Stem-Loop RT-qPCR as an Efficient Tool for the Detection and Quantification of Small RNAs in Giardia lamblia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Experimental Conditions
2.2. RNA Samples: Total RNA and Small RNA
2.3. Primers and Endpoint PCR
2.4. Stem-Loop RT-qPCR Assay Design
2.5. Synthesis of First-Strand cDNA
2.6. RT-qPCR Assay
3. Results
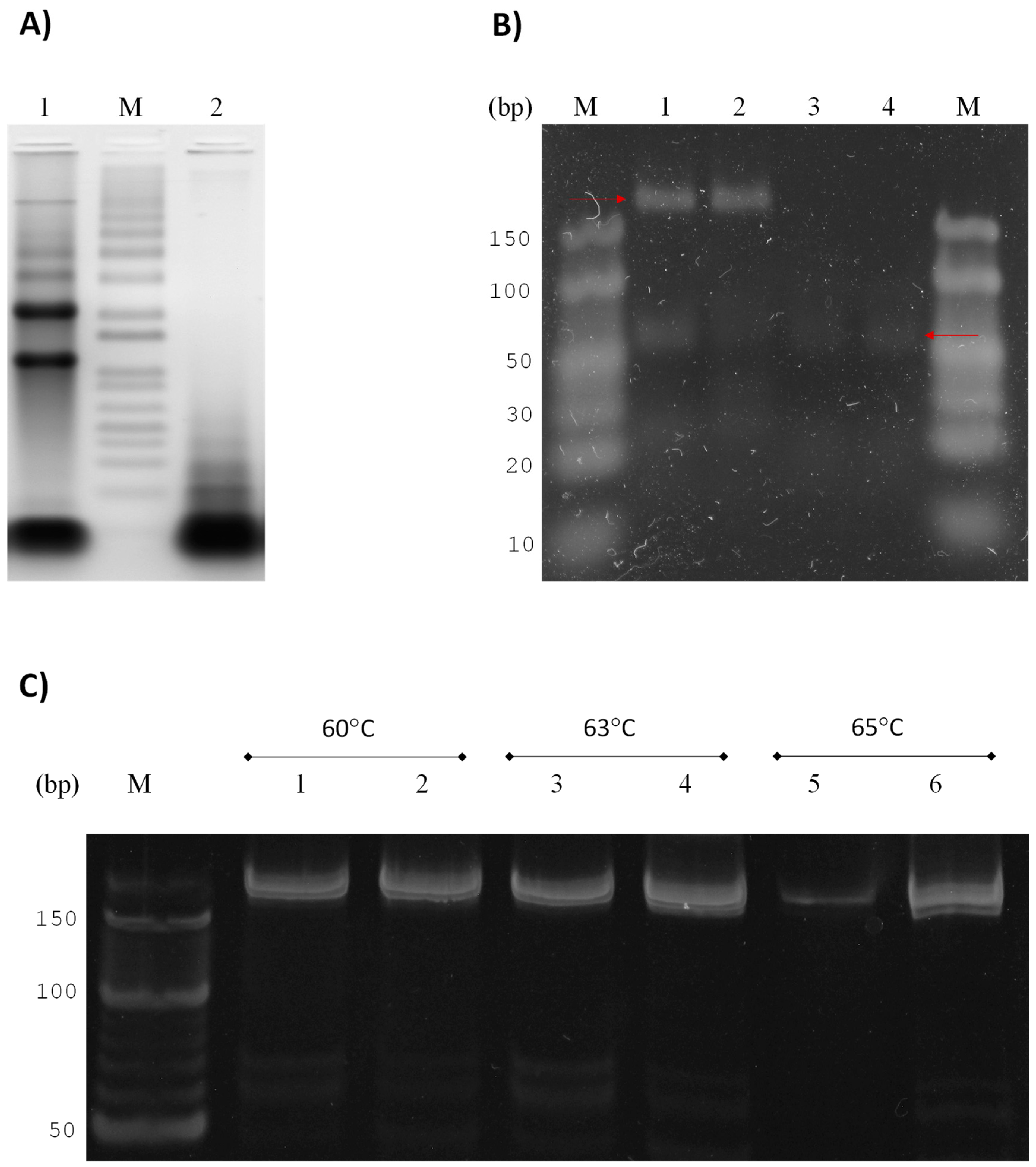
3.1. Strategy for Detecting Small RNAs of G. lamblia
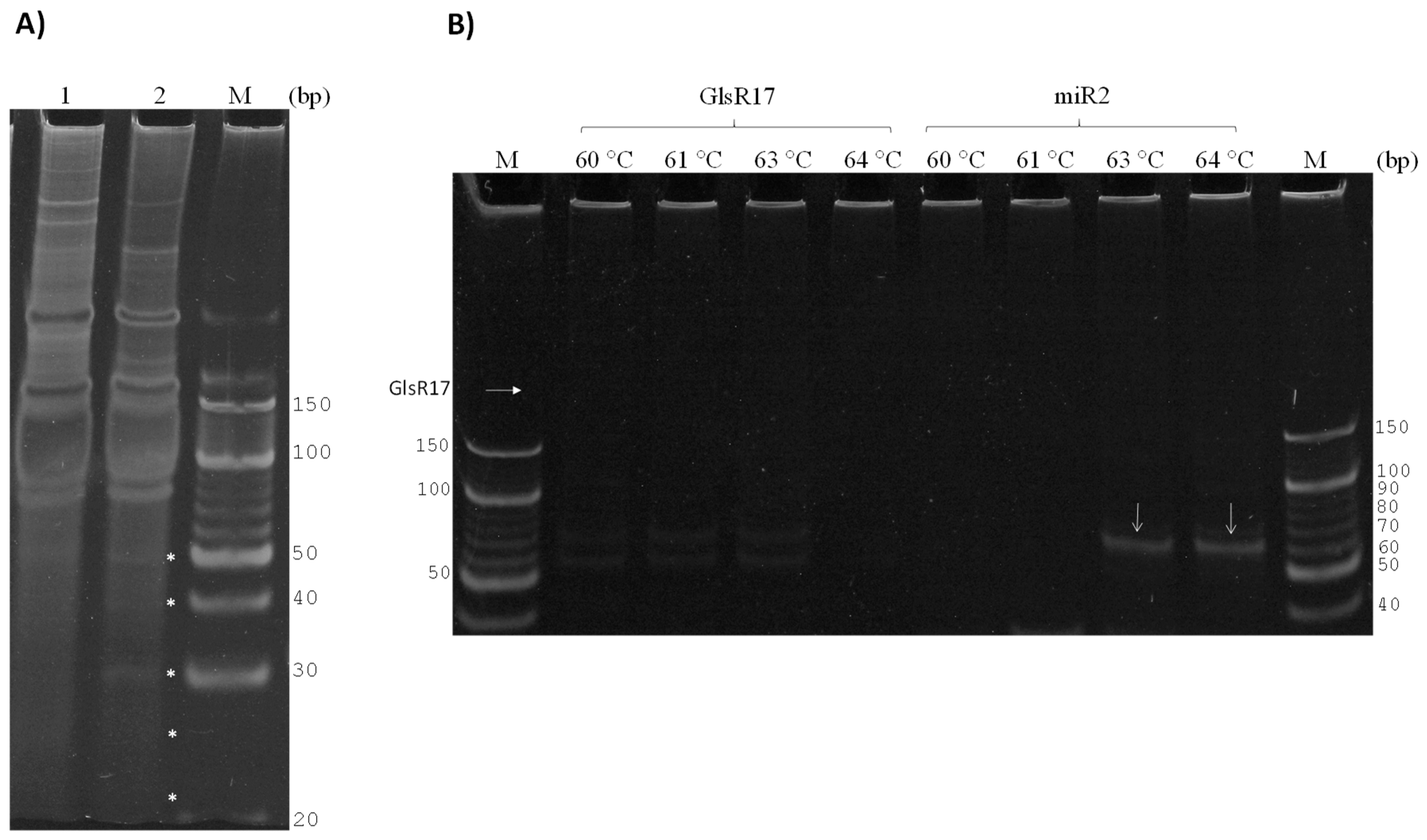
3.2. GlsR17 and miR2 Amplification by Stem-Loop RT-PCR
