Worldwide Population Dynamics of Salmonella Saintpaul: Outbreaks, Epidemiology, and Genome Structure
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic Characterization of SSa Strain PP_BR059
2.2. WGS of SSa Strain PP_BR059
2.3. Comparative Genomic Analysis of Salmonella Saintpaul
3. Results and Discussion
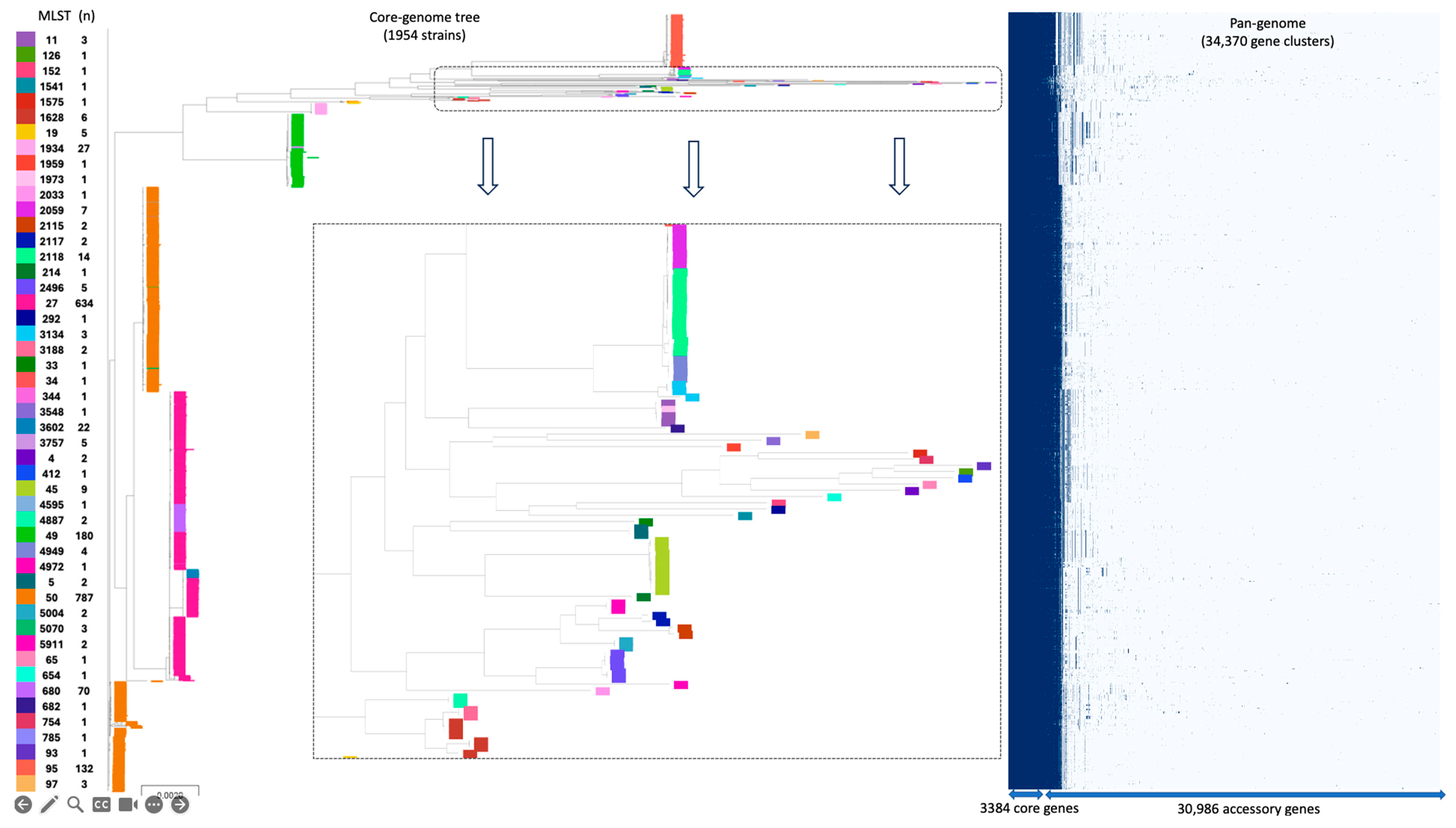
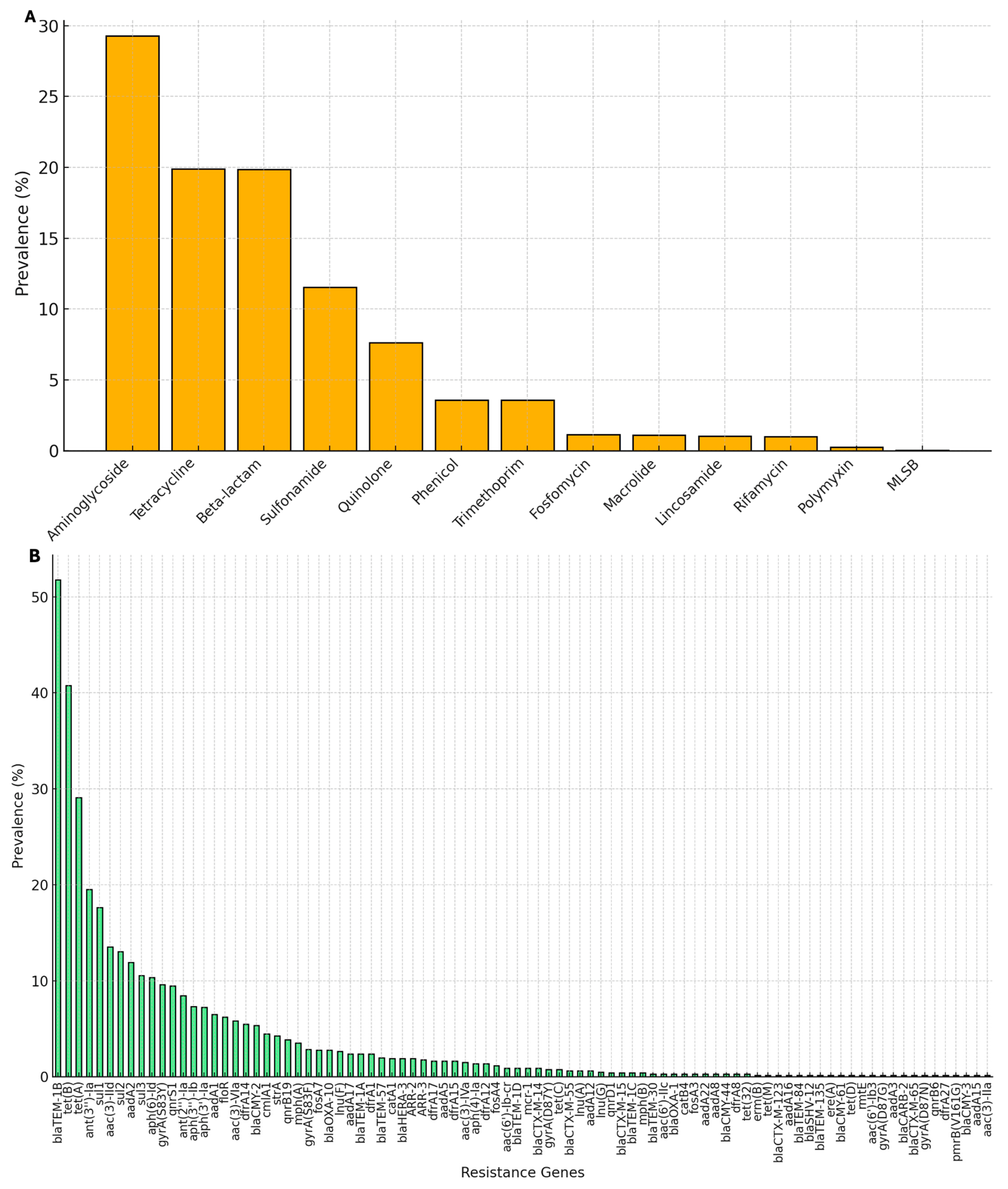
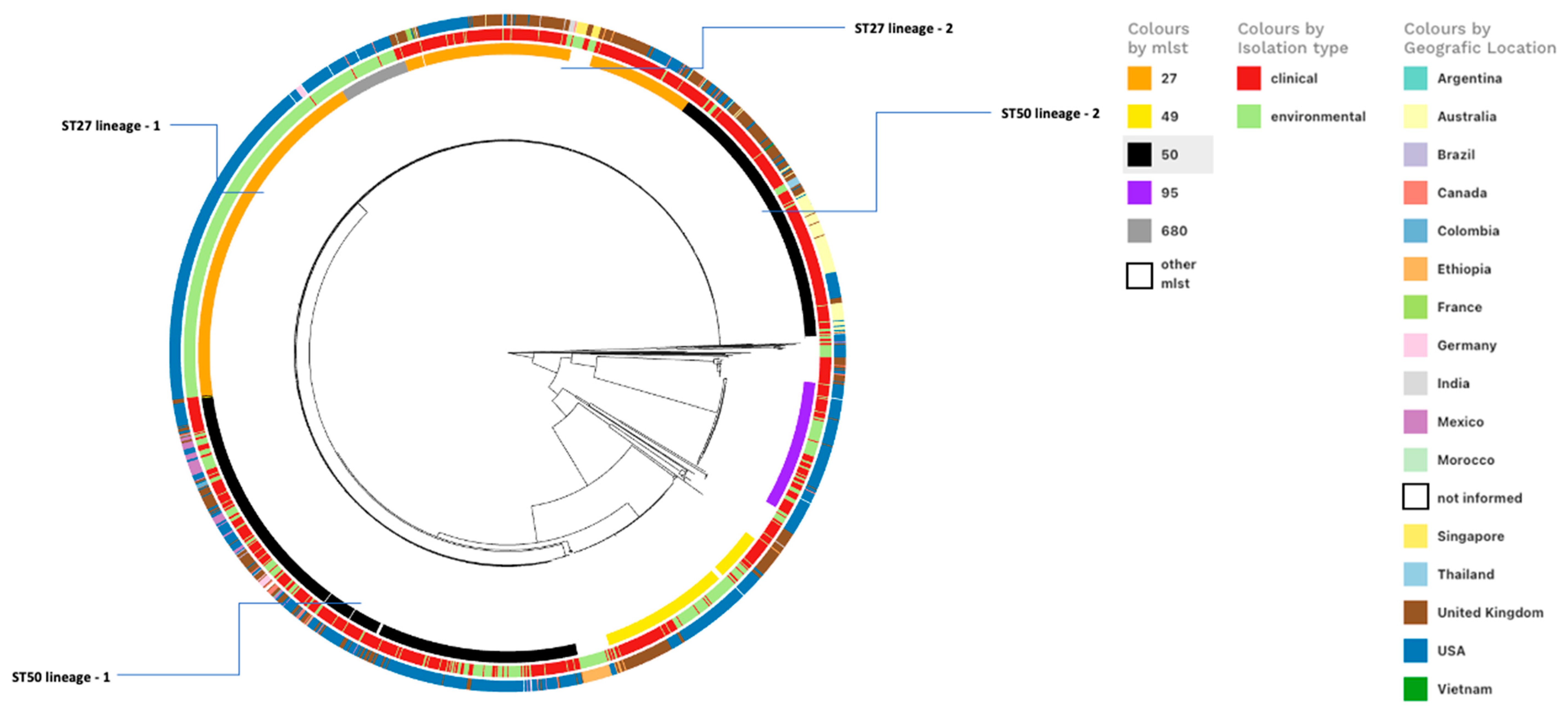
3.1. Comparative Genomic Analysis of SSa
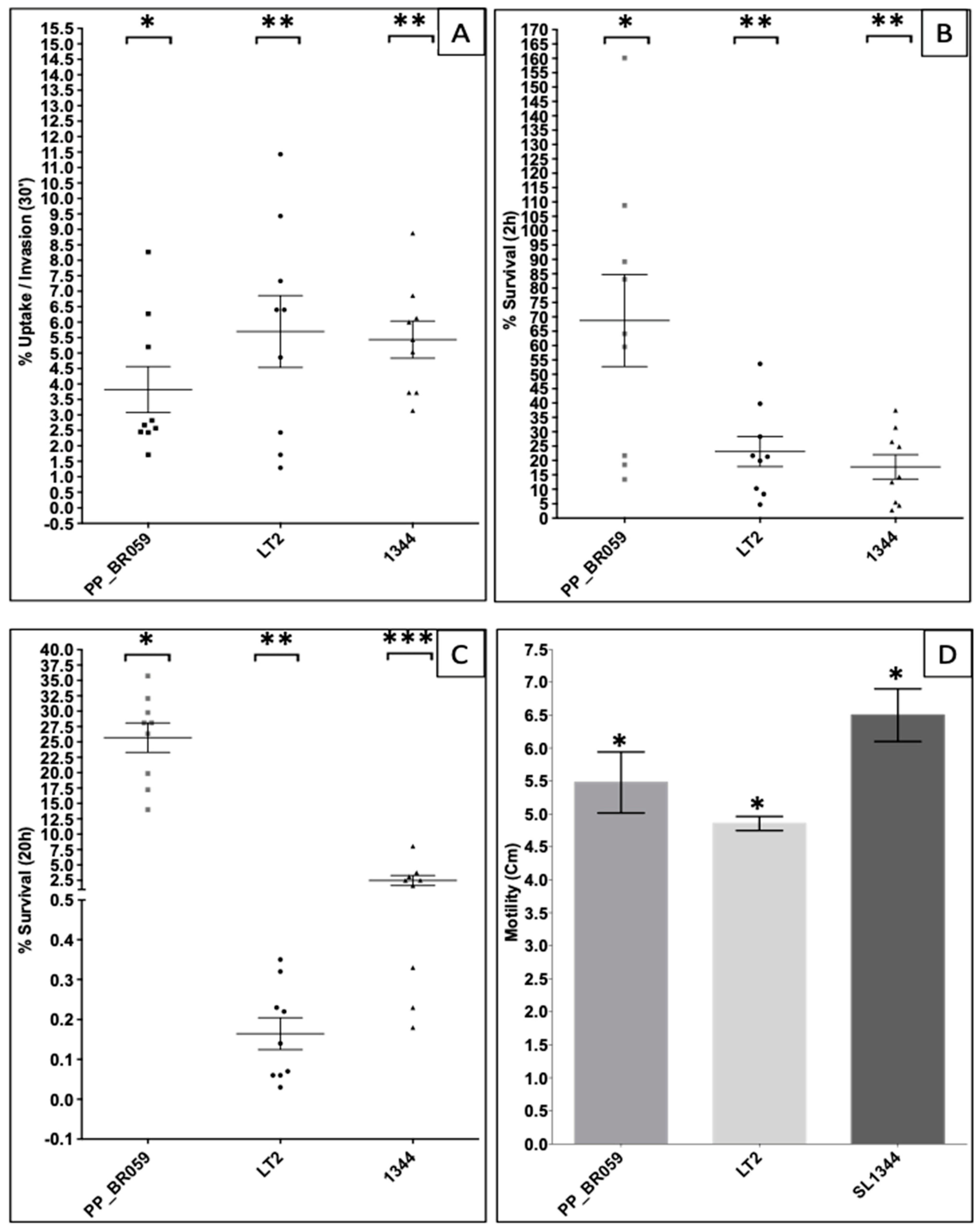
3.2. Invasiveness and Virulence Potential of the SSa Strain PP_BR059
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Balasubramanian, R.; Im, J.; Lee, J.-S.; Jeon, H.J.; Mogeni, O.D.; Kim, J.H.; Rakotozandrindrainy, R.; Baker, S.; Marks, F. The Global Burden and Epidemiology of Invasive Non-Typhoidal Salmonella Infections. Hum. Vaccines Immunother. 2019, 15, 1421–1426. [Google Scholar] [CrossRef]
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Crump, J.A. Global Burden of Invasive Nontyphoidal Salmonella Disease, 2010. Emerg. Infect. Dis. 2015, 21, 941. [Google Scholar] [CrossRef] [PubMed]
- CDC. Outbreaks Involving Salmonella. In Reports of Selected Salmonella Outbreak Investigations; CDC: Atlanta, GA, USA, 2018. [Google Scholar]
- Fey, P.D.; Iwen, P.C.; Zentz, E.B.; Briska, A.M.; Henkhaus, J.K.; Bryant, K.A.; Larson, M.A.; Noel, R.K.; Hinrichs, S.H. Assessment of Whole-Genome Mapping in a Well-Defined Outbreak of Salmonella Enterica Serotype Saintpaul. J. Clin. Microbiol. 2012, 50, 3063–3065. