Establishment and Validation of a Method for the Identification of Recessive Mastitis Resistance Genes in Dairy Cows
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animals and Samples Collection
2.3. Blood DNA Extraction and Detection
2.4. KASP Genotyping Design
2.5. Design of Multi-Locus Penetrance Variance Analysis (MPVA)
2.6. Statistical Analysis
2.7. Construction and Evaluation of KASP Genotyping Kit for Occult Mastitis in Dairy Cows
2.7.1. Test Animals
2.7.2. The Determination of Resistance to Occult Mastitis in Dairy Cows
2.7.3. Construction of KASP Genotyping Kit and Evaluation of Detection Effects
- (1)
- Composition and storage condition of the KASP genotyping kit
- (2)
- Operation procedure
- (3)
- The PCR reaction system is presented in Table 4. After the reaction is completed, the PCR plate can be stored at 4 °C for up to one week, during which the fluorescence signal remains relatively stable. Additional cycles or data re-reads can be performed within this one-week period.
3. Results
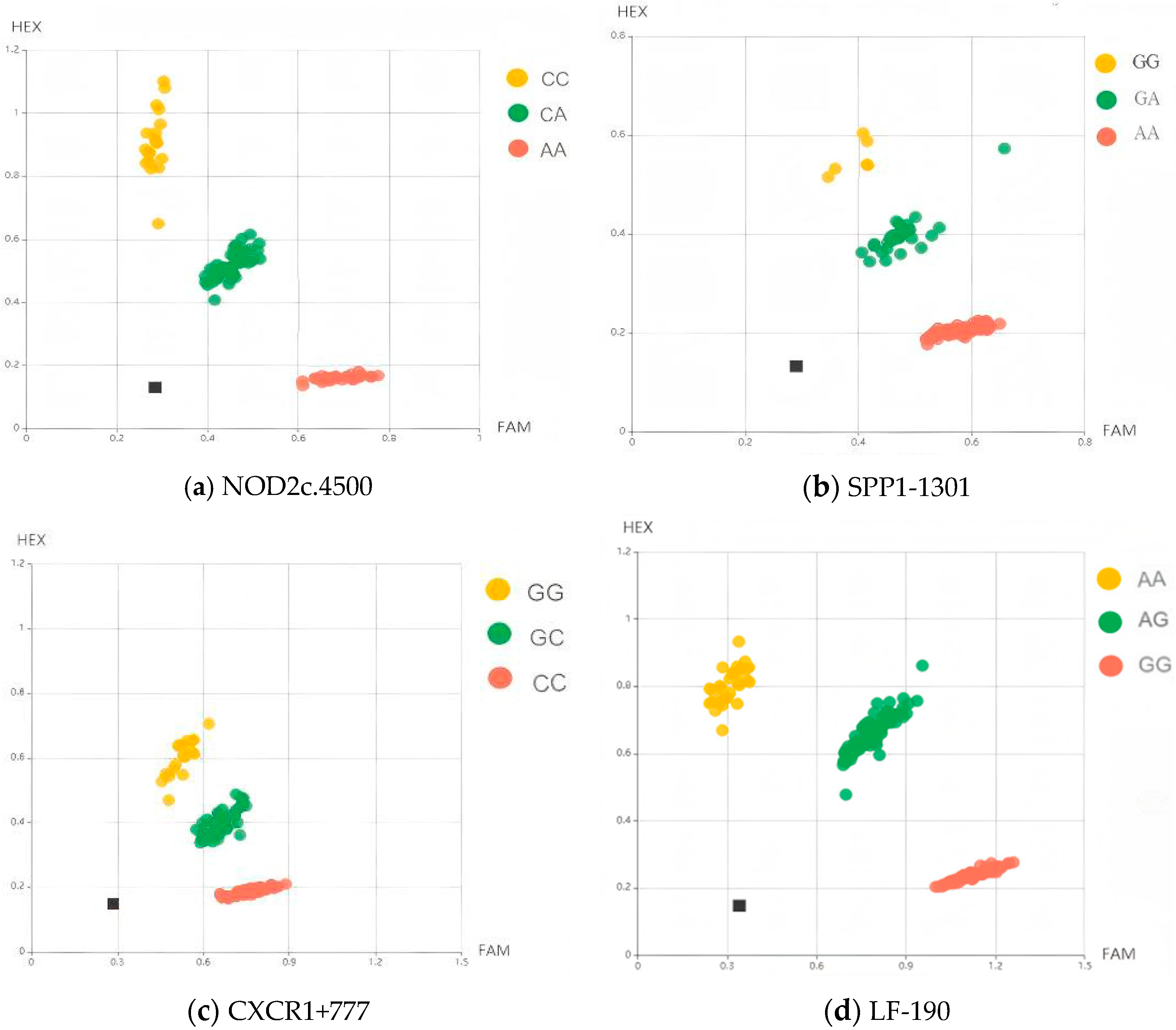
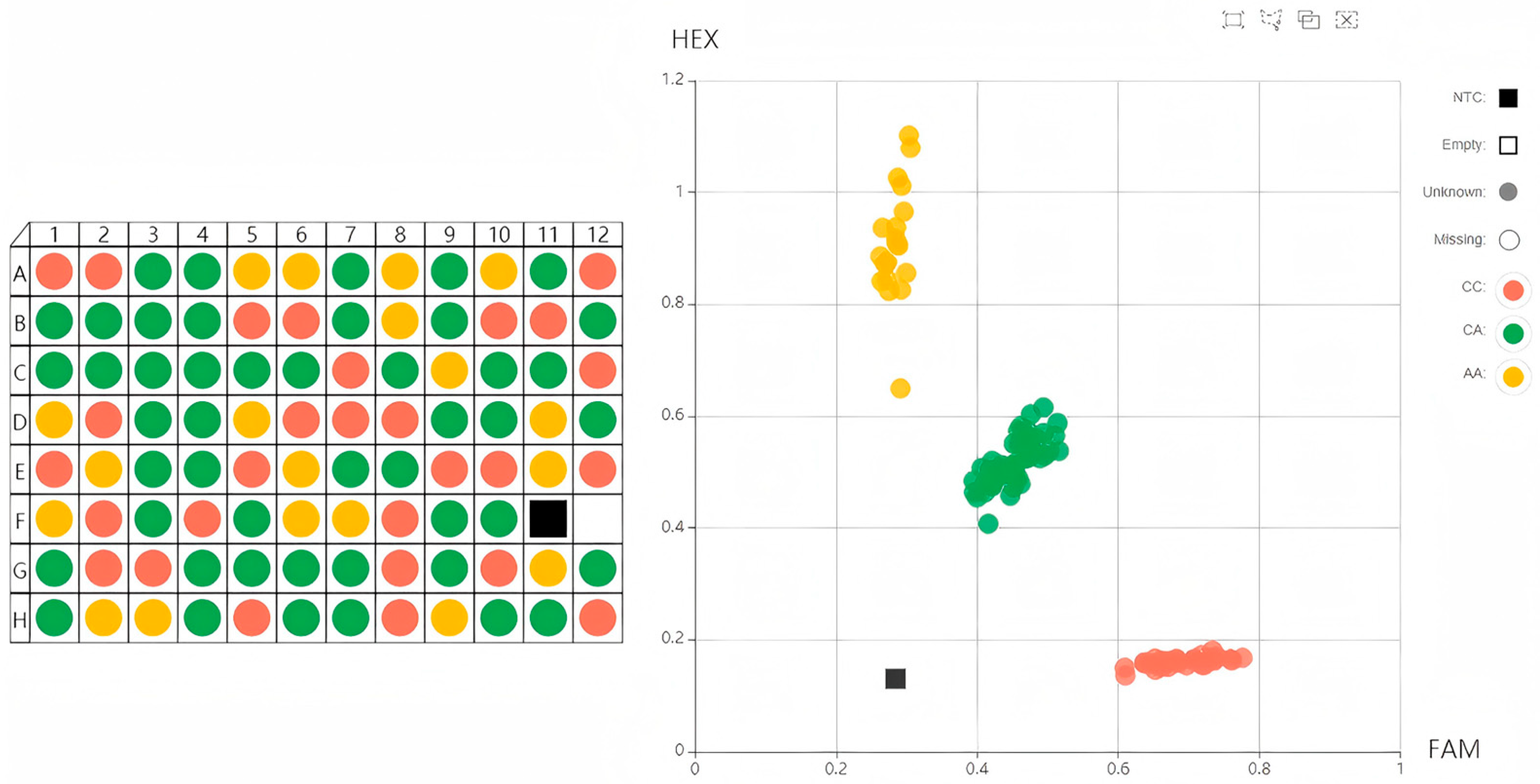
3.1. KASP Genotyping
3.2. Genetic Diversity Analysis of NOD2c.4500, CXCR1+777, SPP1-1303, and LF-190 Genes
3.2.1. Analysis of Genetic Characteristics of Different SNPs in Holstein Dairy Cows
3.2.2. Association Analysis of Different Genes and SNPs on Somatic Cell Score of Holstein Dairy Cows
3.2.3. Mufti-Locus Penetrance Variance Analysis
3.3. Validation of KASP Genotyping Kit for Occult Mastitis in Dairy Cows
3.3.1. KASP Genotypes and Classification of Resistance to Occult Mastitis for 300 Dairy Cows
3.3.2. Genotyping Accuracy of KASP Kit for Recessive Mastitis in Dairy Cows
4. Discussion
4.1. The Influence of NOD2 Gene Polymorphism on Occult Mastitis in Dairy Cows
4.2. The Influence of CXCR1 Gene Polymorphism on Occult Mastitis in Dairy Cows
4.3. The Influence of SPP1 Gene Polymorphism on Occult Mastitis in Dairy Cows
4.4. The Influence of LF Gene Polymorphism on Occult Mastitis in Dairy Cows
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SCC | Somatic Cell Count |
| KASP | Kompetitive Allele-Specific |
| MPVA | Multi-Locus Penetrance Variance Analysis |
| DHI | Dairy Herd Improvement |
| SNP | Single Nucleotide Polymorphisms |
| NOD2 | Nucleotide-binding Oligomerization Domain Protein 2 |
| CXCR1 | CXC-chemokine receptor 1 |
| SPP1 | Secreted Pyrophosphoprotein-1 |
| LF | Lactoferrin |
| InDels | Insertions/Deletions |
| PIC | Polymorphism Information Content |
| MSE | Minimum Mean Squared Error |
| SCS | Somatic Cell Score |
| KBP | Kilobase Pairs |
| LPS | Lipopolysaccharides |
Appendix A
| SNPs | Genotype Frequency | Allele Frequency | Ho | He | Ne | PIC | p-Value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| NOD2c.4500 C>A | CC 22.22% (n = 86) | CA 52.59% (n = 205) | AA 25.16% (n = 98) | C 48.52% | A 51.48% | 0.50 | 0.50 | 1.86 | 0.38 | 0.65 |
| CXCR1+777 G>C | GG 27.41% (n = 107) | GC 24.44% (n = 95) | CC 48.15% (n = 187) | G 60.37% | C 39.63% | 0.52 | 0.48 | 1.72 | 0.36 | 0.53 |
| SPP1-1301 G>A | GG 22.22% (n = 86) | GA 33.33% (n = 130) | AA 44.44% (n = 173) | G 21.11% | A 78.89% | 0.67 | 0.33 | 1.54 | 0.25 | 0.84 |
| LF-190 G>A | GG 40.74% (n = 158) | GA 41.48% (n = 161) | AA 17.78% (n = 70) | G 61.21% | A 38.79% | 0.62 | 0.38 | 1.74 | 0.37 | 0.51 |
| Locus | Genotype | Number of Records | p-Value | Somatic Cell Score |
|---|---|---|---|---|
| NOD2c.4500 C>A | CC | 86 | 0.038 | 5.58 ± 2.17 a |
| AA | 205 | 5.26 ± 2.24 a | ||
| CA | 98 | 3.68 ± 2.38 b | ||
| CXCR1+777 G>C | GG | 107 | 0.045 | 4.7 ± 2.26 a |
| CG | 95 | 4.71 ± 2.55 a | ||
| CC | 187 | 3.25 ± 1.35 b | ||
| SPP1-1301 G>A | GA | 86 | 0.013 | 5.83 ± 1.79 a |
| GG | 130 | 6.76 ± 1.13 a | ||
| AA | 173 | 2.71 ± 2.44 b | ||
| LF-190G>A | GG | 158 | 0.025 | 6.54 ± 2.45 a |
| GA | 161 | 5.15 ± 1.85 a | ||
| AA | 70 | 4.56 ± 2.45 b |
| Gene | MSE Value | Somatic Cell Count (Ten Thousand/mL) | |||
|---|---|---|---|---|---|
| SPP1 (GG) | CXCR1 (CG) | NOD2 (CC) | LF (GG) | 1.00 × 10−4 | 58.87 ± 5.92 |
| SPP1 (GG) | CXCR1 (CC) | NOD2 (CC) | LF (GG) | 9.98 × 10−3 | 52.10 ± 4.72 |
| SPP1 (GG) | CXCR1 (GG) | NOD2 (CC) | LF (GG) | 1.00 × 10−4 | 56.86 ± 5.63 |
| SPP1 (AG) | CXCR1 (CG) | NOD2 (CC) | LF (GG) | 1.02 × 10−2 | 55.94 ± 6.58 |
| SPP1 (AG) | CXCR1 (CC) | NOD2 (CC) | LF (GG) | 1.05 × 10−−3 | 48.17 ± 5.38 |
| SPP1 (AG) | CXCR1 (GG) | NOD2 (CC) | LF (GG) | 1.02 × 10−4 | 55.93 ± 6.29 |
