Chromosome Morphology and Heterochromatin Patterns in Paspalum notatum: Insights into Polyploid Genome Structure
Highlights
- A detailed karyotype analysis was conducted for the first time on diploid (2n= 2x= 20) and tetraploid (2n= 4x= 40) accessions of Paspalum notatum.
- Constitutive heterochromatin patterns were revealed using CMA-DA-DAPI staining, showing distinct banding differences between diploids and tetraploids.
- Genome size analysis indicated a reduction in monoploid genome size in tetraploids compared to diploids, consistent with an autopolyploid origin.
- The karyotype analysis and heterochromatin patterns provide novel insights into the structure and evolution of the N genome in Paspalum notatum.
- The findings support the hypothesis of an autopolyploid origin of tetraploid cytotypes, with only minor structural changes in the genome following polyploidization.
- These cytogenetic insights can guide future studies integrating genomic and cytogenetic maps, contributing to a deeper understanding of this economically and ecologically significant species.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Chromosome Preparation
2.3. Feulgen Staining and Karyotype Analyses
2.4. Karyotype Morphometry
2.5. Idiograms Construction
2.6. Karyotype Asymmetry
2.7. Chromosome Preparation for Constitutive Heterochromatin Detection
2.8. Chromosome Sequential Tri-Staining with Fluorochromes CMA/DA/DAPI
2.9. Genome Size Measurements
3. Results
3.1. Karyotype of P. notatum
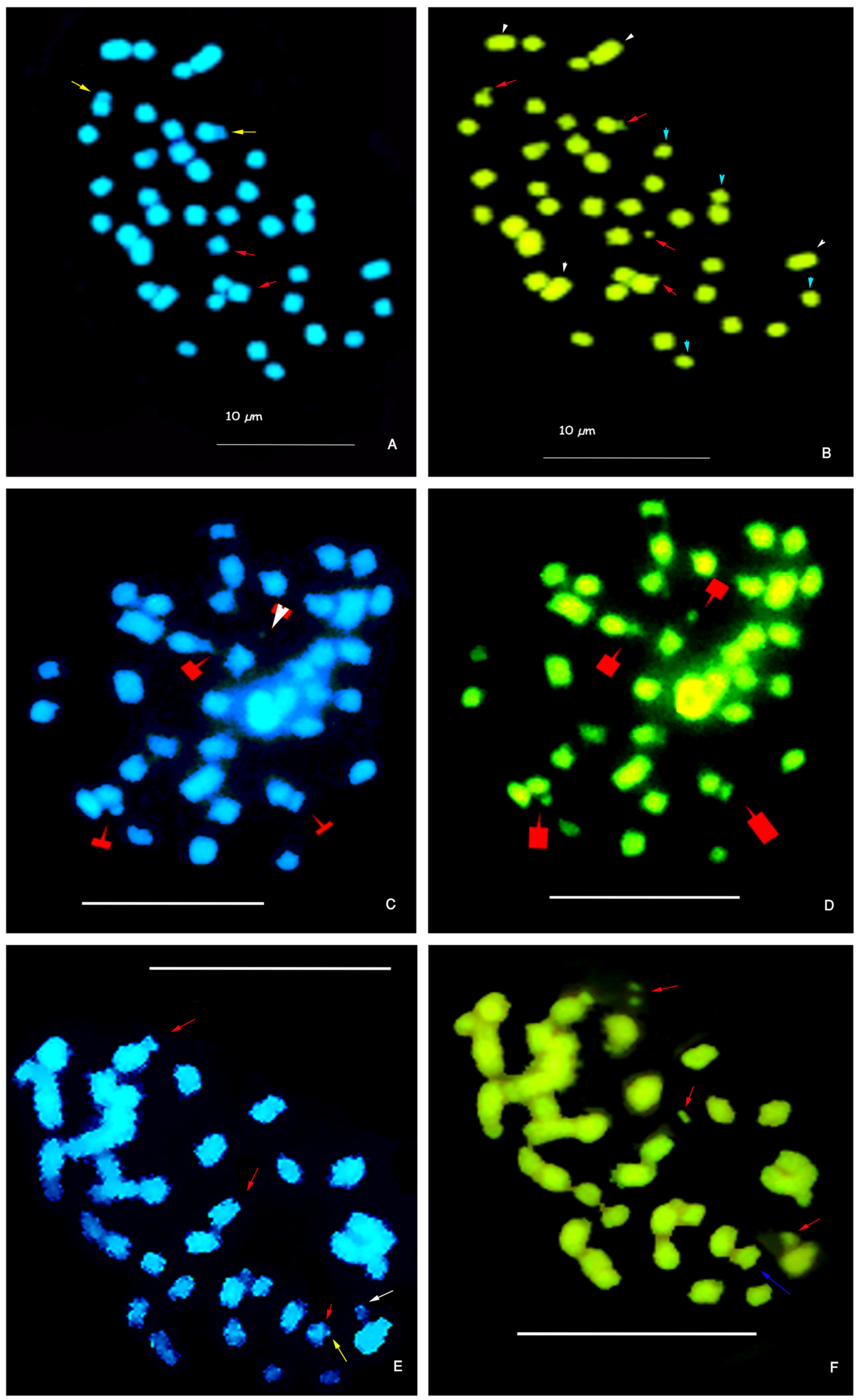
3.2. Constitutive Heterochromatin in Paspalum notatum
3.3. Genome Size of P. notatum
4. Discussion
4.1. Chromosome Evidence of Remodeling and Downsizing of N Genome of Paspalum notatum
4.2. Genome Evidence of Remodeling and Downsizing of N Genome of P. notatum Polyploid Complex
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TCL | Total chromosome length of the complement |
| HTCL | Haploid total chromosome length |
| i | Centromeric Index |
| r | Arm ratio |
| R | Ratio of the shortest to the longest chromosome pair |
| A1 | Intra-karyotypic asymmetry index |
| A2. | Inter-karyotypic asymmetry index |
| CMA | Chromomicyn A3 |
| DA | Distamicyn A |
| DAPI | 4′-6-diamidino-2-phenilindol |
| Het-C | Constitutive heterochromatin |
| C-DNAabs | Absolute DNA content |
| 1Cx | Monoploid genome size |
References
- Bennett, M.D. The genome, the natural karyotype and biosystematics. In Plant Biosystematics; Grant, W.F., Ed.; Academic Press: New York, NY, USA, 1984; pp. 41–66. [Google Scholar]
- Brazier, T.; Glémin, S. Diversity and determinants of recombination landscapes in flowering plants. PLoS Genet. 2022, 18, e1010141. [Google Scholar] [CrossRef] [PubMed]
- Johnston, S.E. Understanding the Genetic Basis of Variation in Meiotic Recombination: Past, Present, and Future. Mol. Biol. Evol. 2024, 41, 7. [Google Scholar] [CrossRef]
- Vimala, Y.; Lavania, S.; Chandra Lavania, U. Chromosome change and karyotype differentiation implications in speciation and plant systematics. Nucleus 2021, 64, 33–54. [Google Scholar] [CrossRef]
- Hamon, P.; Siljak-Yakovlev, S.; Srisuwan, S.; Robin, O.; Poncet, V.; Hamon, S.; de Kochko, A. Physical mapping of rDNA and heterochromatin in chromosomes of 16 Coffea species: A revised view of species differentiation. Chromosome Res. 2009, 17, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.; Barrela, R.M.; Bergès, H.; Marques, C.; Loureiro, J.; Morais-Cecílio, L.; Paiva, J.A.P. Advancing Eucalyptus Genomics: Cytogenomics Reveals Conservation of Eucalyptus Genomes. Front. Plant Sci. 2016, 7, 510. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M. Patterns of heterochromatin distribution in plant chromosomes. Gen. Mol. Biol. 2000, 23, 1029–1041. [Google Scholar] [CrossRef]
- Roa, F.; Guerra, M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- Gianini Aquino, A.C.; González Flores, M.; Honfi, A.I.; Daviña, J.R. Heterochromatin patterns in four diploid Zephyranthes species with different basic chromosome number (Amaryllidaceae). Darwiniana 2023, 11, 705–718. [Google Scholar] [CrossRef]
