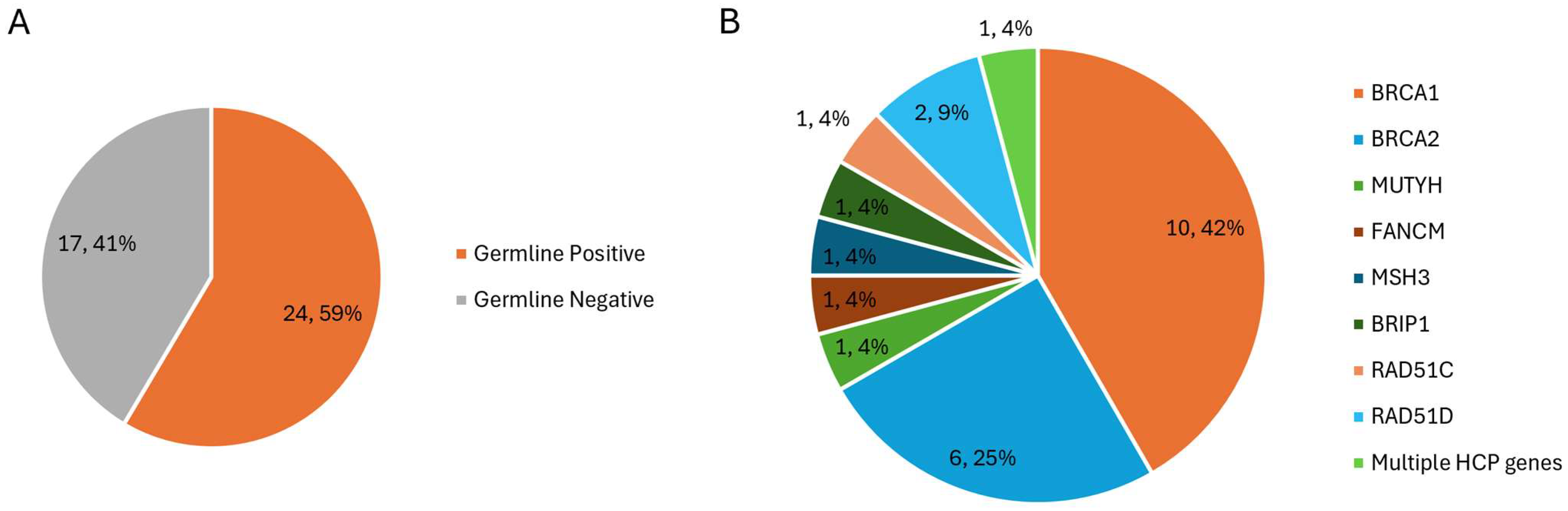
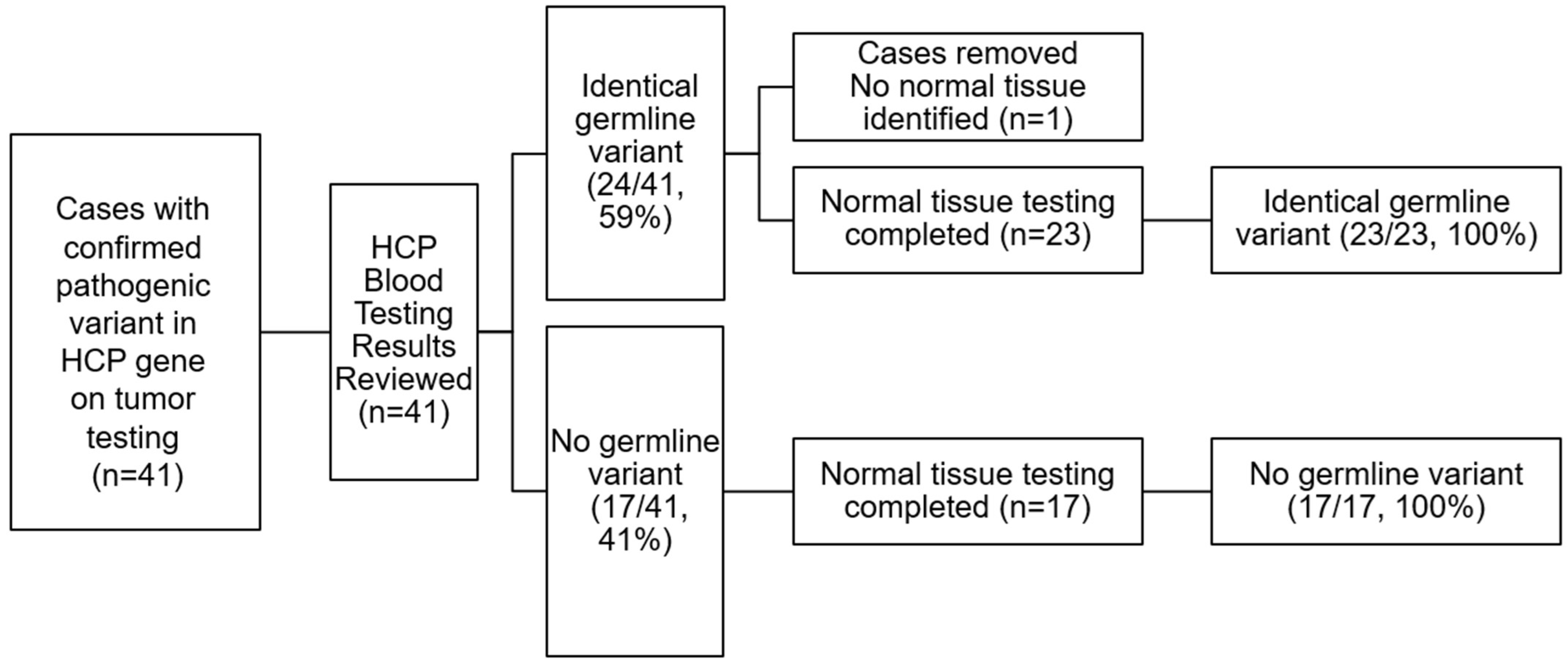
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Konstantinopoulos, P.A.; Norquist, B.; Lacchetti, C.; Armstrong, D.; Grisham, R.N.; Goodfellow, P.J.; Kohn, E.C.; Levine, D.A.; Liu, J.F.; Lu, K.H.; et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J. Clin. Oncol. 2020, 38, 1222–1245. [Google Scholar] [CrossRef]
- Capoluongo, E.; Ellison, G.; López-Guerrero, J.A.; Penault-Llorca, F.; Ligtenberg, M.J.; Banerjee, S.; Singer, C.; Friedman, E.; Markiefka, B.; Schirmacher, P.; et al. Guidance Statement on BRCA1/2 Tumor Testing in Ovarian Cancer Patients. Semin. Oncol. 2017, 44, 189–197. [Google Scholar] [CrossRef]
- Metcalfe, K.A.; Fan, I.; McLaughlin, J.; Risch, H.A.; Rosen, B.; Murphy, J.; Bradley, L.; Armel, S.; Sun, P.; Narod, S.A. Uptake of clinical genetic testing for ovarian cancer in Ontario: A population-based study. Gynecol. Oncol. 2009, 112, 68–72. [Google Scholar] [CrossRef]
- McGee, J.; Panabaker, K.; Leonard, S.; Ainsworth, P.; Elit, L.; Shariff, S.Z. Genetics Consultation Rates Following a Diagnosis of High-Grade Serous Ovarian Carcinoma in the Canadian Province of Ontario. Int. J. Gynecol. Cancer 2017, 27, 437–443. [Google Scholar] [CrossRef]
- McCuaig, J.M.; Stockley, T.L.; Shaw, P.; Fung-Kee-Fung, M.; Altman, A.D.; Bentley, J.; Bernardini, M.Q.; Cormier, B.; Hirte, H.; Kieser, K.; et al. Evolution of genetic assessment for BRCA-associated gynaecologic malignancies: A Canadian multisociety roadmap. J. Med. Genet. 2018, 55, 571–577. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, M.M.; Ruano, D.; van Eijk, R.; van der Stoep, N.; Nielsen, M.; Wijnen, J.T.; Ter Haar, N.T.; Baalbergen, A.; Bos, M.E.; Kagie, M.J.; et al. Validation and Implementation of BRCA1/2 Variant Screening in Ovarian Tumor Tissue. J. Mol. Diagn. 2018, 20, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Goebel, E.A.; Kerkhof, J.; Dzyubak, O.; McLachlin, C.M.; McGee, J.; Sadikovic, B. Examining the Diagnostic Yield of Tumour Testing and Qualifying Germline Concordance for Hereditary Cancer Variants in Patients with High-Grade Serous Carcinoma. Genes 2022, 13, 1398. [Google Scholar] [CrossRef]
