LncRNAs Ride the Storm of Epigenetic Marks
Abstract
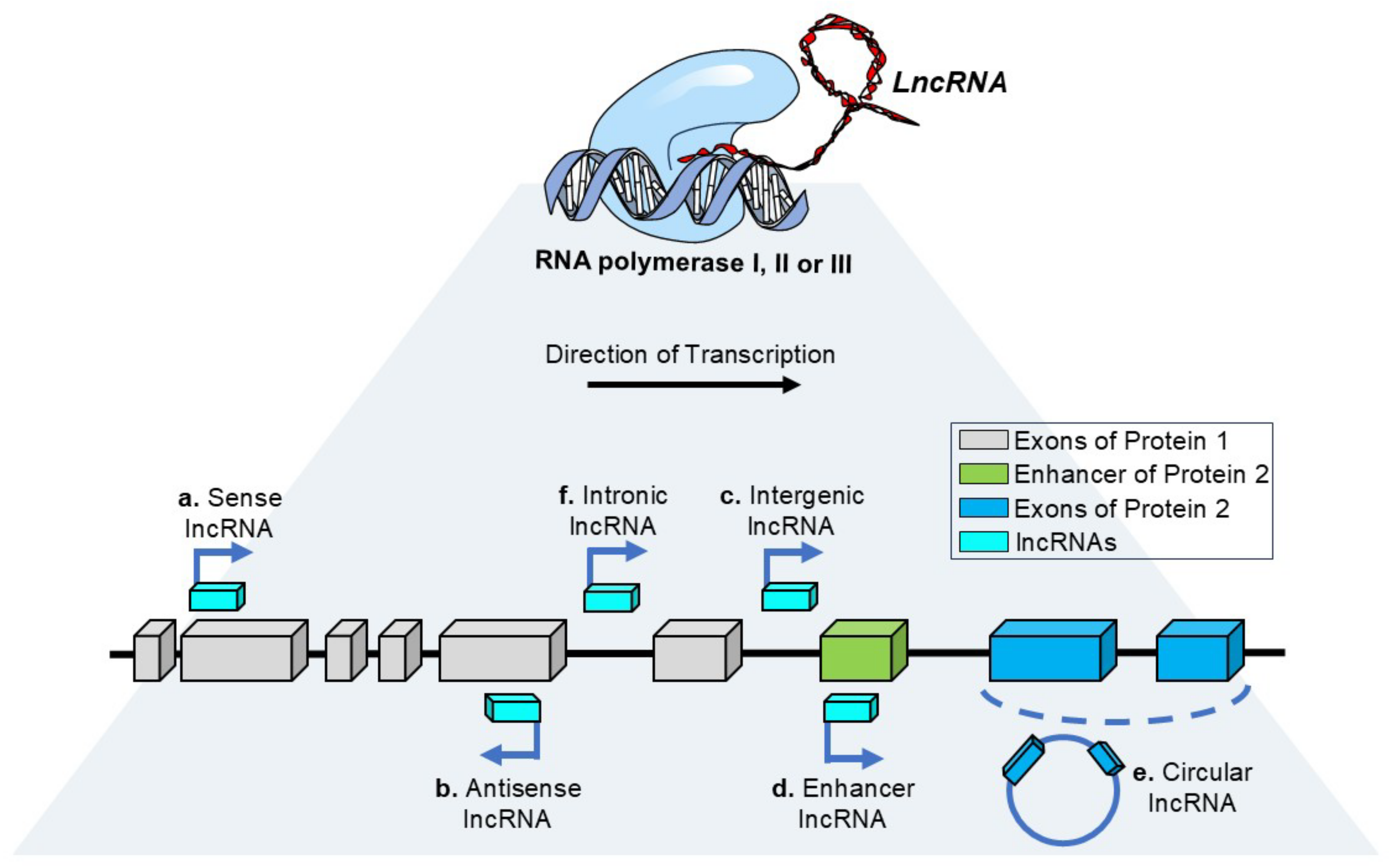
1. Introduction
2. LncRNA Interaction with the Epigenetic Machinery
2.1. Role of lncRNA in DNA Methylation
2.2. Role of lncRNA in DNA Hydroxymethylation
2.3. Role of lncRNA in Histone Methylation
2.4. Role of lncRNA in Histone Acetylation
2.5. Epigenetic Memory of Histone Acetylation and Chromatin Replication
3. The Multifaceted Roles of lncRNAs in Epigenetic Regulation and Disease Development
3.1. Role of lncRNAs in Cellular Senescence and Aging: Guardians and Regulators of Genome Stability
3.2. Long Non-Coding RNAs in Age-Related Cardiovascular Diseases: Molecular Regulators of Heart Health and Dysfunction
4. Long Non-Coding RNAs in the Epigenetic of Cancers
4.1. Age-Related Factors and lncRNAs in Epigenetic Cell Reprogramming Leading to Malignant Transformation
4.2. Role of LncRNAs as Epigenetic Regulators of Acute Myeloid Leukemia (AML)
4.3. LncRNAs Regulate the Epigenetics of Non-Small-Cell Lung Cancer
4.4. LncRNAs in Breast Cancer: Mediators of Tamoxifen Resistance and Disease Advancement
4.5. LncRNAs in the Epigenetic Modulation of Molecular Mechanisms in Thyroid Cancer
5. LncRNAs in Inflammatory and Infectious Diseases (COVID-19)
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hanly, D.J.; Esteller, M.; Berdasco, M. Interplay between long non-coding RNAs and epigenetic machinery: Emerging targets in cancer? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170074. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Kim, V.N. Small and long non-coding RNAs: Past, present, and future. Cell 2024, 187, 6451–6485. [Google Scholar] [CrossRef] [PubMed]
- Antonazzo, G.; Gaudet, P.; Lovering, R.C.; Attrill, H. Representation of non-coding RNA-mediated regulation of gene expression using the Gene Ontology. RNA Biol. 2024, 21, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Kugel, J.F.; Goodrich, J.A. Non-coding RNAs: Key regulators of mammalian transcription. Trends Biochem. Sci. 2012, 37, 144–151. [Google Scholar] [CrossRef]
- Turowski, T.W.; Boguta, M. Specific Features of RNA Polymerases I and III: Structure and Assembly. Front. Mol. Biosci. 2021, 8, 680090. [Google Scholar] [CrossRef]
- Goodrich, J.A.; Kugel, J.F. Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell Biol. 2006, 7, 612–616. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.A.; Alba, M.M. Long non-coding RNAs as a source of new peptides. Elife 2014, 3, e03523. [Google Scholar] [CrossRef]
- Poliseno, L.; Lanza, M.; Pandolfi, P.P. Coding, or non-coding, that is the question. Cell Res. 2024, 34, 609–629. [Google Scholar] [CrossRef]
- Dasgupta, A.; Prensner, J.R. Upstream open reading frames: New players in the landscape of cancer gene regulation. NAR Cancer 2024, 6, zcae023. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Song, Q.; Zhang, W.; Geng, B.; Cai, J. Roles of long noncoding RNAs in aging and aging complications. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.W.; Faulkner, G.J.; Dinger, M.E. Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat. Rev. Genet. 2020, 21, 191–201. [Google Scholar] [CrossRef]
- Frith, M.C.; Wilming, L.G.; Forrest, A.; Kawaji, H.; Tan, S.L.; Wahlestedt, C.; Bajic, V.B.; Kai, C.; Kawai, J.; Carninci, P.; et al. Pseudo-messenger RNA: Phantoms of the transcriptome. PLoS Genet. 2006, 2, e23. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. Gencode 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- St Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef]
- Lozano-Vidal, N.; Bink, D.I.; Boon, R.A. Long noncoding RNA in cardiac aging and disease. J. Mol. Cell Biol. 2019, 11, 860–867. [Google Scholar] [CrossRef]
- Ferrer, J.; Dimitrova, N. Transcription regulation by long non-coding RNAs: Mechanisms and disease relevance. Nat. Rev. Mol. Cell Biol. 2024, 25, 396–415. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Author Correction: Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 159. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, H.; Wang, H. Long noncoding RNAs in DNA methylation: New players stepping into the old game. Cell Biosci. 2016, 6, 45. [Google Scholar] [CrossRef]
- Anastasiadi, D.; Esteve-Codina, A.; Piferrer, F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenet. Chromatin 2018, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, J.; Huang, S.; He, X. Genome-wide analysis reveals that exon methylation facilitates its selective usage in the human transcriptome. Brief. Bioinform. 2018, 19, 754–764. [Google Scholar] [CrossRef]
- Di Ruscio, A.; Ebralidze, A.K.; Benoukraf, T.; Amabile, G.; Goff, L.A.; Terragni, J.; Figueroa, M.E.; De Figueiredo Pontes, L.L.; Alberich-Jorda, M.; Zhang, P.; et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013, 503, 371–376. [Google Scholar] [CrossRef]