- Reutemann, A.V.; Martínez, E.J.; Daviña, J.R.; Hojsgaard, D.H.; Honfi, A.I. El cariotipo de Paspalum cromyorrhizon diploide y tetraploide (Poaceae, Panicoideae, Paspaleae). Darwiniana 2021, 9, 375–386. [Google Scholar] [CrossRef]
- Rosselló, J.A.; Maravilla, A.J.; Rosato, M. The nuclear 35S rDNA world in plant systematics and evolution: A primer of cautions and common misconceptions in cytogenetic studies. Front. Plant Sci. 2022, 13, 788911. [Google Scholar] [CrossRef]
- Luo, X.; Liu, Y.; Gong, X.; Ye, M.; Xiao, Q.; Zeng, Z. Karyotype description and comparative chromosomal mapping of 5S rDNA in 42 Species. Genes 2024, 15, 647. [Google Scholar] [CrossRef] [PubMed]
- Las Peñas, M.L.; Urdampilleta, J.D.; Bernardello, G.; Forni-Martins, E.R. Karyotypes, heterochromatin, and physical mapping of 18S-26S rDNA in Cactaceae. Cytogenet. Genome Res. 2009, 124, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Hasterok, R.; Wang, K.; Jenkins, G. Progressive refinement of the karyotyping of Brachypodium genomes. New Phytol. 2020, 227, 1668–1675. [Google Scholar] [CrossRef]
- Galdeano, F.; Urbani, M.H.; Sartor, M.E.; Honfi, A.I.; Espinoza, F.; Quarin, C.L. Relative DNA content in diploid, polyploid, and multiploid species of Paspalum (Poaceae) with relation to reproductive mode and taxonomy. J. Pl. Res. 2016, 129, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J.; Urbiš, P.; Said, M.; Lucretti, S.; Molnár, I. Flow cytometric analysis and sorting of plant chromosomes. Nucleus 2023, 66, 355–369. [Google Scholar] [CrossRef]
- Stace, C.A. Plant Taxonomy and Biosystematics; Edward Arnold Limited: London, UK, 1980; pp. 74–83. [Google Scholar]
- Heng, J.; Heng, H.H. Karyotype coding: The creation and maintenance of system information for complexity and biodiversity. BioSystem 2021, 208, 104476. [Google Scholar] [CrossRef]
- Stebbins, G.L. Types of polyploids: Their classification and significance. Adv. Genet. 1947, 1, 403–429. [Google Scholar] [CrossRef] [PubMed]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar] [CrossRef]
- Lv, Z.; Nyarko, C.A.; Ramtekey, V.; Mason, A.S. Defining autopolyploidy: Cytology, genetics, and taxonomy. Am. J. Bot. 2024, 111, 16292. [Google Scholar] [CrossRef] [PubMed]
- Chalup, L.; Grabiele, M.; Solís Neffa, V.; Seijo, G. Structural karyotypic variability and polyploidy in natural populations of the South American Lathyrus nervosus Lam. (Fabaceae). Plant Syst. Evol. 2012, 298, 761–773. [Google Scholar] [CrossRef]
- Reis, A.C.; Chester, M.; de Sousa, S.M. Chromosomal view of Lippia alba, a tropical polyploid complex under genome stabilization process. Protoplasma 2022, 259, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Giacò, A.; De Giorgi, P.; Astuti, G.; Varaldo, L.; Sáez, L.; Carballal, R.; Serrano, M.; Casazza, G.; Caputo, P.; Bacchetta, G.; et al. Diploids and polyploids in the Santolina chamaecyparissus complex (Asteraceae) show different karyotype asymmetry. Plant Biosyst. 2022, 156, 1237–1246. [Google Scholar] [CrossRef]
- Maluszynska, J.; Heslop-Harrison, J.S. Localization of tandemly repetead DNA sequences in Arabidopsis thaliana. Plant J. 1993, 2, 159–166. [Google Scholar] [CrossRef]
- Sochorová, J.; Coriton, O.; Kuderová, A.; Lunerová, J.; Chèvre, A.M.; Kovařik, A. Gene conversion events and variable degree of homogenization of rDNA loci in cultivars of Brassica napus. Ann. Bot. 2017, 119, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Shi, S.; Shen, H.; Xie, Q.; Li, H. Haplotype-specific chromosome painting provides insights into the chromosomal characteristics in self-duplicating autotetraploid sugarcane. Ind. Crop. Prod. 2023, 202, 117085. [Google Scholar] [CrossRef]
- Sumner, A.T. Chromosome Banding; Unwin Hyman: London, UK, 1990; 434p. [Google Scholar]
- Barros e Silva, A.E.; Guerra, M. The meaning of DAPI bands observed after C-banding and FISH procedures. Biotech. Histochem. 2010, 85, 115–125. [Google Scholar] [CrossRef]
- Chase, A. The North American species of Paspalum. Contr. U. S. Natl. Herb. 1929, 28, 1310. [Google Scholar]