| SPP1 (AA) | CXCR1 (CG) | NOD2 (CC) | LF (GG) | 1.01 × 10−3 | 52.82 ± 7.23 |
| SPP1 (AA) | CXCR1 (CC) | NOD2 (CC) | LF (GG) | 1.03 × 10−2 | 25.05 ± 6.03 |
| SPP1 (AA) | CXCR1 (GG) | NOD2 (CC) | LF (GG) | 1.00 × 10−3 | 52.81 ± 6.94 |
| SPP1 (GG) | CXCR1 (CG) | NOD2 (CA) | LF (GA) | 1.02 × 10−2 | 35.56 ± 4.91 |
| SPP1 (GG) | CXCR1 (CC) | NOD2 (CA) | LF (GA) | 1.58 × 10−1 | 19.70 ± 3.71 |
| SPP1 (GG) | CXCR1 (GG) | NOD2 (CA) | LF (GA) | 1.61 × 10−2 | 29.46 ± 4.62 |
| SPP1 (AG) | CXCR1 (CG) | NOD2 (CA) | LF (GA) | 6.21 × 10−2 | 28.54 ± 5.57 |
| SPP1 (AG) | CXCR1 (CC) | NOD2 (CA) | LF (GA) | 5.78 × 10−2 | 19.77 ± 4.37 |
| SPP1 (AG) | CXCR1 (GG) | NOD2 (CA) | LF (GA) | 6.37 × 10−2 | 23.53 ± 5.28 |
| SPP1 (AA) | CXCR1 (CG) | NOD2 (CA) | LF (GA) | 6.45 × 10−2 | 29.42 ± 6.22 |
| SPP1 (AA) | CXCR1 (CC) | NOD2 (CA) | LF (GA) | 1.01 × 10−4 | 17.45 ± 5.02 |
| SPP1 (AA) | CXCR1 (GG) | NOD2 (CA) | LF (GA) | 2.49 × 10−1 | 25.41 ± 5.93 |
| SPP1 (GG) | CXCR1 (CG) | NOD2 (AA) | LF (AA) | 1.32 × 10−4 | 48.57 ± 4.35 |
| SPP1 (GG) | CXCR1 (CC) | NOD2 (AA) | LF (AA) | 1.69 × 10−3 | 50.26 ± 5.26 |
| SPP1 (GG) | CXCR1 (GG) | NOD2 (AA) | LF (AA) | 1.02 × 10−4 | 45.31 ± 5.14 |
| SPP1 (AG) | CXCR1 (CG) | NOD2 (AA) | LF (AA) | 1.56 × 10−4 | 46.94 ± 6.52 |
| SPP1 (AG) | CXCR1 (CC) | NOD2 (AA) | LF (AA) | 4.25 × 10−3 | 50.45 ± 2.56 |
| SPP1 (AG) | CXCR1 (GG) | NOD2 (AA) | LF (AA) | 8.56 × 10−4 | 49.25 ± 6.23 |
| SPP1 (AA) | CXCR1 (CG) | NOD2 (AA) | LF (AA) | 3.25 × 10−4 | 50.05 ± 6.22 |
| SPP1 (AA) | CXCR1 (CC) | NOD2 (AA) | LF (AA) | 1.95 × 10−2 | 36.29 ± 5.13 |
| SPP1 (AA) | CXCR1 (GG) | NOD2 (AA) | LF (AA) | 5.26 × 10−3 | 54.58 ± 3.56 |
References
- Essa, B.; Al-Sharif, M.; Abdo, M.; Fericean, L.; Ateya, A. New Insights on Nucleotide Sequence Variants and mRNA Levels of Candidate Genes Assessing Resistance/Susceptibility to Mastitis in Holstein and Montbéliarde Dairy Cows. Vet. Sci. 2023, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Samad, M.A. Review on Mastitis in Dairy Lactating Animals and Their Public Health Importance: The 56 Years Bangladesh Perspective. J. Vet. Med. One Health Res. 2022, 4, 33–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhou, C.; Basang, W.; Zhu, Y.; Wang, X.; Li, C.; Chen, L.; Zhou, X. Mechanisms by Which Mastitis Affects Reproduction in Dairy Cow: A Review. Reprod. Domest. Anim. 2021, 56, 1165–1175. [Google Scholar] [CrossRef]
- Hogeveen, H.; Steeneveld, W.; Wolf, C.A. Production Diseases Reduce the Efficiency of Dairy Production: A Review of the Results, Methods, and Approaches Regarding the Economics of Mastitis. Annu. Rev. Resour. Econ. 2019, 11, 289–312. [Google Scholar] [CrossRef]
- Harmon, R.J. Physiology of Mastitis and Factors Affecting Somatic Cell Counts. J. Dairy Sci. 1994, 77, 2103–2112. [Google Scholar] [CrossRef]
- Chen, Y.; Zeng, B.; Shi, P.; Xiao, H.; Chen, S. Comparative Analysis of the Liver and Spleen Transcriptomes between Holstein and Yunnan Humped Cattle. Animals 2019, 9, 527. [Google Scholar] [CrossRef]
- Sodeland, M.; Grove, H.; Kent, M.; Taylor, S.; Svendsen, M.; Hayes, B.J.; Lien, S. Molecular Characterization of a Long Range Haplotype Affecting Protein Yield and Mastitis Susceptibility in Norwegian Red Cattle. BMC Genet. 2011, 12, 70. [Google Scholar] [CrossRef]
- Park, Y.H.; Joo, Y.S.; Park, J.Y.; Moon, J.S.; Kim, S.H.; Kwon, N.H.; Ahn, J.S.; Davis, W.C.; Davies, C.J. Characterization of Lymphocyte Subpopulations and Major Histocompatibility Complex Haplotypes of Mastitis-Resistant and Susceptible Cows. J. Vet. Sci. 2004, 5, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.; Günther, J.; Talbot, R.; Petzl, W.; Zerbe, H.; Schuberth, H.-J.; Seyfert, H.-M.; Glass, E.J. Escherichia Coli- and Staphylococcus Aureus-Induced Mastitis Differentially Modulate Transcriptional Responses in Neighbouring Uninfected Bovine Mammary Gland Quarters. BMC Genom. 2013, 14, 36. [Google Scholar] [CrossRef]
- Pumipuntu, N.; Kulpeanprasit, S.; Santajit, S.; Tunyong, W.; Kong-ngoen, T.; Hinthong, W.; Indrawattana, N. Screening Method for Staphylococcus Aureus Identification in Subclinical Bovine Mastitis from Dairy Farms. Vet. World 2017, 10, 721–726. [Google Scholar] [CrossRef]
- Garcia-Erill, G.; Hanghøj, K.; Heller, R.; Wiuf, C.; Albrechtsen, A. Estimating Admixture Pedigrees of Recent Hybrids without a Contiguous Reference Genome. Mol. Ecol. Resour. 2023, 23, 1604–1619. [Google Scholar] [CrossRef]