- Esposito, C.L.; Autiero, I.; Sandomenico, A.; Li, H.; Bassal, M.A.; Ibba, M.L.; Wang, D.; Rinaldi, L.; Ummarino, S.; Gaggi, G.; et al. Targeted systematic evolution of an RNA platform neutralizing DNMT1 function and controlling DNA methylation. Nat. Commun. 2023, 14, 99. [Google Scholar] [CrossRef]
- Chalei, V.; Sansom, S.N.; Kong, L.; Lee, S.; Montiel, J.F.; Vance, K.W.; Ponting, C.P. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 2014, 3, e04530. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, Y.; Bao, X.; Zhu, X.; Kwok, Y.K.; Sun, K.; Chen, X.; Huang, Y.; Jauch, R.; Esteban, M.A.; et al. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015, 25, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Savell, K.E.; Gallus, N.V.; Simon, R.C.; Brown, J.A.; Revanna, J.S.; Osborn, M.K.; Song, E.Y.; O’Malley, J.J.; Stackhouse, C.T.; Norvil, A.; et al. Extra-coding RNAs regulate neuronal DNA methylation dynamics. Nat. Commun. 2016, 7, 12091. [Google Scholar] [CrossRef]
- Merry, C.R.; Forrest, M.E.; Sabers, J.N.; Beard, L.; Gao, X.H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253. [Google Scholar] [CrossRef]
- Fink, J.J.; Delaney-Busch, N.; Dawes, R.; Nanou, E.; Folts, C.; Harikrishnan, K.; Hempel, C.; Upadhyay, H.; Nguyen, T.; Shroff, H.; et al. Deep functional measurements of Fragile X syndrome human neurons reveal multiparametric electrophysiological disease phenotype. Commun. Biol. 2024, 7, 1447. [Google Scholar] [CrossRef]
- Peschansky, V.J.; Pastori, C.; Zeier, Z.; Motti, D.; Wentzel, K.; Velmeshev, D.; Magistri, M.; Bixby, J.L.; Lemmon, V.P.; Silva, J.P.; et al. Changes in expression of the long non-coding RNA FMR4 associate with altered gene expression during differentiation of human neural precursor cells. Front. Genet. 2015, 6, 263. [Google Scholar] [CrossRef][Green Version]
- Huang, G.; Zhu, H.; Wu, S.; Cui, M.; Xu, T. Long Noncoding RNA Can Be a Probable Mechanism and a Novel Target for Diagnosis and Therapy in Fragile X Syndrome. Front. Genet. 2019, 10, 446. [Google Scholar] [CrossRef] [PubMed]
- Liaci, C.; Prandi, L.; Pavinato, L.; Brusco, A.; Maldotti, M.; Molineris, I.; Oliviero, S.; Merlo, G.R. The Emerging Roles of Long Non-Coding RNAs in Intellectual Disability and Related Neurodevelopmental Disorders. Int. J. Mol. Sci. 2022, 23, 6118. [Google Scholar] [CrossRef]
- Ladd, P.D.; Smith, L.E.; Rabaia, N.A.; Moore, J.M.; Georges, S.A.; Hansen, R.S.; Hagerman, R.J.; Tassone, F.; Tapscott, S.J.; Filippova, G.N. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 2007, 16, 3174–3187. [Google Scholar] [CrossRef]
- Richa, R.; Sinha, R.P. Hydroxymethylation of DNA: An epigenetic marker. EXCLI J. 2014, 13, 592–610. [Google Scholar]
- Solary, E.; Bernard, O.A.; Tefferi, A.; Fuks, F.; Vainchenker, W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia 2014, 28, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Kunimoto, H. TET2 as an epigenetic master regulator for normal and malignant hematopoiesis. Cancer Sci. 2014, 105, 1093–1099. [Google Scholar] [CrossRef]
- Gu, T.P.; Guo, F.; Yang, H.; Wu, H.P.; Xu, G.F.; Liu, W.; Xie, Z.G.; Shi, L.; He, X.; Jin, S.G.; et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011, 477, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Ruzov, A.; Tsenkina, Y.; Serio, A.; Dudnakova, T.; Fletcher, J.; Bai, Y.; Chebotareva, T.; Pells, S.; Hannoun, Z.; Sullivan, G.; et al. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011, 21, 1332–1342. [Google Scholar] [CrossRef] [PubMed]
- de la Rica, L.; Rodriguez-Ubreva, J.; Garcia, M.; Islam, A.B.; Urquiza, J.M.; Hernando, H.; Christensen, J.; Helin, K.; Gomez-Vaquero, C.; Ballestar, E. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013, 14, R99. [Google Scholar] [CrossRef]
- Costa, Y.; Ding, J.; Theunissen, T.W.; Faiola, F.; Hore, T.A.; Shliaha, P.V.; Fidalgo, M.; Saunders, A.; Lawrence, M.; Dietmann, S.; et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 2013, 495, 370–374. [Google Scholar] [CrossRef]
- Chen, L.L.; Lin, H.P.; Zhou, W.J.; He, C.X.; Zhang, Z.Y.; Cheng, Z.L.; Song, J.B.; Liu, P.; Chen, X.Y.; Xia, Y.K.; et al. SNIP1 Recruits TET2 to Regulate c-MYC Target Genes and Cellular DNA Damage Response. Cell Rep. 2018, 25, 1485–1500. [Google Scholar] [CrossRef] [PubMed]
- Sardina, J.L.; Collombet, S.; Tian, T.V.; Gomez, A.; Di Stefano, B.; Berenguer, C.; Brumbaugh, J.; Stadhouders, R.; Segura-Morales, C.; Gut, M.; et al. Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem Cell 2018, 23, 905–906. [Google Scholar] [CrossRef]
- Arab, K.; Park, Y.J.; Lindroth, A.M.; Schafer, A.; Oakes, C.; Weichenhan, D.; Lukanova, A.; Lundin, E.; Risch, A.; Meister, M.; et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 2014, 55, 604–614. [Google Scholar] [CrossRef]
- Zhou, L.; Ren, M.; Zeng, T.; Wang, W.; Wang, X.; Hu, M.; Su, S.; Sun, K.; Wang, C.; Liu, J.; et al. TET2-interacting long noncoding RNA promotes active DNA demethylation of the MMP-9 promoter in diabetic wound healing. Cell Death Dis. 2019, 10, 813. [Google Scholar] [CrossRef] [PubMed]