- Acuña, C.A.; Martínez, E.J.; Zilli, A.L.; Brugnoli, E.A.; Espinoza, F.; Marcón, F.; Urbani, M.H.; Quarin, C.L. Reproductive systems in Paspalum: Relevance for germplasm collection and conservation, breeding techniques, and adoption of released cultivars. Front. Plant Sci. 2019, 10, 1377. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, J.P.A.; Pupilli, F.; Acuña, C.A.; Leblanc, O.; Pessino, S.C. How to become an apomixis model: The multifaceted case of Paspalum. Genes 2020, 11, 974. [Google Scholar] [CrossRef]
- Martínez, E.J.; Urbani, M.H.; Quarín, C.L.; Ortiz, J.P.A. Inheritance of apospory in bahiagrass, Paspalum notatum. Hereditas 2001, 135, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Martínez, E.J.; Hopp, H.; Stein, J.; Ortiz, J.P.A.; Quarin, C.L. Genetic characterization of apospory in tetraploid Paspalum notatum based on the identification of linked molecular markers. Mol. Breed. 2003, 12, 319–327. [Google Scholar] [CrossRef]
- D’Aurelio, L.D.; Espinoza, F.; Quarin, C.L.; Pessino, S.C. Genetic diversity in sexual diploid and apomictic tetraploid populations of Paspalum notatum situated in sympatry or allopatry. Plant Syst. Evol. 2004, 244, 189–199. [Google Scholar] [CrossRef]
- Burton, G.W. Breeding Pensacola bahiagrass, Paspalum notatum: I. Method of reproduction. Agron. J. 1955, 47, 311–314. [Google Scholar] [CrossRef]
- Quarin, C.L.; Norrmann, G.A.; Urbani, M.H. Polyploidization in aposporous Paspalum species. Apomixis Newsl. 1989, 2, 44–46. [Google Scholar]
- Tischler, C.R.; Burson, B.L. Evaluating different bahiagrass cytotypes for heat tolerance and leaf epicular wax content. Euphytica 1995, 84, 229–235. [Google Scholar] [CrossRef]
- Forbes, I.J.R.; Burton, G.W. Cytology of diploids, natural and Induced tetraploids, and intra-species hybrids of Bahiagrass, Paspalum notatum Flügge. Crop Sci. 1961, 1, 402–406. [Google Scholar] [CrossRef]
- Quarin, C.L.; Burson, B.L.; Burton, G.W. Cytology of intra-and interspecific hybrids between two cytotypes of Paspalum notatum and P. cromyorrhizon. Bot. Gaz. 1984, 145, 420–426. [Google Scholar] [CrossRef]
- Quarin, C.L.; Norrmann, G.A. Cytology and reproductive behavior of Paspalum equitans, P. ionanthum, and their hybrids with diploid and tetraploid cytotypes of P. cromyorrhizon. Bot. Gaz. 1987, 148, 386–391. Available online: http://www.jstor.org/stable/2995357 (accessed on 2 December 2018). [CrossRef]
- Stein, J.; Quarin, C.L.; Martnez, E.J.; Pessino, S.C.; Ortiz, J.P.A. Tetraploid races of Paspalum notatum show polysomic inheritance and preferential chromosome pairing around the apospory-controlling locus. Theor. Appl. Genet. 2004, 109, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Stein, J.; Pessino, S.C.; Martínez, E.J.; Pıa, M.; Rodriguez, L.; Siena, A.; Quarin, C.L.; Ortiz, J.P.A. A genetic map of tetraploid Paspalum notatum Fluggé (bahiagrass) based on single-dose molecular markers. Mol. Breed. 2007, 20, 153–166. [Google Scholar] [CrossRef]
- Ortiz, J.P.A.; Pessino, S.C.; Bhat, V.; Hayward, M.D.; Quarin, C.L. A genetic linkage map of diploid Paspalum notatum. Crop Sci. 2001, 41, 823–830. [Google Scholar] [CrossRef]
- Quarin, C.L.; Espinoza, F.; Martínez, E.J.; Pessino, S.C.; Bovo, O.A. A rise of ploidy level induces the expression of apomixis in Paspalum notatum. Sex Plant Reprod. 2001, 13, 243–249. [Google Scholar] [CrossRef]
- Yan, Z.; Yan, Z.; Cong, L.; Liu, H.; Chen, Y.; Sun, J.; Ma, L.; Wang, A.; Miao, F.; Song, H.; et al. High-quality chromosome-scale de novo assembly of the Paspalum notatum ‘Flugge’ genome. BMC Genom. 2022, 23, 293. [Google Scholar] [CrossRef] [PubMed]
- Vega, J.M.; Podio, M.; Orjuela, J.; Siena, L.A.; Pessino, S.C.; Combes, M.C.; Mariac, C.; Albertini, E.; Pupilli, F.; Ortiz, J.P.A.; et al. Chromosome-scale genome assembly and annotation of Paspalum notatum Flüggé var. saurae. Sci. Data 2024, 11, 891. [Google Scholar] [CrossRef] [PubMed]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Reeves, A. MicroMeasure: A new computer program for the collection and analysis of cytogenetic data. Genome 2001, 44, 439–443. [Google Scholar] [CrossRef]
- Honfi, A.I.; Bolzán, A.D.; Daviña, J.R. Dimensión Cromosómica. Cienc. Investig. AAPC 2017, 67, 5–24. Available online: http://hdl.handle.net/11336/68433 (accessed on 2 December 2018).