- Sun, X.; Zhao, R.; Wang, N.; Zhang, J.; Xiao, B.; Huang, F.; Chen, A. Milk Somatic Cell Count: From Conventional Microscope Method to New Biosensor-Based Method. Trends Food Sci. Technol. 2023, 135, 102–114. [Google Scholar] [CrossRef]
- Li, N.; Richoux, R.; Boutinaud, M.; Martin, P.; Gagnaire, V. Role of Somatic Cells on Dairy Processes and Products: A Review. Dairy Sci. Technol. 2014, 94, 517–538. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, A.; Imran, M. Causes, Types, Etiological Agents, Prevalence, Diagnosis, Treatment, Prevention, Effects on Human Health and Future Aspects of Bovine Mastitis. Anim. Health Res. Rev. 2020, 21, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Ghulam Mohyuddin, S.; Liang, Y.; Ni, W.; Adam Idriss Arbab, A.; Zhang, H.; Li, M.; Yang, Z.; Karrow, N.A.; Mao, Y. Polymorphisms of the IL-17A Gene Influence Milk Production Traits and Somatic Cell Score in Chinese Holstein Cows. Bioengineering 2022, 9, 448. [Google Scholar] [CrossRef]
- Sindhu, S.; Saini, T.; Rawat, H.K.; Chahar, M.; Grover, A.; Ahmad, S.; Mohan, H. Beyond Conventional Antibiotics Approaches: Global Perspectives on Alternative Therapeutics Including Herbal Prevention, and Proactive Management Strategies in Bovine Mastitis. Microb. Pathog. 2024, 196, 106989. [Google Scholar] [CrossRef]
- Luo, S.; Wang, Y.; Kang, X.; Liu, P.; Wang, G. Research Progress on the Association between Mastitis and Gastrointestinal Microbes in Dairy Cows and the Effect of Probiotics. Microb. Pathogen. 2022, 173, 105809. [Google Scholar] [CrossRef]
- Tiezzi, F.; Parker-Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. A Genome-Wide Association Study for Clinical Mastitis in First Parity US Holstein Cows Using Single-Step Approach and Genomic Matrix Re-Weighting Procedure. PLoS ONE 2015, 10, e0114919. [Google Scholar] [CrossRef]
- Meredith, B.K.; Berry, D.P.; Kearney, F.; Finlay, E.K.; Fahey, A.G.; Bradley, D.G.; Lynn, D.J. A Genome-Wide Association Study for Somatic Cell Score Using the Illumina High-Density Bovine Beadchip Identifies Several Novel QTL Potentially Related to Mastitis Susceptibility. Front. Genet. 2013, 4, 229. [Google Scholar] [CrossRef]
- Campos, B.; Pickering, A.C.; Rocha, L.S.; Aguilar, A.P.; Fabres-Klein, M.H.; De Oliveira Mendes, T.A.; Fitzgerald, J.R.; De Oliveira Barros Ribon, A. Diversity and Pathogenesis of Staphylococcus Aureus from Bovine Mastitis: Current Understanding and Future Perspectives. BMC Vet. Res. 2022, 18, 115. [Google Scholar] [CrossRef]
- Muslimova, Z.; Abdualiyeva, A.; Shaugimbayeva, N.; Orynkhanov, K.; Ussenbekov, Y. Genotyping of Holstein Cows by SELL, MX1 and CXCR1 Gene Loci Associated with Mastitis Resistance. Reprod. Domest. Anim. 2024, 59, e14713. [Google Scholar] [CrossRef] [PubMed]
- Axford, M.M.; Khansefid, M.; Chamberlain, A.J.; Haile-Mariam, M.; Goddard, M.E.; Pryce, J.E. Genetic Variation in Novel Calf Traits Using a Farmer-Centred, Co-Design Approach to Data Collection. J. Dairy Sci. 2025, S0022030225001717. [Google Scholar] [CrossRef] [PubMed]
- Vinet, A.; Mattalia, S.; Vallée, R.; Bertrand, C.; Cuyabano, B.C.D.; Boichard, D. Estimation of Genotype by Temperature-Humidity Index Interactions on Milk Production and Udder Health Traits in Montbeliarde Cows. Genet. Sel. Evol. 2023, 55, 4. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Wang, J.; Luoreng, Z.; Wang, X.; Wei, D.; Yang, J.; Hu, Q.; Ma, Y. Progress in Expression Pattern and Molecular Regulation Mechanism of LncRNA in Bovine Mastitis. Animals 2022, 12, 1059. [Google Scholar] [CrossRef]
- Welderufael, B.G.; Løvendahl, P.; De Koning, D.-J.; Janss, L.L.G.; Fikse, W.F. Genome-Wide Association Study for Susceptibility to and Recoverability from Mastitis in Danish Holstein Cows. Front. Genet. 2018, 9, 141. [Google Scholar] [CrossRef]
- Wagner, P.; Yin, T.; Brügemann, K.; Engel, P.; Weimann, C.; Schlez, K.; König, S. Genome-Wide Associations for Microscopic Differential Somatic Cell Count and Specific Mastitis Pathogens in Holstein Cows in Compost-Bedded Pack and Cubicle Farming Systems. Animals 2021, 11, 1839. [Google Scholar] [CrossRef]
- Usman, T.; Wang, Y.; Liu, C.; Wang, X.; Zhang, Y.; Yu, Y. Association Study of Single Nucleotide Polymorphisms in JAK2 and STAT5B Genes and Their Differential mRNA Expression with Mastitis Susceptibility in Chinese Holstein Cattle. Anim. Genet. 2015, 46, 371–380. [Google Scholar] [CrossRef]