- Song, C.X.; Szulwach, K.E.; Fu, Y.; Dai, Q.; Yi, C.; Li, X.; Li, Y.; Chen, C.H.; Zhang, W.; Jian, X.; et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011, 29, 68–72. [Google Scholar] [CrossRef]
- Shi, D.Q.; Ali, I.; Tang, J.; Yang, W.C. New Insights into 5hmC DNA Modification: Generation, Distribution and Function. Front. Genet. 2017, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Kienhofer, S.; Musheev, M.U.; Stapf, U.; Helm, M.; Schomacher, L.; Niehrs, C.; Schafer, A. GADD45a physically and functionally interacts with TET1. Differentiation 2015, 90, 59–68. [Google Scholar] [CrossRef]
- Li, Z.; Gu, T.P.; Weber, A.R.; Shen, J.Z.; Li, B.Z.; Xie, Z.G.; Yin, R.; Guo, F.; Liu, X.; Tang, F.; et al. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res. 2015, 43, 3986–3997. [Google Scholar] [CrossRef]
- Arab, K.; Karaulanov, E.; Musheev, M.; Trnka, P.; Schafer, A.; Grummt, I.; Niehrs, C. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 2019, 51, 217–223. [Google Scholar] [CrossRef]
- Arivazhagan, L.; Lopez-Diez, R.; Shekhtman, A.; Ramasamy, R.; Schmidt, A.M. Glycation and a Spark of ALEs (Advanced Lipoxidation End Products)—Igniting RAGE/Diaphanous-1 and Cardiometabolic Disease. Front. Cardiovasc. Med. 2022, 9, 937071. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Miller, K.M. Histone methylation and the DNA damage response. Mutat. Res. Rev. Mutat. Res. 2019, 780, 37–47. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef]
- Terranova, R.; Yokobayashi, S.; Stadler, M.B.; Otte, A.P.; van Lohuizen, M.; Orkin, S.H.; Peters, A.H. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell 2008, 15, 668–679. [Google Scholar] [CrossRef]
- Pasmant, E.; Laurendeau, I.; Heron, D.; Vidaud, M.; Vidaud, D.; Bieche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Butler, A.A.; Johnston, D.R.; Kaur, S.; Lubin, F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 2019, 12, eaaw9277. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am. J. Cancer Res. 2018, 8, 81–90. [Google Scholar] [PubMed]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef]
- Chen, X.; Xie, R.; Gu, P.; Huang, M.; Han, J.; Dong, W.; Xie, W.; Wang, B.; He, W.; Zhong, G.; et al. Long Noncoding RNA LBCS Inhibits Self-Renewal and Chemoresistance of Bladder Cancer Stem Cells through Epigenetic Silencing of SOX2. Clin. Cancer Res. 2019, 25, 1389–1403. [Google Scholar] [CrossRef] [PubMed]
- Mersfelder, E.L.; Parthun, M.R. The tale beyond the tail: Histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006, 34, 2653–2662. [Google Scholar] [CrossRef]
- Wang, Z.; Zang, C.; Cui, K.; Schones, D.E.; Barski, A.; Peng, W.; Zhao, K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009, 138, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Policarpi, C.; Munafo, M.; Tsagkris, S.; Carlini, V.; Hackett, J.A. Systematic epigenome editing captures the context-dependent instructive function of chromatin modifications. Nat. Genet. 2024, 56, 1168–1180. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, J.S. A short guide to histone deacetylases including recent progress on class II enzymes. Exp. Mol. Med. 2020, 52, 204–212. [Google Scholar] [CrossRef]
- Camblong, J.; Iglesias, N.; Fickentscher, C.; Dieppois, G.; Stutz, F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 2007, 131, 706–717. [Google Scholar] [CrossRef]
- Hegazy, Y.A.; Cloutier, S.C.; Utturkar, S.M.; Das, S.; Tran, E.J. The genomic region of the 3’ untranslated region (3’UTR) of PHO84, rather than the antisense RNA, promotes gene repression. Nucleic Acids Res. 2023, 51, 7900–7913. [Google Scholar] [CrossRef]
- Ding, H.; Wang, F.; Shi, X.; Ma, H.; Du, Y.; Hou, L.; Xing, N. LncRNA MALAT1 induces the dysfunction of beta cells via reducing the histone acetylation of the PDX-1 promoter in type 1 diabetes. Exp. Mol. Pathol. 2020, 114, 104432. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Feng, B.; Chakrabarti, S. ANRIL: A Regulator of VEGF in Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Poltronieri, P. Regulatory RNAs: Role as scaffolds assembling protein complexes and their epigenetic deregulation. Explor. Target. Antitumor Ther. 2024, 5, 841–876. [Google Scholar] [CrossRef]
- Zhang, C.; Ge, S.; Gong, W.; Xu, J.; Guo, Z.; Liu, Z.; Gao, X.; Wei, X.; Ge, S. LncRNA ANRIL acts as a modular scaffold of WDR5 and HDAC3 complexes and promotes alteration of the vascular smooth muscle cell phenotype. Cell Death Dis. 2020, 11, 435. [Google Scholar] [CrossRef]
- Tong, P.; Peng, Q.H.; Gu, L.M.; Xie, W.W.; Li, W.J. LncRNA-MEG3 alleviates high glucose induced inflammation and apoptosis of retina epithelial cells via regulating miR-34a/SIRT1 axis. Exp. Mol. Pathol. 2019, 107, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.; Zhu, C.; Wang, K. A review of current evidence about lncRNA MEG3: A tumor suppressor in multiple cancers. Front. Cell Dev. Biol. 2022, 10, 997633. [Google Scholar] [CrossRef]