- Battaglia, E. Chromosome morphology and terminology. Caryologia 1955, 8, 179–187. [Google Scholar] [CrossRef]
- Battaglia, E. The chromosome satellite (Navashin’s Sputnik or Satelles): A terminological comment. Acta Biol. Cracov. Ser. Bot. 1999, 41, 15–18. [Google Scholar]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Addison-Wesley Publishing Co.: Reading, MA, USA, 1971; 216p. [Google Scholar]
- Romero Zarco, C. A new method for estimating karyotype asymmetry. Taxon 1986, 35, 526–531. [Google Scholar] [CrossRef]
- Schwarzacher, T.; Ambros, P.; Schweizer, D. Application of Giemsa banding to orchid karyotype analysis. Plant Syst. Evol. 1980, 134, 293–297. [Google Scholar] [CrossRef]
- Daviña, J.R.; Gianini Aquino, A.C.; Rodríguez Mata, O.A.; Tapia Campos, E.; Barba-Gonzalez, R.; Honfi, A.I. Chromosomic studies in Zephyranthes citrina Baker (Amaryllidaceae), a polyploid ornamental. BAG J. Basic Appl. Genet. 2022, 33, 89–95. [Google Scholar] [CrossRef]
- Schweizer, D. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 1976, 58, 307–324. [Google Scholar] [CrossRef]
- Dolezel, J.; Bartos, J.; Voglmayr, H.; Greilhuber, J. Nuclear DNA content and genome size of trout and human. Cytometry 2003, A51, 127–128. [Google Scholar] [CrossRef]
- Dolezel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Pozzobon, M.T.; Valls, J.F.M. Chromosome number in germplasm accessions of Paspalum notatum (Gramineae). Braz. J. Genet. 1997, 20, 29–34. [Google Scholar] [CrossRef]
- Gates, R.N.; Quarin, C.L.; Pedreira, C.G.S. Bahiagrass, in Warm-Season (C4) Grasses; Moser, L.E., Burson, B.L., Sollenberger, L.E., Eds.; Agronomy Monography 45; ASA, CSSA, SSSA: Madison, WI, USA, 2004; pp. 651–680. [Google Scholar] [CrossRef]
- Hojsgaard, D.H.; Honfi, A.I.; Rua, G.; Daviña, J.R. Chromosome numbers and ploidy levels of Paspalum species from subtropical South America (Poaceae) Genet. Resour. Crop Evol. 2009, 56, 533–545. [Google Scholar] [CrossRef]
- Reutemann, A.V.; Rua, G.H.; Daviña, J.R.; Honfi, A.I. IAPT chromosome data 31/11. In Marhold, K. & Kucera, J. (eds.) & al., IAPT chromosome data 31. Taxon 2019, 68, 1379–1380, E39–E42. [Google Scholar] [CrossRef]
- Burson, B.L. Cytogenetic relationships between Paspalum jurgensii and P. intermedium, P. vaginatum, and P. setaceum var. ciliatifolium. Crop Sci. 1981, 21, 515–519. [Google Scholar] [CrossRef]
- Quarin, C.L.; Burson, B.L. Cytogenetic relations among Paspalum notatum var. saurae, P. pumilum, P. indecorum, and P. vaginatum. Bot. Gaz. 1983, 144, 433–438. [Google Scholar] [CrossRef]
- Bonilla, J.; Quarin, C.L. Diplosporous and aposporous apomixis in a pentaploid race of Paspalum minus. Plant Sci. 1997, 127, 97–104. [Google Scholar] [CrossRef]
- Reutemann, A.V.; Martínez, E.J.; Schedler, M.; Daviña, J.R.; Hojsgaard, D.H.; Honfi, A.I. Uniparentality: Advantages for range expansion in diploid and diploid-autopolyploid species. Bot. J. Linn. Soc. 2022, 200, 563–585. [Google Scholar] [CrossRef]
- Schedler, M.; Reutemann, A.V.; Hojsgaard, D.H.; Zilli, A.L.; Brugnoli, E.A.; Galdeano, F.; Acuña, C.A.; Honfi, A.I.; Martínez, E.J. Alternative evolutionary pathways in Paspalum involving allotetraploidy, sexuality, and varied mating systems. Genes 2023, 14, 1137. [Google Scholar] [CrossRef] [PubMed]