- Gernand, E.; König, S. Random Regression Test-Day Model for Clinical Mastitis: Genetic Parameters, Model Comparison, and Correlations with Indicator Traits. J. Dairy Sci. 2014, 97, 3953–3963. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, J.; Gao, Y.; Ju, Z.; Huang, J. A SNP in Intron 8 of CD46 Causes a Novel Transcript Associated with Mastitis in Holsteins. BMC Genom. 2014, 15, 630. [Google Scholar]
- Khan, M.Z.; Wang, D.; Liu, L.; Usman, T.; Wen, H.; Zhang, R.; Liu, S.; Shi, L.; Mi, S.; Xiao, W.; et al. Significant Genetic Effects of JAK2 and DGAT1 Mutations on Milk Fat Content and Mastitis Resistance in Holsteins. J. Dairy Res. 2019, 86, 388–393. [Google Scholar] [CrossRef]
- Hasan, M.U.; Ceyhan, A. The Investigation of SNP in SOCS2 Gene and Its Effect on Milk Yield, Fat, Protein, and Somatic Cell Count in Awassi Ewes. Trop. Anim. Health Prod. 2024, 56, 272. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.R.; Bhat, N.N.; Shabir, A.; Mir, M.U.R.; Ahmad, S.B.; Hussain, I.; Hussain, S.A.; Ali, A.; Shamim, K.; Rehman, M.U. SNP Analysis of TLR4 Promoter and Its Transcriptional Factor Binding Profile in Relevance to Bovine Subclinical Mastitis. Biochem. Genet. 2024, 62, 3605–3623. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Chu, G.; Dan, Y.; Li, J.; Zhang, L.; Gao, X.; Gao, H.; Li, J.; Xu, S.; Liu, Z. BRCA1: A New Candidate Gene for Bovine Mastitis and Its Association Analysis between Single Nucleotide Polymorphisms and Milk Somatic Cell Score. Mol. Biol. Rep. 2012, 39, 6625–6631. [Google Scholar] [CrossRef]
- Shin, S.C.; Chung, E.R. Association of SNP Marker in the Thyroglobulin Gene with Carcass and Meat Quality Traits in Korean Cattle. Asian-Australas. J. Anim. Sci. 2006, 20, 172–177. [Google Scholar] [CrossRef]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A Review on SNP and Other Types of Molecular Markers and Their Use in Animal Genetics. Genet. Sel. Evol. 2002, 34, 275. [Google Scholar] [CrossRef] [PubMed]
- Pighetti, G.M.; Elliott, A.A. Gene Polymorphisms: The Keys for Marker Assisted Selection and Unraveling Core Regulatory Pathways for Mastitis Resistance. J. Mammary Gland Biol. Neoplasia 2011, 16, 421–432. [Google Scholar] [CrossRef]
- Moretti, R.; Soglia, D.; Chessa, S.; Sartore, S.; Finocchiaro, R.; Rasero, R.; Sacchi, P. Identification of SNPs Associated with Somatic Cell Score in Candidate Genes in Italian Holstein Friesian Bulls. Animals 2021, 11, 366. [Google Scholar] [CrossRef]
- Panigrahi, M.; Kumar, H.; Nayak, S.S.; Rajawat, D.; Parida, S.; Bhushan, B.; Sharma, A.; Dutt, T. Molecular Characterization of CRBR2 Fragment of TLR4 Gene in Association with Mastitis in Vrindavani Cattle. Microb. Pathogen. 2022, 165, 105483. [Google Scholar] [CrossRef]
- Worku, D.; Gowane, G.R.; Mukherjee, A.; Alex, R.; Joshi, P.; Verma, A. Associations between Polymorphisms of LAP3 and SIRT1 Genes with Clinical Mastitis and Milk Production Traits in Sahiwal and Karan Fries Dairy Cattle. Vet. Med. Sci. 2022, 8, 2593–2604. [Google Scholar] [CrossRef]
- Chang, C.-C.; Silva, B.B.I.; Huang, H.-Y.; Tsai, C.-Y.; Flores, R.J.D.; Tayo, L.L.; Tyan, Y.-C.; Tsai, M.-A.; Catulin, G.E.M.; Chuang, K.-P.; et al. Development and Validation of KASP Assays for the Genotyping of Racing Performance-Associated Single Nucleotide Polymorphisms in Pigeons. Genes 2021, 12, 1383. [Google Scholar] [CrossRef]
- Ilie, D.E.; Gavojdian, D.; Kusza, S.; Neamț, R.I.; Mizeranschi, A.E.; Mihali, C.V.; Cziszter, L.T. Kompetitive Allele Specific PCR Genotyping of 89 SNPs in Romanian Spotted and Romanian Brown Cattle Breeds and Their Association with Clinical Mastitis. Animals 2023, 13, 1484. [Google Scholar] [CrossRef] [PubMed]
- Sender, G.; Korwin-Kossakowska, A.; Pawlik, A.; Hameed, K.G.A.; Oprządek, J. Genetic Basis of Mastitis Resistance in Dairy Cattle—A Review/Podstawy Genetyczne Odporności Krów Mlecznych Na Zapalenie Wymienia—Artykuł Przeglądowy. Ann. Anim. Sci. 2013, 13, 663–673. [Google Scholar] [CrossRef]
- Jaiswal, S.; Jagannadham, J.; Kumari, J.; Iquebal, M.A.; Gurjar, A.K.S.; Nayan, V.; Angadi, U.B.; Kumar, S.; Kumar, R.; Datta, T.K.; et al. Genome Wide Prediction, Mapping and Development of Genomic Resources of Mastitis Associated Genes in Water Buffalo. Front. Vet. Sci. 2021, 8, 593871. [Google Scholar] [CrossRef]