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Klontz, K.C.; Klontz, J.C.; Mody, R.K.; Hoekstra, R.M. Analysis of Tomato and Jalapeño and Serrano Pepper Imports into the United States from Mexico before and during a National Outbreak of Salmonella Serotype Saintpaul Infections in 2008. J. Food Prot. 2010, 73, 1967–1974. [Google Scholar] [CrossRef] [PubMed]
- Mody, R.K.; Greene, S.A.; Gaul, L.; Sever, A.; Pichette, S.; Zambrana, I.; Dang, T.; Gass, A.; Wood, R.; Herman, K.; et al. National Outbreak of Salmonella Serotype Saintpaul Infections: Importance of Texas Restaurant Investigations in Implicating Jalapeño Peppers. PLoS ONE 2011, 6, e16579. [Google Scholar] [CrossRef]
- Taylor, E.; Kastner, J.; Renter, D. Challenges Involved in the Salmonella Saintpaul Outbreak and Lessons Learned. J. Public Health Manag. Pract. 2010, 16, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Muruvanda, T.; Allard, M.W.; Hoffmann, M. Complete Genome Sequences of Three Salmonella Enterica Subsp. Enterica Serovar Saintpaul Isolates Associated with a 2013 Multistate Outbreak in the United States. Genome Announc. 2017, 5, e00456-17. [Google Scholar] [CrossRef] [PubMed]
- Boore, A.L.; Jungk, J.; Russo, E.T.; Redd, J.T.; Angulo, F.J.; Williams, I.T.; Cheek, J.E.; Gould, L.H. Added Value of a Household-Level Study during an Outbreak Investigation of Salmonella Serotype Saintpaul Infections, New Mexico 2008. Epidemiol. Infect. 2013, 141, 2068–2073. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dyda, A.; Nguyen, P.-Y.; Chughtai, A.A.; MacIntyre, C.R. Changing Epidemiology of Salmonella Outbreaks Associated with Cucumbers and Other Fruits and Vegetables. Glob. Biosecurity 2020, 2, 1–13. [Google Scholar]