- Zuloaga, F.O.; Morrone, O. Revisión de las especies de Paspalum para América del Sur austral (Argentina, Bolivia, sur del Brasil, Chile, Paraguay y Uruguay). Ann. Mo. Bot. Gard. Monogr. Syst. Bot. 2005, 102, 304. [Google Scholar]
- Ortiz, J.P.A.; Quarin, C.L.; Pessino, S.C.; Acuna, C.; Martínez, E.J.; Espinoza, F.; Pupilli, F. Harnessing apomictic reproduction in grasses: What we have learned from Paspalum. Ann. Bot. 2013, 112, 767–787. [Google Scholar] [CrossRef]
- Peruzzi, L.; Leitch, I.J.; Caparelli, K.F. Chromosome diversity and evolution in Liliaceae. Ann. Bot. 2009, 103, 459–475. [Google Scholar] [CrossRef] [PubMed]
- Speranza, P.; Vaio, M.; Mazzella, C. Karyotypes of two cytotypes of Paspalum quadrifarium Lam. (Poaceae): An alternative technique for small chromosomes in plants. Genet. Mol. Biol. 2003, 26, 449–503. [Google Scholar] [CrossRef]
- Honfi, A.I.; Morrone, O.; Zuloaga, F.O. Chromosome numbers and ploidy levels of some Paniceae and Paspaleae species (Poaceae, Panicoideae). Ann. Mo. Bot. Gard. 2021, 106, 234–244. [Google Scholar] [CrossRef]
- Wachtler, F.; Stahl, A. The nucleolus: A structural and functional interpretation. Micron 1993, 24, 473–505. [Google Scholar] [CrossRef]
- Vaio, M.; Speranza, P.; Valls, J.F.; Guerra, M.; Mazzella, C. Localization of the 5S and 45S rDNA Sites and cpDNA Sequence Analysis in Species of the Quadrifaria Group of Paspalum (Poaceae, Paniceae). Ann. Bot. 2005, 96, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Vaio, M.; Mazzella, C.; Guerra, M.; Speranza, P. Effects of the diploidisation process upon the 5S and 35S rDNA sequences in the allopolyploid species of the Dilatata group of Paspalum (Poaceae, Paniceae). Aust. J. Bot. 2019, 67, 521–530. [Google Scholar] [CrossRef]
- Greilhuber, J. Trends in der chromosomen evolution von Scilla (Liliaceae). Stapfia 1982, 10, 11–51. [Google Scholar]
- John, B. The biology of heterochromatin. In Heterochromatin, Molecular and Structural Aspects; Verma, R.S., Ed.; Cambridge University Press: Cambridge, UK, 1988; pp. 1–147. [Google Scholar]
- Arantes Chaves, A.L.; Bezerra Chiavegatto, R.; Gandolfi Benites, F.R.; Techio, V.H. Comparative karyotype analysis among cytotypes of Cynodon dactylon (L.) Pers. (Poaceae). Mol. Biol. Rep. 2019, 46, 4873–4881. [Google Scholar] [CrossRef] [PubMed]
- John, B. Meiosis; Cambridge University Press: Cambridge, UK, 1990; 412p. [Google Scholar]
- Ozias-Akins, P.; van Dijk, P.J. Mendelian Genetics of Apomixis in Plants. Annu. Rev. Genet. 2007, 41, 509–537. [Google Scholar] [CrossRef] [PubMed]
- Calderini, O.; Chang, S.B.; de Jong, H.; Busti, A.; Paolocci, F.; Arcioni, S.; de Vries, S.C.; Abma-Henkens, M.H.C.; Lankhorst, R.M.K.; Donnison, L.S.; et al. Molecular cytogenetics and DNA sequence analysis of an apomixis-linked BAC in Paspalum simplex reveal a non pericentromere location and partial microcolinearity with rice. Theor. Appl. Genet. 2006, 112, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.A.; Goel, S.; Gunawan, G.; Cordonnier-Pratt, M.M.; Johnson, V.E.; Liang, C.; Wang, H.; Pratt, L.H.; Mullet, J.E.; DeBarry, J.; et al. Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 2008, 147, 1396–1411. [Google Scholar] [CrossRef] [PubMed]