- Zhang, Y. Analysis of the Relationship Between Candidate Genes and Susceptibility to Mastitis in Holstein Cattle Using Multi-Locus Penetrance Variance Analysis; Yangzhou University: Yangzhou, China, 2012. [Google Scholar]
- Raza, S.H.A.; Khan, R.; Pant, S.D.; Shah, M.A.; Quan, G.; Feng, L.; Cheng, G.; Gui, L.; Zan, L. Genetic Variation in the OPN Gene Affects Milk Composition in Chinese Holstein Cows. Anim. Biotechnol. 2023, 34, 893–899. [Google Scholar] [CrossRef]
- Aasmul-Olsen, K.; Henriksen, N.L.; Nguyen, D.N.; Heckmann, A.B.; Thymann, T.; Sangild, P.T.; Bering, S.B. Milk Osteopontin for Gut, Immunity and Brain Development in Preterm Pigs. Nutrients 2021, 13, 2675. [Google Scholar] [CrossRef]
- Jiang, R.; Lönnerdal, B. Evaluation of Bioactivities of Bovine Milk Osteopontin Using a Knockout Mouse Model. J. Pediatr. Gastroenterol. Nutr. 2020, 71, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.; Zachariae, E.D.; Poulsen, N.A.; Buitenhuis, A.J.; Larsen, L.B.; Sørensen, E.S. Factors Influencing Milk Osteopontin Concentration Based on Measurements from Danish Holstein Cows. J. Dairy Res. 2021, 88, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, A.E.I. Chemokine Receptors: Multifaceted Therapeutic Targets. Nat. Rev. Immunol. 2002, 2, 106–1152. [Google Scholar] [CrossRef]
- Pighetti, G.M.; Kojima, C.J.; Wojakiewicz, L.; Rambeaud, M. The Bovine CXCR1 Gene Is Highly Polymorphic. Vet. Immunol. Immunopathol. 2012, 145, 464–470. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, H.M.; Ju, Z.H.; Zhang, Y.; Huang, J.M.; Qi, C.; Hou, M.H.; An, L.G.; Zhong, J.F.; Wang, C.F. Association of Novel Single Nucleotide Polymorphisms of the CXCR1 Gene with the Milk Performance Traits of Chinese Native Cattle. Genet. Mol. Res. 2013, 12, 2725–2739. [Google Scholar] [CrossRef]
- Caruso, R.; Warner, N.; Inohara, N.; Núñez, G. NOD1 and NOD2: Signaling, Host Defense, and Inflammatory Disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Lyu, H.; Sun, D.; Wang, J.; Wu, R. Staphylococcus Aureus Promotes NOD2 Expression in Bovine Mammary Epithelial Cells. Front. Oncol. 2017, 33, 741–745. [Google Scholar]
- Wang, H.; Yu, G.; Yu, H.; Gu, M.; Zhang, J.; Meng, X.; Liu, Z.; Qiu, C.; Li, J. Characterization of TLR2, NOD2, and Related Cytokines in Mammary Glands Infected by Staphylococcus Aureus in a Rat Model. Acta Vet. Scand. 2015, 57, 25. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Timilsena, Y.P.; Blanch, E.; Adhikari, B. Lactoferrin: Structure, function, denaturation and digestion. Crit. Rev. Food Sci. Nutr. 2019, 59, 580–596. [Google Scholar] [CrossRef]
- Molenaar, A.J.; Kuys, Y.M.; Davis, S.R.; Wilkins, R.J.; Mead, P.E.; Tweedie, J.W. Elevation of Lactoferrin Gene Expression in Developing, Ductal, Resting, and Regressing Parenchymal Epithelium of the Ruminant Mammary Gland. J. Dairy Sci. 1996, 79, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, A.; Murata, E.; Yamamoto, K.; Majima, E.; Sano, E.; Le, Q.T.; Katunuma, N. New Functions of Lactoferrin and B-Casein in Mammalian Milk as Cysteine Protease Inhibitors. Biochem. Biophys. Res. Commun. 2003, 306, 98–103. [Google Scholar] [CrossRef]
- McDermott, M.P.; Erb, H.N.; Natzke, R.P.; Barnes, F.D.; Bray, D. Cost Benefit Analysis of Lactation Therapy with Somatic Cell Counts as Indications for Treatment. J. Dairy Sci. 1983, 66, 1198–1203. [Google Scholar] [CrossRef]
- Suzuki, Y.A.; Lönnerdal, B. Characterization of Mammalian Receptors for Lactoferrin. Biochem. Cell Biol. 2002, 80, 75–80. [Google Scholar] [CrossRef]
- Ryman, V.E.; Packiriswamy, N.; Sordillo, L.M. Role of Endothelial Cells in Bovine Mammary Gland Health and Disease. Anim. Health Res. Rev. 2015, 16, 135–149. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Z.; Zhang, Y.; Zhang, X.; Li, Y. Multi-Locus Penetrance Variance Analysis Method for Association Study in Complex Diseases. Hum. Hered. 2005, 60, 143–149. [Google Scholar] [CrossRef]
- Bekuma, A. Review on Hygienic Milk Products Practice and Occurrence of Mastitis in Cow’s Milk. Agric. Res. Technol. Open Access J. 2018, 18, 1–11. [Google Scholar] [CrossRef]