- Levy, M.A.; Kerkhof, J.; Belmonte, F.R.; Kaufman, B.A.; Bhai, P.; Brady, L.; Bursztyn, L.L.; Tarnopolsky, M.; Rupar, T.; Sadikovic, B. Validation and clinical performance of a combined nuclear-mitochondrial next-generation sequencing and copy number variant analysis panel in a Canadian population. Am. J. Med. Genet. Part A 2021, 185, 486–499. [Google Scholar] [CrossRef] [PubMed]
- Kerkhof, J.; Schenkel, L.C.; Reilly, J.; McRobbie, S.; Aref-Eshghi, E.; Stuart, A.; Rupar, C.A.; Adams, P.; Hegele, R.A.; Lin, H.; et al. Clinical Validation of Copy Number Variant Detection from Targeted Next-Generation Sequencing Panels. J. Mol. Diagn. 2017, 19, 905–920. [Google Scholar] [CrossRef]
- Aref-Eshghi, E.; McGee, J.D.; Pedro, V.P.; Kerkhof, J.; Stuart, A.; Ainsworth, P.J.; Lin, H.; Volodarsky, M.; McLachlin, C.M.; Sadikovic, B. Genetic and epigenetic profiling of BRCA1/2 in ovarian tumors reveals additive diagnostic yield and evidence of a genomic BRCA1/2 DNA methylation signature. J. Hum. Genet. 2020, 65, 865–873. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Nicolussi, A.; Belardinilli, F.; Mahdavian, Y.; Colicchia, V.; D’iNzeo, S.; Petroni, M.; Zani, M.; Ferraro, S.; Valentini, V.; Ottini, L.; et al. Next-generation sequencing of BRCA1 and BRCA2 genes for rapid detection of germline mutations in hereditary breast/ovarian cancer. PeerJ 2019, 7, e6661. [Google Scholar] [CrossRef]
- Feliubadaló, L.; Lopez-Doriga, A.; Castellsagué, E.; del Valle, J.; Menéndez, M.; Tornero, E.; Montes, E.; Cuesta, R.; Gómez, C.; Campos, O.; et al. Next-generation sequencing meets genetic diagnostics: Development of a comprehensive workflow for the analysis of BRCA1 and BRCA2 genes. Eur. J. Hum. Genet. 2013, 21, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.; Amemiya, Y.; Arts, H.; Bayani, J.; Eng, B.; Grafodatskaya, D.; Reid, S.K.; Lariviere, M.; Lo, B.; McClure, R.; et al. Multisite verification of the accuracy of a multi-gene next generation sequencing panel for detection of mutations and copy number alterations in solid tumours. PLoS ONE 2021, 16, e0258188. [Google Scholar] [CrossRef] [PubMed]
- Zannini, G.; Facchini, G.; De Sio, M.; De Vita, F.; Pagliuca, F.; Franco, R.; Marino, F.Z. BRCA1 and BRCA2 mutations testing in prostate cancer: Detection in formalin fixed paraffin embedded (FFPE) and blood samples. Pathol.-Res. Pract. 2025, 266, 155803. [Google Scholar] [CrossRef] [PubMed]