- Podio, M.; Siena, L.A.; Hojsgaard, D.; Stein, J.; Quarin, C.L.; Ortiz, J.P.A. Evaluation of meiotic abnormalities and pollen viability in aposporous and sexual tetraploid Paspalum notatum (Poaceae). Plant Syst. Evol. 2012, 298, 1625–1633. [Google Scholar] [CrossRef]
- Bennett, M.D.; Leitch, Y. Plant Genome Size Research: A Field In Focus. Ann. Bot. 2005, 95, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Greilhuber, J.; Dolezel, J.; Lysák, M.A.; Bennett, M.D. The origin, evolution and proposed stabilization of the terms ’genome size’ and ’C-value’ to describe nuclear DNA contents. Ann. Bot. 2005, 95, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.; James, V.A.; Quesenberry, K.H..; Altpeter, F. Risk assessment of transgenic apomictic tetraploid bahiagrass, cytogenetics, breeding behavior and performance of intra-specific hybrids. Theor. Appl. Genet. 2009, 119, 1383–1395. [Google Scholar] [CrossRef]
- Sandhu, S.; Blount, A.R.; Quesenberry, K.H.; Altpeter, F. Apomixis and ploidy barrier suppress pollen-mediated gene flow in field grown transgenic turf and forage grass (Paspalum notatum Flüggé). Theor. Appl. Genet. 2010, 121, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Vaio, M.; Mazzella, C.; Porro, V.; Speranza, P.; López-Carro, B.; Estramil, E.; Folle, G.A. Nuclear DNA content in allo-polyploid species and synthetic hybrids in the grass genus Paspalum. Pl. Syst. Evol. 2007, 265, 109–121. [Google Scholar] [CrossRef]
- Maretti Gonçalves, T.; Gomes Ferreira, J.R.; Baccili Zanotto Vigna, B.; Sousa Azevedo, A.L.; Toniolo Pozzobon, M.; Pereira Fávero, A. Reproductive mode and DNA content of Paspalum accessions from Plicatula group. Reproduction and DNA content of Paspalum. Flora 2021, 279, 151810. [Google Scholar] [CrossRef]
- Matta, F.P.; Fávero, A.P.; Zanotto Vigna, B.B.; Mattos Cavallari, M.; Alves, F.; Oliveira, F.A.; Pereira de Souza, A.; Pozzobon, M.T.; Sousa Azevedo, A.L.; Gusmão, M.R. Characterization of Paspalum genotypes for turfgrass cultivars development. Crop Sci. 2024, 64, 1–14. [Google Scholar] [CrossRef]
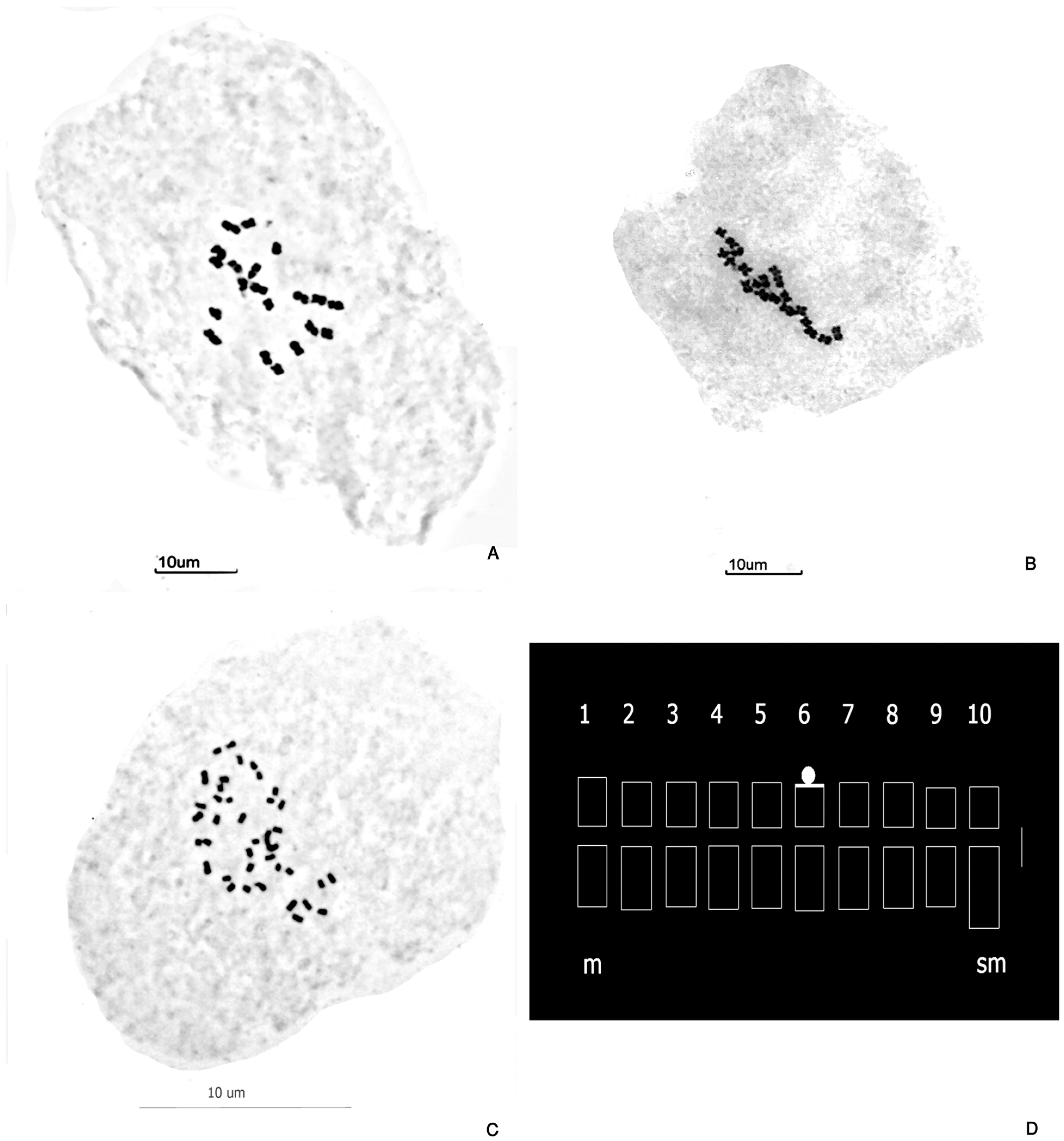

| Species Variety | 2n | Locality and Voucher |
|---|---|---|
| P. notatum Flüggé var. saurae Parodi | 20 | Argentina, Santa Fe, Ruta provincial 19, Colastiné, near to Tunel subfluvial. 28 March 2010. S 31°39′59.3″ W 60°35′10.4″ Honfi & Daviña 1453 #1, #2 (MNES). |
| 20 | Paraguay, Itapúa, Encarnación. 11 March 2015. Honfi 1740 #2 (MNES). S 27°19′12.166″, W 55°51′52.198″ | |
| 20 | Argentina, Entre Ríos, Gualeguaychú, Cerro del Indio. 9 April 2015. Honfi 1961 #11 (MNES). S 33°4′9.199″, W 58°26′28.399″ | |
| P. notatum Flüggé var. notatum | 40 | Argentina, Santa Fe, surroundings access to the subfluvial tunnel. 30 September 2006. Honfi & Daviña 1304 (MNES). |
| 40 | Argentina, Misiones, Departamento Capital, Posadas. 3 April 2012. Honfi 1603 #1, #2, #3 (MNES). |
| Pair | s (µm) ± SE | l (µm) ± SE | c (µm) ± SE | i | Type | % HTCL |
|---|---|---|---|---|---|---|
| 1 | 1.028 ± 0.066 | 1.181 ± 0.094 | 2.209 ± 0.16 | 0.4654 | m | 12.88 |