- Trujano-Chavez, M.Z.; Sánchez-Ramos, R.; Pérez-Rodríguez, P.; Ruíz-Flores, A. Genetic Diversity and Population Structure for Resistance and Susceptibility to Mastitis in Braunvieh Cattle. Vet. Sci. 2021, 8, 329. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Foucras, G.; Boichard, D.; Rupp, R. Invited Review: Low Milk Somatic Cell Count and Susceptibility to Mastitis. J. Dairy Sci. 2018, 101, 6703–6714. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The Biofilm Life Cycle: Expanding the Conceptual Model of Biofilm Formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Koop, G.; Nielen, M.; Van Werven, T. Bulk Milk Somatic Cell Counts Are Related to Bulk Milk Total Bacterial Counts and Several Herd-Level Risk Factors in Dairy Goats. J. Dairy Sci. 2009, 92, 4355–4364. [Google Scholar] [CrossRef]
- Gonzalo, C.; Carriedo, J.A.; García-Jimeno, M.C.; Pérez-Bilbao, M.; De La Fuente, L.F. Factors Influencing Variation of Bulk Milk Antibiotic Residue Occurrence, Somatic Cell Count, and Total Bacterial Count in Dairy Sheep Flocks. J. Dairy Sci. 2010, 93, 1587–1595. [Google Scholar] [CrossRef]
- Bhosale, R.R.; Osmani, R.A.; Ghodake, P.P.; Shaikh, S.M.; Chavan, S.R. Mastitis: An Intensive Crisis in Veterinary Science. Int. J. Pharma Res. Health Sci. 2014, 2, 96–103. [Google Scholar]
- Le Maréchal, C.; Thiéry, R.; Vautor, E.; Le Loir, Y. Mastitis Impact on Technological Properties of Milk and Quality of Milk Products—A Review. Dairy Sci. Technol. 2011, 91, 247–282. [Google Scholar] [CrossRef]
- Cadwallader, K.R.; Singh, T.K. Flavours and Off-Flavours in Milk and Dairy Products. In Advanced Dairy Chemistry; McSweeney, P., Fox, P.F., Eds.; Springer Inc.: New York, NY, USA, 2009. [Google Scholar]
- Ntuli, V.; Sibanda, T.; Elegbeleye, J.A.; Mugadza, D.T.; Seifu, E.; Buys, E.M. Dairy Production: Microbial Safety of Raw Milk and Processed Milk Products. In Present Knowledge in Food Safety; Elsevier Inc.: Amsterdam, The Netherlands, 2023. [Google Scholar]
- Shook, G.E.; Schutz, M.M. Selection on Somatic Cell Score to Improve Resistance to Mastitis in the United States. J. Dairy Sci. 1994, 77, 648–658. [Google Scholar] [CrossRef]
- Shook, G.E. Selection for Disease Resistance. J. Dairy Sci. 1989, 72, 1349–1362. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, L.; Zhang, L.; Gu, S.; Wang, H.; Sui, G.; Zheng, W. A Molecular Identification and Resistance Evaluation of the Blast Resistance Genes in Japonica Rice in Northern China. Agronomy 2023, 13, 2662. [Google Scholar] [CrossRef]
- Li, G.; Zhang, X.; Zhang, X.; Su, Y.; Tu, Y.; Wei, Y. Effect of Single and Pyramiding Genotypes of Three Genes on Egg Production of Baier Chicken. Acta Agric. Univ. Jiangxiensis 2010, 32, 7. [Google Scholar]
- Molnar, T.; Hofner, P.; Nagy, F.; Lakatos, P.L.; Fischer, S.; Lakatos, L.; Kovacs, A.; Altorjay, I.; Papp, M.; Palatka, K.; et al. NOD1 Gene E266K Polymorphism Is Associated with Disease Susceptibility but Not with Disease Phenotype or NOD2/CARD15 in Hungarian Patients with Crohn’s Disease. Dig. Liver Dis. 2007, 39, 1064–1070. [Google Scholar] [CrossRef]
- Wang, W.; Cheng, L.; Yi, J.; Gan, J.; Tang, H.; Fu, M.Z.; Wang, H.; Lai, S.J. Health and Production Traits in Bovine Are Associated with Single Nucleotide Polymorphisms in the NOD2 Gene. Genet. Mol. Res. 2015, 14, 3570–3578. [Google Scholar] [CrossRef] [PubMed]
- Pant, S.D.; Schenkel, F.S.; Leyva-Baca, I.; Sharma, B.S.; Karrow, N.A. Identification of polymorphisms in bovine TLR2 and CARD15, associations between CARD15 polymorphisms and milk somatic cell score in Canadian Holsteins, and functional relevance of SNP c.3020A>T. Dev. Biol. 2008, 132, 247–253. [Google Scholar]
- Grépin, R.; Guyot, M.; Giuliano, S.; Boncompagni, M.; Ambrosetti, D.; Chamorey, E.; Scoazec, J.-Y.; Negrier, S.; Simonnet, H.; Pagès, G. The CXCL7/CXCR1/2 Axis Is a Key Driver in the Growth of Clear Cell Renal Cell Carcinoma. Cancer Res. 2014, 74, 873–883. [Google Scholar] [CrossRef]
- Gallegos-Arreola, M.P.; Briseno-Zuno, C.J.; Figuera, L.E.; Sanchez-Lopez, J.Y.; Zuniga-Gonzalez, G.M.; Puebla-Perez, A.M.; Gomez-Meda, B.C.; Montoya-Fuentes, H.; Delgado-Saucedo, J.I. The rs1008562, rs2234671, and rs3138060 polymorphisms of the CXCR1 gene are associated with breast cancer risk in a Mexican population. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9990–10002. [Google Scholar]
- Verbeke, J.; Piepers, S.; Peelman, L.; Van Poucke, M.; De Vliegher, S. Association of CXCR1 Polymorphisms with Apoptosis, Necrosis and Concentration of Milk Neutrophils in Early Lactating Dairy Heifers. Res. Vet. Sci. 2014, 97, 55–59. [Google Scholar] [CrossRef]
- Youngerman, S.M.; Saxton, A.M.; Oliver, S.P.; Pighetti, G.M. Association of CXCR2 Polymorphisms with Subclinical and Clinical Mastitis in Dairy Cattle. J. Dairy Sci. 2004, 87, 2442–2448. [Google Scholar] [CrossRef]