- Zannini, G.; Facchini, G.; De Sio, M.; De Vita, F.; Ronchi, A.; Orditura, M.; Vietri, M.T.; Ciardiello, F.; Franco, R.; Accardo, M.; et al. Implementation of BRCA mutations testing in formalin-fixed paraffin-embedded (FFPE) samples of different cancer types. Pathol. Res. Pract. 2023, 243, 154336. [Google Scholar] [CrossRef]
- Hertz, D.L.; Kidwell, K.M.; Thibert, J.N.; Gersch, C.; Regan, M.M.; Skaar, T.C.; Henry, N.L.; Hayes, D.F.; Van Poznak, C.H.; Rae, J.M. Genotyping concordance in DNA extracted from formalin-fixed paraffin embedded (FFPE) breast tumor and whole blood for pharmacogenetic analyses. Mol. Oncol. 2015, 9, 1868–1876. [Google Scholar] [CrossRef]
- Bhai, P.; Turowec, J.; Santos, S.; Kerkhof, J.; Pickard, L.; Foroutan, A.; Breadner, D.; Cecchini, M.; Levy, M.A.; Stuart, A.; et al. Molecular profiling of solid tumors by next-generation sequencing: An experience from a clinical laboratory. Front. Oncol. 2023, 13, 1208244. [Google Scholar] [CrossRef]
- Wang, M.; Chen, X.; Dai, Y.; Wu, D.; Liu, F.; Yang, Z.; Song, B.; Xie, L.; Yang, L.; Zhao, W.; et al. Concordance Study of a 520-Gene Next-Generation Sequencing-Based Genomic Profiling Assay of Tissue and Plasma Samples. Mol. Diagn. Ther. 2022, 26, 309–322. [Google Scholar] [CrossRef]
- Wilkins, A.; Chauhan, R.; Rust, A.; Pearson, A.; Daley, F.; Manodoro, F.; Fenwick, K.; Bliss, J.; Yarnold, J.; Somaiah, N. FFPE breast tumour blocks provide reliable sources of both germline and malignant DNA for investigation of genetic determinants of individual tumour responses to treatment. Breast Cancer Res. Treat. 2018, 170, 573–581. [Google Scholar] [CrossRef]
- Oh, E.; Choi, Y.-L.; Kwon, M.J.; Kim, R.N.; Kim, Y.J.; Song, J.-Y.; Jung, K.S.; Shin, Y.K. Comparison of accuracy of whole-exome sequencing with formalin-fixed paraffin-embedded and fresh frozen tissue samples. PLoS ONE 2015, 10, e0144162. [Google Scholar] [CrossRef] [PubMed]
- Youssef, O.; Almangush, A.; Zidi, Y.H.; Loukola, A.; Carpén, O. Nonmalignant Formalin-Fixed Paraffin-Embedded Tissues as a Source to Study Germline Variants and Cancer Predisposition: A Systematic Review. Biopreservation Biobanking 2020, 18, 337–345. [Google Scholar] [CrossRef]