- Mercês, A.F. Top 10 Salmonella Serovars Associated with Human Salmonellosis in Brazil (2011–2020). Gen. Med. 2022, 6, 1306. [Google Scholar]
- Rau, R.B.; de Lima-Morales, D.; Wink, P.L.; Ribeiro, A.R.; Barth, A.L. Salmonella Enterica Mcr-1 Positive from Food in Brazil: Detection and Characterization. Foodborne Pathog. Dis. 2010, 17, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, I.; Silva, R.; Menezes, J.; Machado, S.; Rodrigues, D.; Pomba, C.; Abreu, D.; Nascimento, E.; Aquino, M.; Pereira, V. High Prevalence of Multidrug-Resistant Nontyphoidal Salmonella Recovered from Broiler Chickens and Chicken Carcasses in Brazil. Braz. J. Poult. Sci. 2020, 22, eRBCA-2019-1206. [Google Scholar] [CrossRef]
- Viegas, F.M.; Ramos, C.P.; Xavier, R.G.C.; Lopes, E.O.; Júnior, C.A.O.; Bagno, R.M.; Diniz, A.N.; Lobato, F.C.F.; Silva, R.O.S. Fecal Shedding of Salmonella Spp., Clostridium Perfringens, and Clostridioides Difficile in Dogs Fed Raw Meat-Based Diets in Brazil and Their Owners’ Motivation. PLoS ONE 2020, 15, e0231275. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Shah, D.H.; Zhou, X.; Addwebi, T.; Davis, M.A.; Call, D.R. In Vitro and in Vivo Pathogenicity of Salmonella Enteritidis Clinical Strains Isolated from North America. Arch. Microbiol. 