- Pushpa Magotra, A.; Bangar, Y.C.; Patil, C.S.; Kamaldeep; Sindhu, V.; Malik, D.; Chaudhary, P.; Garg, A.R.; Kumar, S. Association of CXCR1 Gene Polymorphism with Clinical Mastitis, Reproductive Disorders and Performance Traits in Hardhenu (Bos Taurus × Bos Indicus) Cattle. Reprod. Domest. Anim. 2023, 58, 1234–1243. [Google Scholar] [CrossRef]
- Galvão, K.N.; Pighetti, G.M.; Cheong, S.H.; Nydam, D.V.; Gilbert, R.O. Association between Interleukin-8 Receptor-α (CXCR1) Polymorphism and Disease Incidence, Production, Reproduction, and Survival in Holstein Cows. J. Dairy Sci. 2011, 94, 2083–2091. [Google Scholar] [CrossRef]
- Dalal, V.; Kamaldeep, D.; Magotra, A.; Yadav, D.C.; Pushpa, S.; Garg, A.R. Association of CXCR1 Gene Polymorphism with Clinical Mastitis and Performance Traits in Murrah Buffalo. Reprod. Domest. Anim. 2024, 59, e14749. [Google Scholar] [CrossRef] [PubMed]
- Dettori, M.L.; Pazzola, M.; Petretto, E.; Vacca, G.M. Association Analysis between SPP1, POFUT1 and PRLR Gene Variation and Milk Yield, Composition and Coagulation Traits in Sarda Sheep. Animals 2020, 10, 1216. [Google Scholar] [CrossRef] [PubMed]
- De Mello, F.; Cobuci, J.A.; Martins, M.F.; Silva, M.V.G.B.; Neto, J.B. Association of the Polymorphism G.8514C>T in the OSTEOPONTIN Gene (SPP1) with Milk Yield in the Dairy Cattle Breed Girolando. Anim. Genet. 2012, 43, 647–648. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Lu, G.; Pan, G.; Deng, Y.; Liang, J.; Liang, L.; Liu, J.; Tang, Y.; Wei, G. A Case-Control Study of the Association between the SPP1 Gene SNPs and the Susceptibility to Breast Cancer in Guangxi, China. Front. Oncol. 2019, 9, 1415. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, X.; Ma, Y.; Liu, D.; Lu, X.; Zhao, T.; Yang, Z. Molecular Marker-Assisted Selection of ABCG2, CD44, SPP1 Genes Contribute to Milk Production Traits of Chinese Holstein. Animals 2022, 13, 89. [Google Scholar] [CrossRef]
- Al-Huboby, H.A.; Al-Anbari, N.N. Relationship of SPP1 Promoter/Exon 5 Gene Polymorphism G>A/4187 in Some Economic Traits of Awassi Sheep. IOP Conf. Ser. Earth Environ. Sci. 2024, 1325, 12019. [Google Scholar] [CrossRef]
- Bissonnette, N. Short Communication: Genetic Association of Variations in the Osteopontin Gene (SPP1) with Lactation Persistency in Dairy Cattle. J. Dairy Sci. 2018, 101, 456–461. [Google Scholar] [CrossRef]
- Pawlik, A.; Sender, G.; Sobczyńska, M.; Korwin-Kossakowska, A.; Lassa, H.; Oprządek, J. Lactoferrin Gene Variants, Their Expression in the Udder and Mastitis Susceptibility in Dairy Cattle. Anim. Prod. Sci. 2015, 55, 999. [Google Scholar] [CrossRef]
- Zheng, J.; Ather, J.L.; Sonstegard, T.S.; Kerr, D.E. Characterization of the Infection-Responsive Bovine Lactoferrin Promoter. Gene 2005, 353, 107–117. [Google Scholar] [CrossRef]
- Nowier, A.M.; Darwish, H.R.; Ramadan, S.I.; Othman, O.E. Polymorphism of Lactoferrin Gene in Egyptian Goats and Its Association with Milk Composition Traits in Zaraibi Breed. Trop. Anim. Health Prod. 2020, 52, 1065–1071. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, A.M.; Huircan, P.; Lepori, A. Single Nucleotide Polymorphisms in Immunity-Related Genes and Their Association with Mastitis in Chilean Dairy Cattle. Genet. Mol. Res. 2013, 12, 2702–2711. [Google Scholar] [CrossRef] [PubMed]
- O’Halloran, F.; Berry, D.P.; Bahar, B.; Howard, D.J.; Sweeney, T.; Giblin, L. Polymorphisms in the Bovine Lactoferrin Promoter Are Associated with Reproductive Performance and Somatic Cell Count. J. Dairy Sci. 2010, 93, 1253–1259. [Google Scholar] [CrossRef]
- Kinkpe, L.; Khan, R.; Suhail, S.M.; Ahmad, I.; Khan, F.A.; Ayari-Akkari, A.; Siddiqui, S. Polymorphism and Association Study of Lactoferrin (LF) Gene with Milk Yield, Milk Composition, and Somatic Cell Count in Beetal Goats. Trop. Anim. Health Prod. 2023, 55, 415. [Google Scholar] [CrossRef]
- Pawlik, A.; Sender, G.; Sobczynska, M.; Korwin-Kossakowska, A.; Oprzadek, J.; Lukaszewicz, M. Association between Lactoferrin Single Nucleotide Polymorphisms and Milk Production Traits in Polish Holstein Cattle. Arch. Anim. Breed. 2014, 57, 1–12. [Google Scholar] [CrossRef]
- Li, Q.; Ma, S.; Xu, J.; Wen, L.; Tian, L.; Liu, L.; Zhang, L. Genetic polymorphism of LF gene and its associations with milk traits and mastitis of Chinese Holstein cattle. Acta Agric. Zhejiang Ensis 2016, 28, 1142–1147. [Google Scholar]