- Akerman, I.; Kasaai, B.; Bazarova, A.; Sang, P.B.; Peiffer, I.; Artufel, M.; Derelle, R.; Smith, G.; Rodriguez-Martinez, M.; Romano, M.; et al. A predictable conserved DNA base composition signature defines human core DNA replication origins. Nat. Commun. 2020, 11, 4826. [Google Scholar] [CrossRef]
- Tian, M.; Wang, Z.; Su, Z.; Shibata, E.; Shibata, Y.; Dutta, A.; Zang, C. Integrative analysis of DNA replication origins and ORC-/MCM-binding sites in human cells reveals a lack of overlap. Elife 2024, 12, RP89548. [Google Scholar] [CrossRef]
- Vogelauer, M.; Rubbi, L.; Lucas, I.; Brewer, B.J.; Grunstein, M. Histone acetylation regulates the time of replication origin firing. Mol. Cell 2002, 10, 1223–1233. [Google Scholar] [CrossRef]
- Miotto, B.; Struhl, K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell 2010, 37, 57–66. [Google Scholar] [CrossRef]
- Miotto, B.; Struhl, K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes. Dev. 2008, 22, 2633–2638. [Google Scholar] [CrossRef] [PubMed]
- Kemp, M.G.; Ghosh, M.; Liu, G.; Leffak, M. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res. 2005, 33, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishnan, A.; Gafken, P.R.; Tsukiyama, T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat. Struct. Mol. Biol. 2010, 17, 430–437. [Google Scholar] [CrossRef]
- Alabert, C.; Groth, A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012, 13, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Morgan, K.R.; Petryk, N.; Groth, A. Chromatin replication and epigenetic cell memory. Nat. Cell Biol. 2020, 22, 361–371. [Google Scholar] [CrossRef]
- Alabert, C.; Barth, T.K.; Reveron-Gomez, N.; Sidoli, S.; Schmidt, A.; Jensen, O.N.; Imhof, A.; Groth, A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes. Dev. 2015, 29, 585–590. [Google Scholar] [CrossRef]
- Loppin, B.; Berger, F. Histone Variants: The Nexus of Developmental Decisions and Epigenetic Memory. Annu. Rev. Genet. 2020, 54, 121–149. [Google Scholar] [CrossRef]
- Song, Y.; Wang, R.; Li, L.W.; Liu, X.; Wang, Y.F.; Wang, Q.X.; Zhang, Q. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int. J. Oncol. 2019, 54, 77–86. [Google Scholar] [CrossRef]
- Long, H.; Zhang, L.; Lv, M.; Wen, Z.; Zhang, W.; Chen, X.; Zhang, P.; Li, T.; Chang, L.; Jin, C.; et al. Author Correction: H2A.Z facilitates licensing and activation of early replication origins. Nature 2020, 578, E8. [Google Scholar] [CrossRef]
- Ebralidze, A.K.; Ummarino, S.; Bassal, M.A.; Zhang, H.; Budnik, B.; Monteleone, E.; Kappei, D.; Liu, Y.V.; Tenen, D.E.; Coffey, R.; et al. Formation and recycling of an active epigenetic mark mediated by cell cycle-specific RNAs. bioRxiv 2021. [Google Scholar] [CrossRef]
- Grammatikakis, I.; Panda, A.C.; Abdelmohsen, K.; Gorospe, M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging 2014, 6, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, H.; Hu, Q.; Wang, L.; Liu, J.; Zheng, Z.; Zhang, W.; Ren, J.; Zhu, F.; Liu, G.H. Epigenetic regulation of aging: Implications for interventions of aging and diseases. Signal Transduct. Target. Ther. 2022, 7, 374. [Google Scholar] [CrossRef] [PubMed]
- Ovadya, Y.; Krizhanovsky, V. Senescent cells: SASPected drivers of age-related pathologies. Biogerontology 2014, 15, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Anver, S.; Sumit, A.F.; Sun, X.M.; Hatimy, A.; Thalassinos, K.; Marguerat, S.; Alic, N.; Bahler, J. Ageing-associated long non-coding RNA extends lifespan and reduces translation in non-dividing cells. EMBO Rep. 2024, 25, 4921–4949. [Google Scholar] [CrossRef]
- Ji, Y.; Zuo, C.; Liao, N.; Yao, L.; Yang, R.; Chen, H.; Wen, F. Identification of key lncRNAs in age-related macular degeneration through integrated bioinformatics and experimental validation. Aging 2024, 16, 5435–5451. [Google Scholar] [CrossRef]
- He, J.; Tu, C.; Liu, Y. Role of lncRNAs in aging and age-related diseases. Aging Med. 2018, 1, 158–175. [Google Scholar] [CrossRef]
- Coppe, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes. Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef]
- Calado, R.T.; Young, N.S. Telomere diseases. N. Engl. J. Med. 2009, 361, 2353–2365. [Google Scholar] [CrossRef]
- Porro, A.; Feuerhahn, S.; Delafontaine, J.; Riethman, H.; Rougemont, J.; Lingner, J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014, 5, 5379. [Google Scholar] [CrossRef]
- Kwapisz, M.; Morillon, A. Subtelomeric Transcription and its Regulation. J. Mol. Biol. 2020, 432, 4199–4219. [Google Scholar] [CrossRef] [PubMed]
- Shirai, A.; Kawaguchi, T.; Shimojo, H.; Muramatsu, D.; Ishida-Yonetani, M.; Nishimura, Y.; Kimura, H.; Nakayama, J.I.; Shinkai, Y. Correction: Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly. Elife 2017, 6, e25317. [Google Scholar] [CrossRef]