2011, 193, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Panzenhagen, P.H.N.; Paul, N.C.; Conte, C.A.; Costa, R.G.; Rodrigues, D.P.; Shah, D.H. Genetically Distinct Lineages of Salmonella Typhimurium ST313 and ST19 Are Present in Brazil. Int. J. Med. Microbiol. 2018, 308, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.C.; Folster, J.P.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between Phenotypic and In Silico Detection of Antimicrobial Resistance in Salmonella Enterica in Canada Using Staramr. Microorganisms 2022, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and Precise Alignment of Raw Reads against Redundant Databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Zhou, Z.; Sergeant, M.J.; Achtman, M. A Genomic Overview of the Population Structure of Salmonella. PLoS Genet. 2018, 14, e1007261. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A Novel Web Tool for WGS-Based Detection of Antimicrobial Resistance Associated with Chromosomal Point Mutations in Bacterial Pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; Von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An Interactive Viewer for Bacterial Population Genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-Sites: Rapid Efficient Extraction of SNPs from Multi-FASTA Alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [PubMed]
- CDC. Multistate Outbreak of Salmonella Reading Infections Linked to Raw Turkey Products. Available online: https://archive.cdc.gov/#/details?url=https://www.cdc.gov/salmonella/reading-07-18/index.html (accessed on 4 January 2025).