- Spencer, D.H.; Sehn, J.K.; Abel, H.J.; Watson, M.A.; Pfeifer, J.D.; Duncavage, E.J. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J. Mol. Diagn. 2013, 15, 623–633. [Google Scholar] [CrossRef]
- Marsh, S.; Mallon, M.A.; Goodfellow, P.; McLeod, H.L. Concordance of pharmacogenetic markers in germline and colorectal tumor DNA. Pharmacogenomics 2005, 6, 873–877. [Google Scholar] [CrossRef]
- Yap, S.Q. Germline Variant Calling in Formalin-Fixed Paraffin-Embedded Tumours. Master’s Thesis, University of British Columbia, Vancouver, BC, Canada, 2017. [Google Scholar] [CrossRef]
- Adank, M.A.; Brogi, E.; Bogomolniy, F.; Wadsworth, E.A.; Lafaro, K.J.; Yee, C.J.; Kirchhoff, T.; Meijers-Heijboer, E.J.; Kauff, N.D.; Boyd, J.; et al. Accuracy of BRCA1 and BRCA2 founder mutation analysis in formalin-fixed and paraffin-embedded (FFPE) tissue. Fam. Cancer 2006, 5, 337–342. [Google Scholar] [CrossRef]
- Cannon-Albright, L.A.; Cooper, K.G.; Georgelas, A.; Bernard, P.S. High quality and quantity Genome-wide germline genotypes from FFPE normal tissue. BMC Res. Notes 2011, 4, 159. [Google Scholar] [CrossRef]
- Sjöholm, M.I.; Hoffmann, G.; Lindgren, S.; Dillner, J.; Carlson, J. Comparison of archival plasma and formalin-fixed paraffin-embedded tissue for genotyping in hepatocellular carcinoma. Cancer Epidemiol. Biomark. Prev. 2005, 14, 251–255. [Google Scholar] [CrossRef]
- McGee, J.; Peart, T.M.; Foley, N.; Bertrand, M.; Prefontaine, M.; Sugimoto, A.; Ettler, H.; Welch, S.; Panabaker, K. Direct Genetics Referral Pathway for High-Grade Serous Ovarian Cancer Patients: The ‘opt-Out’ Process. J. Oncol. 2019, 2019, 1–7. [Google Scholar] [CrossRef]
- Petersen, A.H.; Aagaard, M.M.; Nielsen, H.R.; Steffensen, K.D.; Waldstrøm, M.; Bojesen, A. Post-mortem testing; germline BRCA1/2 variant detection using archival FFPE non-tumor tissue. A new paradigm in genetic counseling. Eur. J. Hum. Genet. 2016, 24, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Canadian Association of Pathologists. The Retention and Use of Human Biologic Material. 2014. Available online: https://cap-acp.org/general/custom.asp?page=The-Retention-and-Use-of-Human-Biologic-Material (accessed on 24 October 2024).