- Tao, M.; Li, Z.; Liu, M.; Ma, H.; Liu, W. Association Analysis of Polymorphisms in SLK, ARHGEF9, WWC2, GAB3, and FSHR Genes with Reproductive Traits in Different Sheep Breeds. Front. Genet. 2024, 15, 1371872. [Google Scholar] [CrossRef]
- Yang, L.Y.; Wang, L.P.; Rong, R. Establishment of an ABO blood group detection method in Macaca fascicularis and Macaca mulatta using competitive allele specific PCR(KASP). Chin. J. Comp Med. 2019, 29, 31–36. [Google Scholar]

| Mutation Site | Fluorescent Signal | Primer | Sequence (5′-3′) |
|---|---|---|---|
| NOD2 C>A site | FAM | F1 | GAGACACTTGGAGAGAATGGAGC |
| HEX | F2 | GGAGACACTTGGAGAGAATGGAGA | |
| / | R | CAGCGTGGAGTTGTAAGTTATAGAGA | |
| CXCR1 G>C site | FAM | F1 | CCAAATGGGGCACAAGCAC |
| HEX | F2 | CCAAATGGGGCACAAGCAG | |
| / | R | GCCCTCATGAGGGTGTCCG | |
| SPP1 G>A site | FAM | F1 | CAGATGCTCTCCACCTACACAGG |
| HEX | F2 | CCAGATGCTCTCCACCTACACAGA | |
| / | R | TTCTGTGACCACAAAACCAGAGG | |
| LF G>A site | FAM | F1 | GGACAACTACAAGGTCTACAACACA |
| HEX | F2 | GGACAACTACAAGGTCTACAACACG | |
| / | R | CTTCTTGGTCCTAATGCCCTCAGA |
| Composition | Quantity and Specifications |
|---|---|
| SNP Primer Mix: Upstream primer F1, upstream primer F2, universal downstream primer | 2.2 mL each |
| 2× KASP Master mix | 2.5 mL each |
| ddH2O | 1.5 mL each |
| Composition | 96-Well Plates (μL) | 384-Well Plates (μL) |
|---|---|---|
| DNA | 1 | 1 |
| SNP Primer Mix | 0.14 | 0.07 |
| 2× KASP Master mix | 4 | 2 |
| ddH2O | 2.86 | 0.93 |
| total volume | 8 | 4 |
| Steps | Process | Temperature/°C | Time | Number of Cycles |
|---|---|---|---|---|
| 1 | Pre-degeneration | 95 | 10 min | 1 cycle |
| 2 | Denaturation | 95 | 20 s | 10 cycles |
| Annealing/Extension | 61–55 °C (drop 0.6 °C per cycle) | 45 s | ||
| 3 | Denaturation | 95 | 20 s | 38 cycles |
| Annealing/Extension | 55 | 45 s | ||
| 4 | End | 25 | Forever | 1 |
| Genotype | MSE Value | Somatic Cell Count (Ten Thousand/mL) |
|---|---|---|
| SPP1(AA) | 5.45 × 10−1 | Susceptible type |
| CXCR1(CC) | 3.89 × 10−1 | Susceptible type |
| NOD2(CA) | 4.55 × 10−1 | Susceptible type |
| LF(GA) | 3.25 × 10−1 | Susceptible type |
| SPP1(AA)- CXCR1(CC) | 7.78 × 10−2 | Intermediate-resistant type |
| SPP1(AA)- NOD2(CA) | 7.56 × 10−2 | Intermediate-resistant type |
| CXCR1(CC)- NOD2(CA) | 6.56 × 10−2 | Intermediate-resistant type |
| LF(GA)-SPP1(AA) | 4.56 × 10−2 | Intermediate-resistant type |
| LF(GA)-CXCR1(CC) | 4.12 × 10−2 | Intermediate-resistant type |
| LF(GA)-NOD2(CA) | 5.26 × 10−2 | Intermediate-resistant type |
| CXCR1(CC)-SPP1(AA)-NOD2(CA) | 5.52 × 10−3 | Highly-resistant type |
| CXCR1(CC)-SPP1(AA)-LF(GA) | 4.52 × 10−3 | Highly-resistant type |
| SPP1(AA)-NOD2(CA)-LF(GA) | 6.25 × 10−3 | Highly-resistant type |
| CXCR1(CC)-NOD2(CA)-LF(GA) | 7.15 × 10−3 | Highly-resistant type |
| CXCR1(CC)-SPP1(AA)-NOD2(CA)-LF(GA) | 3.23 × 10−4 | Highly-resistant type |
| Grouping | Kit Results | Somatic Cell Count (Million per Milliliter) | Accuracy | ||
|---|---|---|---|---|---|
| <20 | 20–50 | >50 | |||
| High resistance/each | 92 | 89 | 3 | 0 | (89/92) × 100% = 96.74% |
| Medium resistance/each | 148 | 4 | 142 | 2 | (142/148) × 100% = 95.95% |
| Susceptible/each | 60 | 1 | 2 | 57 | (57/60) × 100% = 95.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, W.; Wu, P.; Zhu, M.; Ullah, Y.; Zhao, Z.; Cao, S.; Li, G.; Ou, S.; He, K.; Xu, Y. Establishment and Validation of a Method for the Identification of Recessive Mastitis Resistance Genes in Dairy Cows. Genes 2025, 16, 485. https://doi.org/10.3390/genes16050485
Zheng W, Wu P, Zhu M, Ullah Y, Zhao Z, Cao S, Li G, Ou S, He K, Xu Y. Establishment and Validation of a Method for the Identification of Recessive Mastitis Resistance Genes in Dairy Cows. Genes. 2025; 16(5):485. https://doi.org/10.3390/genes16050485
Chicago/Turabian StyleZheng, Wei, Pei Wu, Mengting Zhu, Yaseen Ullah, Zongsheng Zhao, Shaoqi Cao, Guang Li, Sihai Ou, Kaibing He, and Ye Xu. 2025. "Establishment and Validation of a Method for the Identification of Recessive Mastitis Resistance Genes in Dairy Cows" Genes 16, no. 5: 485. https://doi.org/10.3390/genes16050485
APA StyleZheng, W., Wu, P., Zhu, M., Ullah, Y., Zhao, Z., Cao, S., Li, G., Ou, S., He, K., & Xu, Y. (2025). Establishment and Validation of a Method for the Identification of Recessive Mastitis Resistance Genes in Dairy Cows. Genes, 16(5), 485. https://doi.org/10.3390/genes16050485