- Beutlich, J.; Rodríguez, I.; Schroeter, A.; Käsbohrer, A.; Helmuth, R.; Guerra, B. A Predominant Multidrug-Resistant Salmonella Enterica Serovar Saintpaul Clonal Line in German Turkey and Related Food Products. Appl. Environ. Microbiol. 2010, 76, 3657–3667. [Google Scholar] [CrossRef]
- Cavaco, L.M.; Korsgaard, H.; Sørensen, G.; Aarestrup, F.M. Plasmid-Mediated Quinolone Resistance Due to qnrB5 and qnrS1 Genes in Salmonella Enterica Serovars Newport, Hadar and Saintpaul Isolated from Turkey Meat in Denmark. J. Antimicrob. Chemother. 2008, 62, 632–634. [Google Scholar] [CrossRef]
- Cargnel, M.; Filippitzi, M.; Van Cauteren, D.; Mattheus, W.; Botteldoorn, N.; Cambier, L.; Welby, S. Assessing Evidence of a Potential Salmonella Transmission across the Poultry Food Chain. Zoonoses Public Health 2023, 70, 22–45. [Google Scholar] [CrossRef] [PubMed]
- Rockett, R.J.; Arnott, A.; Wang, Q.; Howard, P.; Sintchenko, V. Genomic Surveillance Enables Suitability Assessment of Salmonella Gene Targets Used for Culture-Independent Diagnostic Testing. J. Clin. Microbiol. 2020, 58, e00038-20. [Google Scholar] [CrossRef] [PubMed]
- Thomson, R.M.; Henderson, H.J.; Smith-Palmer, A. An Outbreak of Salmonella Saintpaul in a Scottish Childcare Facility: The Influence of Parental under-Reporting. BMC Infect. Dis. 2019, 19, 847. [Google Scholar] [CrossRef] [PubMed]
- Barton Behravesh, C.; Mody, R.K.; Jungk, J.; Gaul, L.; Redd, J.T.; Chen, S.; Cosgrove, S.; Hedican, E.; Sweat, D.; Chávez-Hauser, L.; et al. 2008 Outbreak of Salmonella Saintpaul Infections Associated with Raw Produce. N. Engl. J. Med. 2011, 364, 918–927. [Google Scholar] [CrossRef]
- Beatty, M.E.; LaPorte, T.N.; Phan, Q.; Van Duyne, S.V.; Braden, C. A Multistate Outbreak of Salmonella Enterica Serotype Saintpaul Infections Linked to Mango Consumption: A Recurrent Theme. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2004, 38, 1337–1338. [Google Scholar] [CrossRef] [PubMed]
- Bornemann, R.; Zerr, D.M.; Heath, J.; Koehler, J.; Grandjean, M.; Pallipamu, R.; Duchin, J. An Outbreak of Salmonella Serotype Saintpaul in a Children’s Hospital. Infect. Control Hosp. Epidemiol. 2002, 23, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Draper, A.D.; Morton, C.N.; Heath, J.N.; Lim, J.A.; Markey, P.G. An Outbreak of Salmonella Saintpaul Gastroenteritis after Attending a School Camp in the Northern Territory, Australia. Commun. Dis. Intell. 2017, 41, E10–E15. [Google Scholar]
- Jain, S.; Bidol, S.A.; Austin, J.L.; Berl, E.; Elson, F.; Williams, M.L.; Deasy, M.; Moll, M.E.; Rea, V.; Vojdani, J.D.; et al. Multistate Outbreak of Salmonella Typhimurium and Saintpaul Infections Associated with Unpasteurized Orange Juice—United States, 2005. Clin. Infect. Dis. 2009, 48, 1065–1071. [Google Scholar] [CrossRef]
- Munnoch, S.A.; Ward, K.; Sheridan, S.; Fitzsimmons, G.J.; Shadbolt, C.T.; Piispanen, J.P.; Wang, Q.; Ward, T.J.; Worgan, T.L.M.; Oxenford, C.; et al. A Multi-State Outbreak of Salmonella Saintpaul in Australia Associated with Cantaloupe Consumption. Epidemiol. Infect. 2009, 137, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global Epidemiology of CTX-M β-Lactamases: Temporal and Geographical Shifts in Genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-Lactamase (NDM): A Threat to Public Health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A Novel Gene, optrA, That Confers Transferable Resistance to Oxazolidinones and Phenicols and Its Presence in Enterococcus Faecalis and Enterococcus Faecium of Human and Animal Origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Amajoud, N.; Bouchrif, B.; El Maadoudi, M.; Skalli Senhaji, N.; Karraouan, B.; El Harsal, A.; El Abrini, J. Prevalence, Serotype Distribution, and Antimicrobial Resistance of Salmonella Isolated from Food Products in Morocco. J. Infect. Dev. Ctries. 2017, 11, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Eguale, T.; Asrat, D.; Alemayehu, H.; Nana, I.; Gebreyes, W.A.; Gunn, J.S.; Engidawork, E. Phenotypic and Genotypic Characterization of Temporally Related Nontyphoidal Salmonella Strains Isolated from Humans and Food Animals in Central Ethiopia. Zoonoses Public Health 2018, 65, 766–776. [Google Scholar] [CrossRef]