- Wong, C.; DiCioccio, R.A.; Allen, H.J.; Werness, B.A.; Piver, M. Mutations in BRCA1 from fixed, paraffin-embedded tissue can be artifacts of preservation. Cancer Genet. Cytogenet. 1998, 107, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Ademà, V.; Torres, E.; Solé, F.; Serrano, S.; Bellosillo, B. Paraffin Treasures: Do They Last Forever? Biopreservation Biobanking 2014, 12, 281–283. [Google Scholar] [CrossRef] [PubMed]
| Patient | Reportable Variant Detected in Tumour | Variant Detected in Blood | Variant Detected in Normal Tissue |
|---|---|---|---|
| 1 | NM_007294.3(BRCA1):c.(4185+21_4186-21)_(4357+21_4358-21)del (50%) | Yes | Yes |
| 2 | NM_000059.3(BRCA2):c.7069_7070delCT, p.(Leu2357Valfs*2) (18.7%) | Yes | Yes |
| 3 | NM_000059.3(BRCA2):c.3545_3546delTT, p.(Phe1182*) (34%) | Yes | Yes |
| 4 | NM_007294.3(BRCA1):c.4327C>T, p.(Arg1443*) (46.1%) | Yes | Yes |
| 5 | NM_007294.3(BRCA1):c.3748G>T, p.(Glu1250*) (52%) | Yes | Yes |
| 6 | NM_000059.3(BRCA2):c.7988A>T, p.(Glu2663Val) (55.7%) | Yes | Yes |
| 7 | NM_007294.3(BRCA1):c.2071delA, p.(Arg691Aspfs*10) (66.5%) | Yes | Yes |
| 8 | NM_000059.3(BRCA2):c.5303_5304delTT, p.(Leu1768Argfs*5) (76%) | Yes | Yes |
| 9 | NM_007294.3(BRCA1):c.4689C>G, p.(Tyr1563*) (88.1%) | Yes | Yes |
| 10 | NM_007294.3(BRCA1):c.427G>T, p.(Glu143*) (96.3%) | Yes | Yes |
| 11 | NM_007294.3(BRCA1):c.3254_3255dupGA, p.(Leu1086Aspfs*2) (57.6%) | Yes | Yes |
| 12 | NM_000059.3(BRCA2):c.7958T>C, p.(Leu2653Pro) (99.4%) | Yes | Yes |
| 13 | NM_007294.3(BRCA1):c.5266dupC, p.(Gln1756Profs*74) (58.4%) | Yes | Yes |
| 14 | NM_000059.3(BRCA2):c.3170_3174del, p.(Lys1057Thrfs*8) (71.4%) | Yes | Yes |
| 15 | NM_007294.3(BRCA1):c.68_69delAG, p.(Glu23Valfs*17) (76.3%) | Yes | Yes |
| 16 | NM_000059.3(BRCA2):c.(?_-21)_(*21_?)del (40%) | Yes—MUTYH only | Yes—MUTYH only |
| NM_001128425.1(MUTYH):c.1187G>A, p.(Gly396Asp) (57%) | |||
| 17 | NM_007294.3(BRCA1):c.(?_-21)_(*21_?)del (35%) | Yes—FANCM only | Yes—FANCM only |
| NM_020937.2(FANCM):c.5101C>T, p.(Gln1701*) (52.4%) | |||
| 18 | NM_000059.3(BRCA2):c.8164dupA, p.(Thr2722Asnfs*8) (37.3%) | Yes—MUTYH and RAD51D only | Yes—MUTYH and RAD51D only |
| NM_001128425.1(MUTYH):c.1187G>A, p.(Gly396Asp) (64.4%) | |||
| NM_002878.3(RAD51D):c.620C>T, p.(Ser207Leu) (61%) | |||
| 19 | NM_000314.4(PTEN):c.741dup, p.(Pro248Thrfs*5) (33.5%) | Yes—MSH3 only | Yes—MSH3 only |
| NM_002439.4(MSH3):c.703C>T, p.(Gln235*) (36.2%) | |||
| 20 | NM_032043.2(BRIP1):c.2255_2256del, p.(Lys752Argfs*12) (57.6%) | Yes | Yes |
| 21 | NM_058216.1(RAD51C):c.224dup, p.(Tyr75*) (77.1%) | Yes | Yes |
| 22 | NM_002878.3(RAD51D):c.668-2A>C (82.1%) | Yes | Yes |
| 23 | NM_000077.4(CDKN2A):c.341C>A, p.(Pro114His) (26.6%) | Yes—RAD51D only | Yes—RAD51D only |