- Kim, H.S.; Roche, B.; Bhattacharjee, S.; Todeschini, L.; Chang, A.Y.; Hammell, C.; Verdel, A.; Martienssen, R.A. Clr4(SUV39H1) ubiquitination and non-coding RNA mediate transcriptional silencing of heterochromatin via Swi6 phase separation. Nat. Commun. 2024, 15, 9384. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, W.; Jang, Y.; Sommers, J.A.; Yi, G.; Puligilla, C.; Croteau, D.L.; Yang, Y.; Kai, M.; Liu, Y. The RNA-binding motif protein 14 regulates telomere integrity at the interface of TERRA and telomeric R-loops. Nucleic Acids Res. 2023, 51, 12242–12260. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.P.; Cifuentes-Rojas, C.; Kesner, B.; Aeby, E.; Lee, H.G.; Wei, C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 2017, 170, 86–101.e16. [Google Scholar] [CrossRef]
- Aguilera, P.; Lopez-Contreras, A.J. ATRX, a guardian of chromatin. Trends Genet. 2023, 39, 505–519. [Google Scholar] [CrossRef]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef]
- Sharma, V.; Khurana, S.; Kubben, N.; Abdelmohsen, K.; Oberdoerffer, P.; Gorospe, M.; Misteli, T. A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep. 2015, 16, 1520–1534. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Garcia, J.T.; Hung, T.; Flynn, R.A.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef]
- Zhang, A.; Zhou, N.; Huang, J.; Liu, Q.; Fukuda, K.; Ma, D.; Lu, Z.; Bai, C.; Watabe, K.; Mo, Y.Y. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013, 23, 340–350. [Google Scholar] [CrossRef]
- Paneni, F.; Diaz Canestro, C.; Libby, P.; Luscher, T.F.; Camici, G.G. The Aging Cardiovascular System: Understanding It at the Cellular and Clinical Levels. J. Am. Coll. Cardiol. 2017, 69, 1952–1967. [Google Scholar] [CrossRef]
- Li, H.; Zhao, Q.; Chang, L.; Wei, C.; Bei, H.; Yin, Y.; Chen, M.; Wang, H.; Liang, J.; Wu, Y. LncRNA MALAT1 modulates ox-LDL induced EndMT through the Wnt/beta-catenin signaling pathway. Lipids Health Dis. 2019, 18, 62. [Google Scholar] [CrossRef]
- Quinodoz, S.; Guttman, M. Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014, 24, 651–663. [Google Scholar] [CrossRef]
- Wang, K.; Liu, F.; Zhou, L.Y.; Long, B.; Yuan, S.M.; Wang, Y.; Liu, C.Y.; Sun, T.; Zhang, X.J.; Li, P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef]
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017, 9, eaai9118. [Google Scholar] [CrossRef]
- Ounzain, S.; Pezzuto, I.; Micheletti, R.; Burdet, F.; Sheta, R.; Nemir, M.; Gonzales, C.; Sarre, A.; Alexanian, M.; Blow, M.J.; et al. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell. Cardiol. 2014, 76, 55–70. [Google Scholar] [CrossRef]
- Feridooni, H.A.; Dibb, K.M.; Howlett, S.E. How cardiomyocyte excitation, calcium release and contraction become altered with age. J. Mol. Cell. Cardiol. 2015, 83, 62–72. [Google Scholar] [CrossRef]
- Shen, C.; Kong, B.; Liu, Y.; Xiong, L.; Shuai, W.; Wang, G.; Quan, D.; Huang, H. YY1-induced upregulation of lncRNA KCNQ1OT1 regulates angiotensin II-induced atrial fibrillation by modulating miR-384b/CACNA1C axis. Biochem. Biophys. Res. Commun. 2018, 505, 134–140. [Google Scholar] [CrossRef]
- Finkel, T.; Serrano, M.; Blasco, M.A. The common biology of cancer and ageing. Nature 2007, 448, 767–774. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013, 75, 685–705. [Google Scholar] [CrossRef]
- Fulop, T.; Larbi, A.; Dupuis, G.; Le Page, A.; Frost, E.H.; Cohen, A.A.; Witkowski, J.M.; Franceschi, C. Immunosenescence and Inflamm-Aging As Two Sides of the Same Coin: Friends or Foes? Front. Immunol. 2017, 8, 1960. [Google Scholar] [CrossRef]
- Solana, R.; Tarazona, R.; Gayoso, I.; Lesur, O.; Dupuis, G.; Fulop, T. Innate immunosenescence: Effect of aging on cells and receptors of the innate immune system in humans. Semin. Immunol. 2012, 24, 331–341. [Google Scholar] [CrossRef]
- Esposito, R.; Bosch, N.; Lanzos, A.; Polidori, T.; Pulido-Quetglas, C.; Johnson, R. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell 2019, 35, 545–557. [Google Scholar] [CrossRef]
- Fang, K.; Xu, H.; Yuan, S.; Li, X.; Chen, X.; Fan, X.; Gao, X.; Zhang, L.; Sun, S.; Zhu, X. LncRNA mediated metabolic reprogramming: The chief culprits of solid tumor malignant progression: An update review. Nutr Metab 2024, 21, 89. [Google Scholar] [CrossRef]
- Lin, Y.H. Crosstalk of lncRNA and Cellular Metabolism and Their Regulatory Mechanism in Cancer. Int. J. Mol. Sci. 2020, 21, 2947. [Google Scholar] [CrossRef]
- Xin, X.; Li, Q.; Fang, J.; Zhao, T. LncRNA HOTAIR: A Potential Prognostic Factor and Therapeutic Target in Human Cancers. Front. Oncol. 2021, 11, 679244. [Google Scholar] [CrossRef]
- Chen, X.; Wang, S.; Jiang, X.; Zhang, M.; Ding, Y. Long non-coding RNA HOTAIR: From pan-cancer analysis to colorectal cancer-related uridine metabolism. Aging 2024, 16, 7752–7773. [Google Scholar] [CrossRef]
- Potolitsyna, E.; Hazell Pickering, S.; Germier, T.; Collas, P.; Briand, N. Long non-coding RNA HOTAIR regulates cytoskeleton remodeling and lipid storage capacity during adipogenesis. Sci. Rep. 2022, 12, 10157. [Google Scholar] [CrossRef]
- Obaid, M.; Udden, S.M.N.; Alluri, P.; Mandal, S.S. LncRNA HOTAIR regulates glucose transporter Glut1 expression and glucose uptake in macrophages during inflammation. Sci. Rep. 2021, 11, 232. [Google Scholar] [CrossRef]