- Hiko, A.; Irsigler, H.; Bräutigam, L.; Ameni, G.; Fries, R.; Maximilian, B. Antimicrobial Resistance and Genotypic Profiles of Salmonella Saintpaul Isolated along Beef Processing and Distribution Continuum. Heliyon 2018, 4, e01025. [Google Scholar] [CrossRef]
- Molla, B.; Miko, A.; Pries, K.; Hildebrandt, G.; Kleer, J.; Schroeter, A.; Helmuth, R. Class 1 Integrons and Resistance Gene Cassettes among Multidrug Resistant Salmonella Serovars Isolated from Slaughter Animals and Foods of Animal Origin in Ethiopia. Acta Trop. 2007, 103, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Baggesen, D.L.; Wegener, H.C.; Christensen, J.P. Typing of Salmonella Enterica Serovar Saintpaul: An Outbreak Investigation. APMIS 1996, 104, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.A.; Kanki, M.; Nguyen, P.D.; Le, H.T.; Ngo, P.T.; Tran, D.N.M.; Le, N.H.; Dang, C.V.; Kawai, T.; Kawahara, R.; et al. Prevalence, Antibiotic Resistance, and Extended-Spectrum and AmpC β-Lactamase Productivity of Salmonella Isolates from Raw Meat and Seafood Samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 2016, 236, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Lee, K. Diversity of TEM-52 Extended-Spectrum -Lactamase-Producing Non-Typhoidal Salmonella Isolates in Korea. J. Antimicrob. Chemother. 2003, 52, 493–496. [Google Scholar] [CrossRef]
- Singh, B.R.; Singh, P.; Agrawal, S.; Teotia, U.; Verma, A.; Sharma, S.; Chandra, M.; Babu, N.; Kant Agarwal, R. Prevalence of Multidrug Resistant Salmonella in Coriander, Mint, Carrot, and Radish in Bareilly and Kanpur, Northern India. Foodborne Pathog. Dis. 2007, 4, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.R.; Singh, M.; Singh, P.; Babu, N.; Chandra, M.; Agarwal, R.K. Prevalence of Multidrug-Resistant Salmonella on Ready-to-Eat Betel Leaves (Paan) and in Water Used for Soaking Betel Leaves in North Indian Cities. J. Food Prot. 2006, 69, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, E.P.; de Melo, R.T.; Nalevaiko, P.C.; Monteiro, G.P.; Fonseca, B.B.; Galvão, N.N.; Giombelli, A.; Rossi, D.A. Spread of the Serotypes and Antimicrobial Resistance in Strains of Salmonella Spp. Isolated from Broiler. Braz. J. Microbiol. 2019, 50, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Scur, M.C.; da Silva Pinto, F.G.; De Bona, E.A.M.; Weber, L.D.; Alves, L.F.A.; de Moura, A.C. Occurrence and Antimicrobial Resistance of Salmonella Serotypes Isolates Recovered from Poultry of Western Paran, Brazil. Afr. J. Agric. Res. 2014, 9, 823–830. [Google Scholar] [CrossRef]
- Wilson, I.G. Antimicrobial Resistance of Salmonella in Raw Retail Chickens, Imported Chicken Portions, and Human Clinical Specimens. J. Food Prot. 2004, 67, 1220–1225. [Google Scholar] [CrossRef]
- Hayford, A.E.; Brown, E.W.; Zhao, S.; Mammel, M.K.; Gangiredla, J.; Abbott, J.W.; Friedman, S.L.; Ayers, S.L.; Lewis, J.L.; Lacher, D.W.; et al. Genetic and Resistance Phenotypic Subtyping of Salmonella Saintpaul Isolates from Various Food Sources and Humans: Phylogenetic Concordance in Combinatory Analyses. Infect. Genet. Evol. 2015, 36, 92–107. [Google Scholar] [CrossRef]
- Eng, S.-K.; Pusparajah, P.; Ab Mutalib, N.-S.; Ser, H.-L.; Chan, K.-G.; Lee, L.-H. Salmonella: A Review on Pathogenesis, Epidemiology and Antibiotic Resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Hur, J.; Jawale, C.; Lee, J.H. Antimicrobial Resistance of Salmonella Isolated from Food Animals: A Review. Food Res. Int. 2012, 45, 819–830. [Google Scholar] [CrossRef]
- Su, L.-H.; Chiu, C.-H.; Chu, C.; Ou, J.T. Antimicrobial Resistance in Nontyphoid Salmonella Serotypes: A Global Challenge. Clin. Infect. Dis. 2004, 39, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, G.L.; Panzenhagen, P.; Ferrari, R.G.; dos Santos, A.; Paschoalin, V.M.F.; Conte-Junior, C.A. Frequency of Antimicrobial Resistance Genes in Salmonella From Brazil by in Silico Whole-Genome Sequencing Analysis: An Overview of the Last Four Decades. Front. Microbiol. 2020, 11, 1864. [Google Scholar] [CrossRef]