| 2 | 0.867 ± 0.075 | 1.086 ± 0.082 | 1.961 ± 0.15 | 0.4421 | m | 11.44 |
| 3 | 0.809 ± 0.063 | 1.057 ± 0.067 | 1.866 ± 0.13 | 0.4335 | m | 10.88 |
| 4 | 0.751 ± 0.053 | 0.998 ± 0.079 | 1.749 ± 0.12 | 0.4294 | m | 10.20 |
| 5 | 0.736 ± 0.042 | 0.925 ± 0.057 | 1.661 ± 0.096 | 0.4431 | m | 9.69 |
| 6 | 0.656 ± 0.042 | 0.874 ± 0.057 | 1.516 ± 0.109 | 0.4327 | m | 8.84 |
| 7 | 0.685 ± 0.059 | 0.787 ± 0.058 | 1.473 ± 0.115 | 0.465 | m | 8.59 |
| 8 | 0.656 ± 0.063 | 0.78 ± 0.046 | 1.437 ± 0.108 | 0.4565 | m | 8.38 |
| 9 | 0.598 ± 0.037 | 0.729 ± 0.066 | 1.327 ± 0.103 | 0.4506 | m | 7.74 |
| 10 | 0.671 ± 0.045 | 1.262 ± 0.116 | 1.94 ± 0.158 | 0.3459 | sm | 11.32 |
| Chrom | s (µm) ± SE | l (µm) ± SE | c (µm) ± SE | i | Type | % TCL |
|---|---|---|---|---|---|---|
| Q1-C1 | 1.047 ± 0.11 | 1.392 ± 0.04 | 2.411 ± 0.11 | 0.434 | m | 3.77 |
| Q1-C2 | 1.025 ± 0.06 | 1.297 ± 0.12 | 2.323 ± 0.14 | 0.441 | m | 3.63 |
| Q1-C3 | 0.942 ± 0.08 | 1.240 ± 0.11 | 2.180 ± 0.16 | 0.432 | m | 3.41 |
| Q1-C4 | 0.937 ± 0.10 | 1.113 ± 0.03 | 2.051 ± 0.12 | 0.457 | m | 3.20 |
| Q2-C5 | 0.839 ± 0.04 | 1.087 ± 0.10 | 1.975 ± 0.12 | 0.425 | m | 3.08 |
| Q2-C6 | 0.856 ± 0.09 | 1.033 ± 0.05 | 1.889 ± 0.12 | 0.453 | m | 2.95 |
| Q2-C7 | 0.806 ± 0.07 | 1.054 ± 0.07 | 1.860 ± 0.12 | 0.433 | m | 2.91 |
| Q2-C8 | 0.835 ± 0.07 | 0.946 ± 0.06 | 1.813 ± 0.10 | 0.461 | m | 2.83 |
| Q3-C9 | 0.699 ± 0.03 | 1.0255 ± 0.13 | 1.753 ± 0.09 | 0.399 | m | 2.74 |
| Q3-C10 | 0.720 ± 0.06 | 0.944 ± 0.06 | 1.664 ± 0.11 | 0.433 | m | 2.60 |
| Q3-C11 | 0.751 ± 0.05 | 0.884 ± 0.07 | 1.636 ± 0.11 | 0.459 | m | 2.55 |
| Q3-C12 | 0.696 ± 0.05 | 0.939 ± 0.08 | 1.636 ± 0.11 | 0.426 | m | 2.55 |
| Q4-C13 | 0.775 ± 0.06 | 0.861 ± 0.05 | 1.636 ± 0.11 | 0.474 | m | 2.55 |
| Q4-C14 | 0.723 ± 0.04 | 0.913 ± 0.08 | 1.636 ± 0.11 | 0.442 | m | 2.55 |
| Q4-C15 | 0.706 ± 0.06 | 0.913 ± 0.08 | 1.619 ± 0.13 | 0.436 | m | 2.53 |
| Q4-C16 | 0.746± 0.06 | 0.872 ± 0.09 | 1.590 ± 0.12 | 0.469 | m | 2.48 |
| Q5-C17 | 0.699 ± 0.03 | 0.838 ± 0.07 | 1.561 ± 0.11 | 0.448 | m | 2.44 |
| Q5-C18 | 0.694 ± 0.05 | 0.832 ± 0.08 | 1.526 ± 0.11 | 0.455 | m | 2.38 |
| Q5-C19 | 0.667 ± 0.05 | 0.835 ± 0.07 | 1.502 ± 0.10 | 0.444 | m | 2.35 |
| Q5-C20 | 0.723 ± 0.06 | 0.780 ± 0.05 | 1.502 ± 0.10 | 0.481 | m | 2.35 |
| Q6-C21 | 0.694 ± 0.05 | 0.837 ± 0.05 | 1.502 ± 0.10 | 0.462 | m | 2.35 |
| Q6-C22 | 0.639 ± 0.03 | 0.772 ± 0.07 | 1.411 ± 0.10 | 0.453 | m | 2.20 |
| Q6-C23 | 0.639 ± 0.03 | 0.772 ± 0.07 | 1.411 ± 0.10 | 0.453 | m | 2.20 |
| Q6-C24 | 0.639 ± 0.03 | 0.772 ± 0.07 | 1.411 ± 0.10 | 0.453 | m | 2.20 |
| Q7-C25 | 0.663 ± 0.05 | 0.720 ± 0.06 | 1.383 ± 0.09 | 0.479 | m | 2.16 |
| Q7-C26 | 0.605 ± 0.07 | 0.777 ± 0.05 | 1.371 ± 0.10 | 0.442 | m | 2.14 |
| Q7-C27 | 0.627 ± 0.04 | 0.720 ± 0.06 | 1.348 ± 0.08 | 0.466 | m | 2.10 |
| Q7-C28 | 0.610 ± 0.05 | 0.720 ± 0.06 | 1.330 ± 0.10 | 0.459 | m | 2.08 |
| Q8-C29 | 0.582 ± 0.05 | 0.748 ± 0.06 | 1.330 ± 0.10 | 0.437 | m | 2.08 |
| Q8-C30 | 0.569 ± 0.05 | 0.705± 0.05 | 1.275 ± 0.09 | 0.447 | m | 1.99 |
| Q8-C31 | 0.527 ± 0.03 | 0.7440 ± 0.07 | 1.247 ± 0.09 | 0.423 | m | 1.95 |
| Q8-C32 | 0.550 ± 0.05 | 0.696 ± 0.05 | 1.247 ± 0.09 | 0.442 | m | 1.95 |
| Q9-C33 | 0.522 ± 0.05 | 0.753 ± 0.08 | 1.218 ± 0.09 | 0.429 | m | 1.90 |