- Pinton, G.; Perucca, M.; Gigliotti, V.; Mantovani, E.; Clemente, N.; Malecka, J.; Chrostek, G.; Dematteis, G.; Lim, D.; Moro, L.; et al. EZH2-Mediated H3K27 Trimethylation in the Liver of Mice Is an Early Epigenetic Event Induced by High-Fat Diet Exposure. Nutrients 2024, 16, 3260. [Google Scholar] [CrossRef]
- El Said, N.H.; Della Valle, F.; Liu, P.; Paytuvi-Gallart, A.; Adroub, S.; Gimenez, J.; Orlando, V. Malat-1-PRC2-EZH1 interaction supports adaptive oxidative stress dependent epigenome remodeling in skeletal myotubes. Cell Death Dis. 2021, 12, 850. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, L.; Li, H.; Sun, T.; Wen, X.; Li, X.; Meng, Y.; Li, Y.; Liu, M.; Liu, S.; et al. Nuclear-Encoded lncRNA MALAT1 Epigenetically Controls Metabolic Reprogramming in HCC Cells through the Mitophagy Pathway. Mol. Ther. Nucleic Acids 2021, 23, 264–276. [Google Scholar] [CrossRef]
- Yang, X.; Wang, Y.; Pang, S.; Li, X.; Wang, P.; Ma, R.; Ma, Y.; Song, C. LINC00665 promotes the progression of acute myeloid leukemia by regulating the miR-4458/DOCK1 pathway. Sci. Rep. 2021, 11, 5009. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, S.; Chen, X.; Wang, C. HOTAIR suppresses PTEN via DNMT3b and confers drug resistance in acute myeloid leukemia. Hematology 2021, 26, 170–178. [Google Scholar] [CrossRef]
- Song, H.; Chen, L.; Liu, W.; Xu, X.; Zhou, Y.; Zhu, J.; Chen, X.; Li, Z.; Zhou, H. Depleting long noncoding RNA HOTAIR attenuates chronic myelocytic leukemia progression by binding to DNA methyltransferase 1 and inhibiting PTEN gene promoter methylation. Cell Death Dis. 2021, 12, 440. [Google Scholar] [CrossRef]
- Merlo, A.; Herman, J.G.; Mao, L.; Lee, D.J.; Gabrielson, E.; Burger, P.C.; Baylin, S.B.; Sidransky, D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1995, 1, 686–692. [Google Scholar] [CrossRef]
- Kong, Y.; Hsieh, C.H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B Locus With Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef]
- Zemlyakova, V.V.; Zhevlova, A.I.; Zborovskaya, I.B.; Strelnikov, V.V.; Laktionov, K.K.; Zaletaev, D.V.; Nemtsova, M.V. Methylation Profile of Several Tumor Suppressor Genes in Non-Small-Cell Lung Cancer. Mol. Biol. 2003, 37, 836–840. [Google Scholar] [CrossRef]
- Murugan, A.K.; Munirajan, A.K.; Alzahrani, A.S. Long noncoding RNAs: Emerging players in thyroid cancer pathogenesis. Endocr. Relat. Cancer 2018, 25, R59–R82. [Google Scholar] [CrossRef]
- Lan, W.G.; Xu, D.H.; Xu, C.; Ding, C.L.; Ning, F.L.; Zhou, Y.L.; Ma, L.B.; Liu, C.M.; Han, X. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol. Rep. 2016, 36, 263–270. [Google Scholar] [CrossRef]
- Nie, F.Q.; Sun, M.; Yang, J.S.; Xie, M.; Xu, T.P.; Xia, R.; Liu, Y.W.; Liu, X.H.; Zhang, E.B.; Lu, K.H.; et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol. Cancer Ther. 2015, 14, 268–277. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, Y.; Du, L.; Jiang, X.; Yan, S.; Duan, W.; Li, J.; Zhan, Y.; Wang, L.; Zhang, S.; et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol. Oncol. 2018, 12, 648–658. [Google Scholar] [CrossRef]
- Lv, X.; Cui, Z.; Li, H.; Li, J.; Yang, Z.; Bi, Y.; Gao, M.; Zhang, Z.; Wang, S.; Zhou, B.; et al. Association between polymorphism in CDKN2B-AS1 gene and its interaction with smoking on the risk of lung cancer in a Chinese population. Hum. Genom. 2019, 13, 58. [Google Scholar] [CrossRef]
- Chen, B.J.; Wu, Y.L.; Tanaka, Y.; Zhang, W. Small molecules targeting c-Myc oncogene: Promising anti-cancer therapeutics. Int. J. Biol. Sci. 2014, 10, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhou, X.; Xu, L.; Rong, C.; Shen, C.; Bian, W. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. Onco Targets Ther. 2016, 9, 3077–3084. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, R.; Mao, Y.; Chen, K.; He, W.; Shi, W.; Han, Y. The long noncoding RNA ANRIL acts as an oncogene and contributes to paclitaxel resistance of lung adenocarcinoma A549 cells. Oncotarget 2017, 8, 39177–39184. [Google Scholar] [CrossRef]
- Loewen, G.; Jayawickramarajah, J.; Zhuo, Y.; Shan, B. Functions of lncRNA HOTAIR in lung cancer. J. Hematol. Oncol. 2014, 7, 90. [Google Scholar] [CrossRef]
- Nazari, M.; Babakhanzadeh, E.; Mollazadeh, A.; Ahmadzade, M.; Mohammadi Soleimani, E.; Hajimaqsoudi, E. HOTAIR in cancer: Diagnostic, prognostic, and therapeutic perspectives. Cancer Cell Int. 2024, 24, 415. [Google Scholar] [CrossRef]
- Terashima, M.; Tange, S.; Ishimura, A.; Suzuki, T. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J. Biol. Chem. 2017, 292, 82–99. [Google Scholar] [CrossRef]
- Peng, J.C.; Valouev, A.; Swigut, T.; Zhang, J.; Zhao, Y.; Sidow, A.; Wysocka, J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 2009, 139, 1290–1302. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Kim, W.; Fujiwara, Y.; Simon, M.D.; Liu, Y.; Mysliwiec, M.R.; Yuan, G.C.; Lee, Y.; Orkin, S.H. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 2009, 139, 1303–1314. [Google Scholar] [CrossRef]