- Smith, H.; Bossers, A.; Harders, F.; Wu, G.; Woodford, N.; Schwarz, S.; Guerra, B.; Rodríguez, I.; van Essen-Zandbergen, A.; Brouwer, M.; et al. Characterization of Epidemic IncI1-Iγ Plasmids Harboring Ambler Class A and C Genes in Escherichia Coli and Salmonella Enterica from Animals and Humans. Antimicrob. Agents Chemother. 2015, 59, 5357–5365. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Zhang, J.; Gu, Z.; Li, R.; Chan, E.W.; Yan, M.; Wu, C.; Xu, X.; Chen, S. Prevalence and Molecular Characterization of Mcr-1-Positive Salmonella Strains Recovered from Clinical Specimens in China. Antimicrob. Agents Chemother. 2017, 61, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Godbole, G.; Ashton, P.; Larkin, L.; Dallman, T.; Day, M.; Day, M.; Muller-Pebody, B.; Ellington, M.J.; de Pinna, E.; et al. Detection of the Plasmid-Mediated Mcr-1 Gene Conferring Colistin Resistance in Human and Food Isolates of Salmonella Enterica and Escherichia Coli in England and Wales. J. Antimicrob. Chemother. 2016, 71, 2300–2305. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- COMBAT Consortium; Matamoros, S.; van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; de Jong, M.D.; et al. Global Phylogenetic Analysis of Escherichia Coli and Plasmids Carrying the Mcr-1 Gene Indicates Bacterial Diversity but Plasmid Restriction. Sci. Rep. 2017, 7, 15364. [Google Scholar] [CrossRef]
- Laing, C.R.; Whiteside, M.D.; Gannon, V.P.J. Pan-Genome Analyses of the Species Salmonella Enterica, and Identification of Genomic Markers Predictive for Species, Subspecies, and Serovar. Front. Microbiol. 2017, 8, 1345. [Google Scholar] [CrossRef]
- Park, C.J.; Andam, C.P. Distinct but Intertwined Evolutionary Histories of Multiple Salmonella Enterica Subspecies. mSystems 2020, 5, e00515-19. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Liu, J.; Du, B.; Ruan, H.-H.; Huo, Y.-X.; Du, Y.; Qiao, J. Whole-Genome-Based Survey for Polyphyletic Serovars of Salmonella Enterica Subsp. Enterica Provides New Insights into Public Health Surveillance. Int. J. Mol. Sci. 2020, 21, 5226. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.; Hendriksen, R.S.; Aaresturp, F.M.; Ussery, D.W.; Friis, C. The Salmonella Enterica Pan-Genome. Microb. Ecol. 2011, 62, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Hoede, C.; Tenaillon, O.; Barbe, V.; Baeriswyl, S.; Bidet, P.; Bingen, E.; Bonacorsi, S.; Bouchier, C.; Bouvet, O.; et al. Organised Genome Dynamics in the Escherichia Coli Species Results in Highly Diverse Adaptive Paths. PLoS Genet. 2009, 5, e1000344. [Google Scholar] [CrossRef] [PubMed]
- Iveson, J.B.; Bradshaw, S.D.; Smith, D.W. The Movement of Humans and the Spread of Salmonella into Existing and Pristine Ecosystems. Microbiol. Aust. 2017, 38, 201–203. [Google Scholar] [CrossRef]
- Crump, J.A.; Heyderman, R.S. Invasive Salmonella Infections in Africa. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 673–675. [Google Scholar] [CrossRef]
- Feasey, N.A.; Dougan, G.; Kingsley, R.A.; Heyderman, R.S.; Gordon, M.A. Invasive Non-Typhoidal Salmonella Disease: An Emerging and Neglected Tropical Disease in Africa. Lancet 2012, 379, 2489–2499. [Google Scholar] [CrossRef]
- Lan, N.P.H.; Phuong, T.L.T.; Huu, H.N.; Thuy, L.; Mather, A.E.; Park, S.E.; Marks, F.; Thwaites, G.E.; Chau, N.V.V.; Thompson, C.N.; et al. Invasive Non-Typhoidal Salmonella Infections in Asia: Clinical Observations, Disease Outcome and Dominant Serovars from an Infectious Disease Hospital in Vietnam. PLoS Negl. Trop. Dis. 2016, 10, e0004857. [Google Scholar] [CrossRef]
- Okoro, C.K.; Barquist, L.; Connor, T.R.; Harris, S.R.; Clare, S.; Stevens, M.P.; Arends, M.J.; Hale, C.; Kane, L.; Pickard, D.J.; et al. Signatures of Adaptation in Human Invasive Salmonella Typhimurium ST313 Populations from Sub-Saharan Africa. PLoS Negl. Trop. Dis. 2015, 9, e0003611. [Google Scholar] [CrossRef]
- Ramachandran, G.; Aheto, K.; Shirtliff, M.E.; Tennant, S.M. Poor Biofilm-Forming Ability and Long-Term Survival of Invasive Salmonella Typhimurium ST313. Pathog. Dis. 2016, 74, ftw049. [Google Scholar] [CrossRef] [PubMed]
- Vugia, D.J.; Samuel, M.; Farley, M.M.; Marcus, R.; Shiferaw, B.; Shallow, S.; Smith, K.; Angulo, F.J. Invasive Salmonella Infections in the United States, FoodNet, 1996–1999: Incidence, Serotype Distribution, and Outcome. Clin. Infect. Dis. 2004, 38, S149–S156. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Khan, A.A.; Cheng, C.-M.; Stefanova, R. Molecular Characterization of Salmonella Enterica Serovar Saintpaul Isolated from Imported Seafood, Pepper, Environmental and Clinical Samples. Food Microbiol. 2011, 28, 1124–1128. [Google Scholar] [CrossRef]