| Q9-C34 | 0.522 ± 0.05 | 0.636 ± 0.05 | 1.158 ± 0.09 | 0.451 | m | 1.81 |
| Q9-C35 | 0.4985 ± 0.04 | 0.579 ± 0.06 | 1.077 ± 0.08 | 0.462 | m | 1.68 |
| Q9-C36 | 0.517 ± 0.03 | 0.560 ± 0.06 | 1.077 ± 0.08 | 0.480 | m | 1.68 |
| Q10-C37 | 0.655 ± 0.08 | 1.301 ± 0.10 | 2.024 ± 0.24 | 0.324 | sm | 3.16 |
| Q10-C38 | 0.627 ± 0.08 | 1.344 ± 0.14 | 1.971 ± 0.22 | 0.318 | sm | 3.08 |
| Q10-C39 | 0.556 ± 0.05 | 1.187 ± 0.08 | 1.738 ± 0.13 | 0.320 | sm | 2.71 |
| Q10-C40 | 0.550 ± 0.05 | 1.013 ± 0.15 | 1.621 ± 0.16 | 0.340 | sm | 2.53 |
| Parameter | Diploid Cytotype | Tetraploid Cytotype |
|---|---|---|
| 2n | 20 | 40 |
| Karyotype formula | 18 m + 2 sm | 36 m + 4 sm |
| Mean chromosome length | 1.714 ± 0.084 µm | 1.599 ± 0.08 µm |
| cMin | 1.3 µm | 1.1 µm |
| cMax | 2.3 µm | 2.3 µm |
| Stebbins’s category | 1A | 2B |
| A1 | 0.22 | 0.22 |
| A2 | 0.16 | 0.35 |
| Chromosome with SAT | Pair 6 | Quartet 6 |
| Secondary constriction | Pair 6, short arm | Quartet 6, short arm |
| TCL (µm) | 34.28 µm | 63.94 µm |
| Increase in proportion of TCL (µm) | -- | 97.6% |
| Difference in TCL among cytotypes | -- | 4.62 µm |
| Difference in Het-C amount among cytotypes | −0.67% | +0.67% |
| Parameters | Diploid Cytotype | Tetraploid Cytotype |
|---|---|---|
| Het-C bands Position | CMA + DAPI− bands in distal region of short arm and satellite of pair 6. GC-rich Het-C | One chromosome with GC-rich Het-C at distal position of the short arm (CMA + DAPI−), and three with CMA+ + DAPI0. Satellites of quartet 6 GC-rich Het-C. |
| GC-rich Het-C | (+) | (+) |
| AT-rich Het-C | (−) | (−) |
| % Het-C per TCL (µm) | 2.8% | 3.47% |
| Difference in Het-C amount among cytotypes | −0.67% | +0.67% |
| Sample | Ploidy | 2C-Value (pg) ± SE | 1Cx-Value (pg) | 1Cx-Value (Mbp) |
|---|---|---|---|---|
| H1740 #2 | 2x | 1.41 ± 0.008 | 0.701 | 685.504 × 109 bp |
| H1961 #11 | 2x | 1.438 ± 0.017 | 0.719 | 702.871 × 109 bp |
| Mean GS | 2x | 1.424 | 0.713 | 697.314 × 109 bp |
| H1603 #1 | 4x | 2.753 ± 0.015 | 0.678 | 1321.910 × 109 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Honfi, A.I.; Reutemann, A.V.; Schneider, J.S.; Escobar, L.M.; Martínez, E.J.; Daviña, J.R. Chromosome Morphology and Heterochromatin Patterns in Paspalum notatum: Insights into Polyploid Genome Structure. Genes 2025, 16, 242. https://doi.org/10.3390/genes16030242
Honfi AI, Reutemann AV, Schneider JS, Escobar LM, Martínez EJ, Daviña JR. Chromosome Morphology and Heterochromatin Patterns in Paspalum notatum: Insights into Polyploid Genome Structure. Genes. 2025; 16(3):242. https://doi.org/10.3390/genes16030242
Chicago/Turabian StyleHonfi, Ana I., A. Verena Reutemann, Juan S. Schneider, Lucas M. Escobar, Eric J. Martínez, and Julio R. Daviña. 2025. "Chromosome Morphology and Heterochromatin Patterns in Paspalum notatum: Insights into Polyploid Genome Structure" Genes 16, no. 3: 242. https://doi.org/10.3390/genes16030242
APA StyleHonfi, A. I., Reutemann, A. V., Schneider, J. S., Escobar, L. M., Martínez, E. J., & Daviña, J. R. (2025). Chromosome Morphology and Heterochromatin Patterns in Paspalum notatum: Insights into Polyploid Genome Structure. Genes, 16(3), 242. https://doi.org/10.3390/genes16030242