- Pasini, D.; Cloos, P.A.; Walfridsson, J.; Olsson, L.; Bukowski, J.P.; Johansen, J.V.; Bak, M.; Tommerup, N.; Rappsilber, J.; Helin, K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 2010, 464, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Margueron, R.; Ku, M.; Chambon, P.; Bernstein, B.E.; Reinberg, D. Jarid2 and PRC2, partners in regulating gene expression. Genes. Dev. 2010, 24, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Bonasio, R.; Saldana-Meyer, R.; Yoshida, T.; Son, J.; Nishino, K.; Umezawa, A.; Reinberg, D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 2014, 53, 290–300. [Google Scholar] [CrossRef]
- Shou, J.; Massarweh, S.; Osborne, C.K.; Wakeling, A.E.; Ali, S.; Weiss, H.; Schiff, R. Mechanisms of tamoxifen resistance: Increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J. Natl. Cancer Inst. 2004, 96, 926–935. [Google Scholar] [CrossRef]
- Shi, Q.; Li, Y.; Li, S.; Jin, L.; Lai, H.; Wu, Y.; Cai, Z.; Zhu, M.; Li, Q.; Li, Y.; et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat. Commun. 2020, 11, 5513. [Google Scholar] [CrossRef]
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 1996, 379, 131–137. [Google Scholar] [CrossRef]
- Gendrel, A.V.; Heard, E. Fifty years of X-inactivation research. Development 2011, 138, 5049–5055. [Google Scholar] [CrossRef]
- Cerase, A.; Pintacuda, G.; Tattermusch, A.; Avner, P. Xist localization and function: New insights from multiple levels. Genome Biol. 2015, 16, 166. [Google Scholar] [CrossRef]
- Keohane, A.M.; O’Neill L, P.; Belyaev, N.D.; Lavender, J.S.; Turner, B.M. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev. Biol. 1996, 180, 618–630. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Chang, M. Tamoxifen resistance in breast cancer. Biomol. Ther. 2012, 20, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Pike, M.C.; Spicer, D.V.; Dahmoush, L.; Press, M.F. Estrogens, progestogens, normal breast cell proliferation, and breast cancer risk. Epidemiol. Rev. 1993, 15, 17–35. [Google Scholar] [CrossRef]
- The Arimidex, Tamoxifen, Alone or in Combination (ATAC) Trialists’ Group; Forbes, J.F.; Cuzick, J.; Buzdar, A.; Howell, A.; Tobias, J.S.; Baum, M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008, 9, 45–53. [Google Scholar] [CrossRef]
- Li, S.; Ran, M.Y.; Qiao, H. A cell cycle-related lncRNA signature predicts the progression-free interval in papillary thyroid carcinoma. Front. Endocrinol. 2023, 14, 1110987. [Google Scholar] [CrossRef]
- Wang, S.; Lloyd, R.V.; Hutzler, M.J.; Safran, M.S.; Patwardhan, N.A.; Khan, A. The role of cell cycle regulatory protein, cyclin D1, in the progression of thyroid cancer. Mod. Pathol. 2000, 13, 882–887. [Google Scholar] [CrossRef]
- Heydarzadeh, S.; Moshtaghie, A.A.; Daneshpoor, M.; Hedayati, M. Regulators of glucose uptake in thyroid cancer cell lines. Cell Commun. Signal 2020, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Huo, N.; Cong, R.; Sun, Z.J.; Li, W.C.; Zhu, X.; Xue, C.Y.; Chen, Z.; Ma, L.Y.; Chu, Z.; Han, Y.C.; et al. STAT3/LINC00671 axis regulates papillary thyroid tumor growth and metastasis via LDHA-mediated glycolysis. Cell Death Dis. 2021, 12, 799. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.T.; Lee, E.J.; Huang, M.G.; Park, Y.I.; Khullar, A.; Plodkowski, R.A. Diagnosis and treatment of patients with thyroid cancer. Am. Health Drug Benefits 2015, 8, 30–40. [Google Scholar]
- Peng, X.; Gralinski, L.; Armour, C.D.; Ferris, M.T.; Thomas, M.J.; Proll, S.; Bradel-Tretheway, B.G.; Korth, M.J.; Castle, J.C.; Biery, M.C.; et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Vishnubalaji, R.; Shaath, H.; Alajez, N.M. Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response. Genes 2020, 11, 760. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Li, Z.; Chao, T.C.; Chang, K.Y.; Lin, N.; Patil, V.S.; Shimizu, C.; Head, S.R.; Burns, J.C.; Rana, T.M. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef]
- Natarelli, L.; Parca, L.; Mazza, T.; Weber, C.; Virgili, F.; Fratantonio, D. MicroRNAs and Long Non-Coding RNAs as Potential Candidates to Target Specific Motifs of SARS-CoV-2. Non-Coding RNA 2021, 7, 14. [Google Scholar] [CrossRef]
- Shaath, H.; Alajez, N.M. Identification of PBMC-based molecular signature associational with COVID-19 disease severity. Heliyon 2021, 7, e06866. [Google Scholar] [CrossRef]
- Saha, C.; Laha, S.; Chatterjee, R.; Bhattacharyya, N.P. Co-Regulation of Protein Coding Genes by Transcription Factor and Long Non-Coding RNA in SARS-CoV-2 Infected Cells: An In Silico Analysis. Non-Coding RNA 2021, 7, 74. [Google Scholar] [CrossRef]
- Liu, X.; Xiong, W.; Ye, M.; Lu, T.; Yuan, K.; Chang, S.; Han, Y.; Wang, Y.; Lu, L.; Bao, Y. Non-coding RNAs expression in SARS-CoV-2 infection: Pathogenesis, clinical significance, and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 441. [Google Scholar] [CrossRef] [PubMed]