- Daitsu, T.; Maeda, S.; Abe, A.; Kanazawa, C.; Shimizu, Y. Relapse of Salmonella Saintpaul Meningitis in a 4-Month-Old Infant. Pediatr. Int. Off. J. Jpn. Pediatr. Soc. 2017, 59, 233–234. [Google Scholar] [CrossRef]
- Jones, B.D.; Lee, C.A.; Falkow, S. Invasion by Salmonella Typhimurium Is Affected by the Direction of Flagellar Rotation. Infect. Immun. 1992, 60, 2475–2480. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.; Perkins, D.J.; Schmidlein, P.J.; Tulapurkar, M.E.; Tennant, S.M. Invasive Salmonella Typhimurium ST313 with Naturally Attenuated Flagellin Elicits Reduced Inflammation and Replicates within Macrophages. PLoS Negl. Trop. Dis. 2015, 9, e3394. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, C.K.; Ikeda, J.S.; Darnell, S.C.; Watson, P.R.; Bispham, J.; Wallis, T.S.; Weinstein, D.L.; Metcalf, E.S.; O’Brien, A.D. Absence of All Components of the Flagellar Export and Synthesis Machinery Differentially Alters Virulence of Salmonella Enterica Serovar Typhimurium in Models of Typhoid Fever, Survival in Macrophages, Tissue Culture Invasiveness, and Calf Enterocolitis. Infect. Immun. 2001, 69, 5619–5625. [Google Scholar] [CrossRef]
- Tomita, T.; Kanegasaki, S. Enhanced Phagocytic Response of Macrophages to Bacteria by Physical Impact Caused by Bacterial Motility or Centrifugation. Infect. Immun. 1982, 38, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, D.L.; Carsiotis, M.; Lissner, C.R.; O’Brien, A.D. Flagella Help Salmonella Typhimurium Survive within Murine Macrophages. Infect. Immun. 1984, 46, 819–825. [Google Scholar] [CrossRef]
- Yim, L.; Sasías, S.; Martínez, A.; Betancor, L.; Estevez, V.; Scavone, P.; Bielli, A.; Sirok, A.; Chabalgoity, J.A. Repression of Flagella Is a Common Trait in Field Isolates of Salmonella Enterica Serovar Dublin and Is Associated with Invasive Human Infections. Infect. Immun. 2014, 82, 1465–1476. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Antimicrobial Drug | Susceptible 1 (S) | Intermediary Resistance 1 (I) | Resistant 1 (R) | PP_BR059 | E. coli 2 | S. Aureus 2 | P. Aeruginosa 2 |
|---|---|---|---|---|---|---|---|
| Ampicillin (AM—10 µg) | ≥17 | 14–16 | ≤13 | 24 (S) | 19 (16–22) | 31 (27–35) | R |
| Chloramphenicol (C—30 µg) | ≥18 | 13–17 | ≤12 | 26 (S) | 26 (21–27) | 21 (19–26) | R |
| Gentamicin (GM—10 µg) | ≥15 | 13–14 | ≤12 | 15 (S) | 18 (19–26) | 21 (19–27) | 25 (16–21) |
| Amikacin (AN—30 µg) | ≥17 | 15–16 | ≤14 | 22 (S) | 25 (19–26) | 23 (20–26) | 26 (18–26) |
| Ciprofloxacin (CIP—5 µg) | ≥21 | 16–20 | ≤15 | 36 (S) | 34 | 22 | 26 |
| Sulfamethoxazole /Trimethoprim (SXT—23.75 /1.25 µg) | ≥16 | 11–15 | ≤10 | 27 (S) | 26 (23–29) | 27 (24–32) | R |
| Streptomycin (S—10 µg) | ≥15 | 12–14 | ≤11 | 15 (S) | 18 (12–20) | 20 (14–22) | 20 |
| Tetracycline (TE—30 µg) | ≥15 | 12–14 | ≤11 | 25 (S) | 26 (18–25) | 28 (24–30) | 13 |
| Amoxicillin /Clavulanic Acid (Amc—20/10 µg) | ≥18 | 14–17 | ≤13 | 27 (S) | 16 (18–24) | 31 (28–36) | R |
| Nalidixid Acid (NAL—30 µg) | ≥19 | 14–18 | ≤13 | 23 (S) | 26 | 16 | R |
| Sulfisoxazole (G—0.25 µg) | ≥17 | 13–16 | ≤12 | 19 (S) | 18 (15–23) | 27 (24–34) | R |
| Ceftiofur (XNL—30 µg) | ≥21 | 18–20 | ≤17 | 23 (S) | 24 (26–31) | 28 (27–31) | 17 (14–18) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panzenhagen, P.; Shah, D.H.; Rodrigues, D.d.P.; Conte Junior, C.A. Worldwide Population Dynamics of Salmonella Saintpaul: Outbreaks, Epidemiology, and Genome Structure. Genes 2025, 16, 254. https://doi.org/10.3390/genes16030254
Panzenhagen P, Shah DH, Rodrigues DdP, Conte Junior CA. Worldwide Population Dynamics of Salmonella Saintpaul: Outbreaks, Epidemiology, and Genome Structure. Genes. 2025; 16(3):254. https://doi.org/10.3390/genes16030254
Chicago/Turabian StylePanzenhagen, Pedro, Devendra H. Shah, Dalia dos Prazeres Rodrigues, and Carlos Adam Conte Junior. 2025. "Worldwide Population Dynamics of Salmonella Saintpaul: Outbreaks, Epidemiology, and Genome Structure" Genes 16, no. 3: 254. https://doi.org/10.3390/genes16030254
APA StylePanzenhagen, P., Shah, D. H., Rodrigues, D. d. P., & Conte Junior, C. A. (2025). Worldwide Population Dynamics of Salmonella Saintpaul: Outbreaks, Epidemiology, and Genome Structure. Genes, 16(3), 254. https://doi.org/10.3390/genes16030254