| NM_002878.3(RAD51D):c.649_655delinsTGAGGTT, p.(Gly217*) (65%) | |||
| 24 | NM_007294.3(BRCA1):c.212+3A>G (96.7%) | Yes | No Result |
| 25 | NM_007294.3(BRCA1):c.(?_-21)_(*21_?)del (25%) | No | No |
| 26 | NM_000059.3(BRCA2):c.(?_-21)_(*21_?)del (35%) | No | No |
| 27 | NM_007294.3(BRCA1):c.(?_-21)_(*21_?)del (50%) | No | No |
| 28 | NM_000059.3(BRCA2):c.(?_-21)_(*21_?)del (50%) | No | No |
| 29 | NM_007294.3(BRCA1):c.(?_-21)_(*21_?)del (55%) | No | No |
| 30 | NM_000059.3(BRCA2):c.(?_-21)_(*21_?)del (40%) | No | No |
| 31 | NM_000059.3(BRCA2):c.(?_-21)_(*21_?)del (50%) | No | No |
| 32 | NM_007294.3(BRCA1):c.3569delC, p.(Pro1190Leufs*20) (11.4%) | No | No |
| 33 | NM_007294.3(BRCA1):c.3339T>A, p.(Tyr1113*) (35%) | No | No |
| 34 | NM_000059.3(BRCA2):c.7758G>A, p.(Trp2586*) (43.2%) | No | No |
| 35 | NM_007294.3(BRCA1):c.1961dupA, p.(Tyr655Valfs*18) (33.9%) | No | No |
| 36 | NM_007294.3(BRCA1):c.4986+3G>T (69.3%) | No | No |
| 37 | NM_007294.3(BRCA1):c.2603C>A, p.(Ser868*) (90.5%) | No | No |
| 38 | NM_007294.3(BRCA1):c.1912G>T, p.(Glu638*) (65.1%) | No | No |
| NM_000059.3(BRCA2):c.(?_-21)_(*21_?)del (30%) | |||
| 39 | NM_007294.3(BRCA1):c.3394_3406del, p.(Asn1132Leufs*19) (70%) | No | No |
| 40 | NM_007294.3(BRCA1):c.2891delG, p.(Gly964Aspfs*36) (58.4%) | No | No |
| 41 | NM_000314.4(PTEN):c.1004G>A, p.(Arg335Gln) (17%) | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mui, L.; Kerkhof, J.; McLachlin, C.M.; Panabaker, K.; McGee, J.; Sadikovic, B.; Goebel, E.A. Examining the Concordance of Detection of Hereditary Cancer Gene Variants Between Blood, Tumour, and Normal Tissue in Patients with High-Grade Serous Ovarian Carcinoma. Genes 2025, 16, 1260. https://doi.org/10.3390/genes16111260
Mui L, Kerkhof J, McLachlin CM, Panabaker K, McGee J, Sadikovic B, Goebel EA. Examining the Concordance of Detection of Hereditary Cancer Gene Variants Between Blood, Tumour, and Normal Tissue in Patients with High-Grade Serous Ovarian Carcinoma. Genes. 2025; 16(11):1260. https://doi.org/10.3390/genes16111260
Chicago/Turabian StyleMui, L., J. Kerkhof, C. M. McLachlin, K. Panabaker, J. McGee, B. Sadikovic, and E. A. Goebel. 2025. "Examining the Concordance of Detection of Hereditary Cancer Gene Variants Between Blood, Tumour, and Normal Tissue in Patients with High-Grade Serous Ovarian Carcinoma" Genes 16, no. 11: 1260. https://doi.org/10.3390/genes16111260
APA StyleMui, L., Kerkhof, J., McLachlin, C. M., Panabaker, K., McGee, J., Sadikovic, B., & Goebel, E. A. (2025). Examining the Concordance of Detection of Hereditary Cancer Gene Variants Between Blood, Tumour, and Normal Tissue in Patients with High-Grade Serous Ovarian Carcinoma. Genes, 16(11), 1260. https://doi.org/10.3390/genes16111260