- Corell-Sierra, J.; Marquez-Molins, J.; Marques, M.C.; Hernandez-Azurdia, A.G.; Montagud-Martinez, R.; Cebria-Mendoza, M.; Cuevas, J.M.; Albert, E.; Navarro, D.; Rodrigo, G.; et al. SARS-CoV-2 remodels the landscape of small non-coding RNAs with infection time and symptom severity. NPJ Syst. Biol. Appl. 2024, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Natarelli, L.; Virgili, F.; Weber, C. SARS-CoV-2, Cardiovascular Diseases, and Noncoding RNAs: A Connected Triad. Int. J. Mol. Sci. 2021, 22, 12243. [Google Scholar] [CrossRef] [PubMed]


| Name of lncRNA | Target Gene | DNMT’s Association and Activity | Reference |
|---|---|---|---|
| ecCEBPA | CEBPA | ecCEBPA halts DNMT1’s catalytic activity and establishes a functional link with CEBPA expression. | [23] |
| Dali | DLGAP5, HMGB2, NOS1 | Dali interacts with DNMT1 to regulate transcription at the POU3F3 locus. DLGAP5, HMGB2, and NOS1 display increases in DNA methylation levels after Dali knockdown. | [25] |
| Dum | DPPA2 | Dum is involved in the differentiation of skeletal myoblast by recruiting DNMT1, DNMT3a, and DNMTt3b. In the promoter region, it induces DPPA2 silencing. | [26] |
| Fos ecRNA | Fos | Fos ecRNA physically interacts with DNMT1 and DNMT3a, impairing DNA methylation in the promoter region of Fos. | [27] |
| DACOR1 | (CBS) Cystathionine β-synthase (SMAD6), Sma and Mad-related protein 6 | DACOR1 overexpression results in the recruitment of DNMT1 and an increase in DNA methylation in many gene regulatory regions involved in the control of cell metabolism and the TGF-β/BMP signaling pathway. | [28] |
| FMR1-AS1 | FMR1 | FMR1-AS1 transcription affects the methylation status and expression of FMR1. The exact mechanism is still unclear. It is hypothesized to occur by direct interaction with the DNA promoter sequence and/or through DNMT1 inhibition within the FMR1 promoter region. | [33] |
| Name of lncRNA | Target Gene | Function/Mechanism | Up/Downregulated | Associated Disease | Reference |
|---|---|---|---|---|---|
| LINC00665 | DOCK1 | LINC00665/miR-4458/DOCK1 axis: experimental results indicated that LINC00665 exerted a positive function on AML cells by sponging miR-4458 and that miR-4458 influenced the progression of AML by modulating DOCK1 expression. | Upregulated | AML | [136] |
| HOTAIR | PTEN | HOTAIR activates methylation at the PTEN locus by upregulating the expression of DNMT3b, thereby promoting resistance to adriamycin (ADM) in acute myeloid leukemia. | Upregulated | AML | [137] |
| Name of lncRNA | Target Gene | Function/Mechanism | Up/Downregulated | Associated Disease | Reference |
|---|---|---|---|---|---|
| ANRIL | Not yet confirmed (NSCLC) | ANRIL has been hypothesized to recruit PRC2 to the CDKN2A/B locus, resulting in H3K27me3 modifications and transcriptional repression of tumor suppressor genes. Its function in NSCLC has not yet been confirmed. | Upregulated | Non-Small-Cell Lung Cancer | [146] |
| HOTAIR | Not yet confirmed (NSCLC) | HOTAIR acts as a bridging scaffold for PRC2 and LSD1/CoREST/REST, needed for histone demethylation (H3K4me2/3) and gene silencing. | Upregulated | Non-Small-Cell Lung Cancer | [150] |
| MEG3 | CDH1 (Cadherin 1, E-cadherin) microRNA-200 family genes | MEG3’s interaction with JARID2 regulates EZH2 recruitment, thereby facilitating the establishment of H3K27me3. | MEG3 is frequently found to be downregulated in NSCLC. It is significantly downregulated in A549 and LC-2/ad (lung adenocarcinoma cell lines). | Non-Small-Cell Lung Cancer | [152] |
| Name of lncRNA | Target Gene | Function/Mechanism | Up/Downregulated | Associated Disease | Reference |
|---|---|---|---|---|---|
| DILA1 | Cyclin 1 | DILA1 binds directly to Thr 286 of cyclin D1 protein, preventing its ubiquitination and subsequent degradation. | Upregulated | Tamoxifen-resistant ER + breast cancer | [158,159,166,167,168] |
| XIST | Not yet confirmed in breast cancer | HDAC3 plays a role in X-chromosome inactivation by directly interacting with Xist in the mouse ES cell line. | Upregulated | TNBC | [164,165] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaggi, G.; Hausman, C.; Cho, S.; Badalamenti, B.C.; Trinh, B.Q.; Di Ruscio, A.; Ummarino, S. LncRNAs Ride the Storm of Epigenetic Marks. Genes 2025, 16, 313. https://doi.org/10.3390/genes16030313
Gaggi G, Hausman C, Cho S, Badalamenti BC, Trinh BQ, Di Ruscio A, Ummarino S. LncRNAs Ride the Storm of Epigenetic Marks. Genes. 2025; 16(3):313. https://doi.org/10.3390/genes16030313
Chicago/Turabian StyleGaggi, Giulia, Clinton Hausman, Soomin Cho, Brianna C. Badalamenti, Bon Q. Trinh, Annalisa Di Ruscio, and Simone Ummarino. 2025. "LncRNAs Ride the Storm of Epigenetic Marks" Genes 16, no. 3: 313. https://doi.org/10.3390/genes16030313
APA StyleGaggi, G., Hausman, C., Cho, S., Badalamenti, B. C., Trinh, B. Q., Di Ruscio, A., & Ummarino, S. (2025). LncRNAs Ride the Storm of Epigenetic Marks. Genes, 16(3), 313. https://doi.org/10.3390/genes16030313