3.3. Amplification of miR2 by PCR
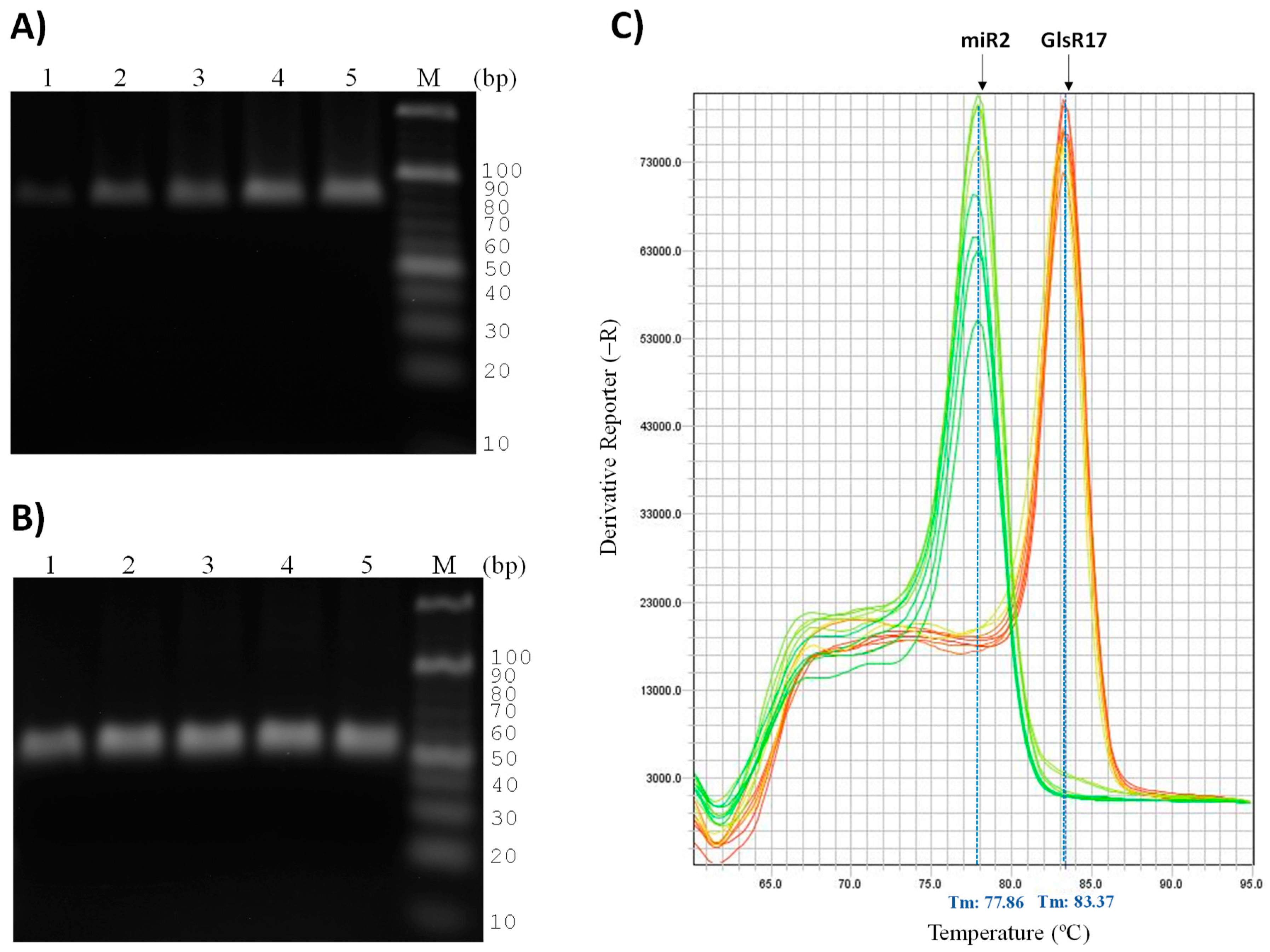
3.4. Validation of Primers for RT-qPCR
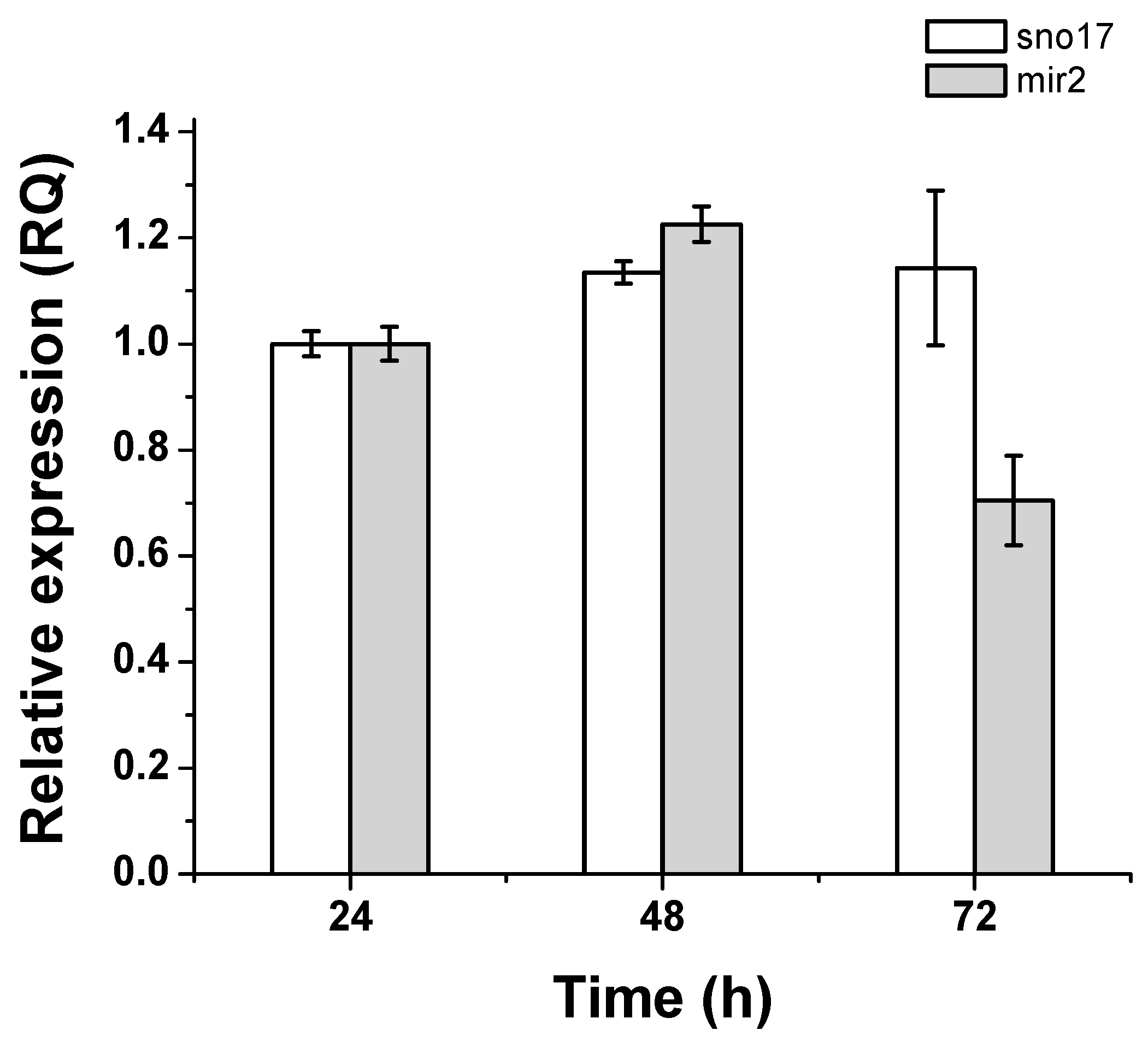
3.5. Quantification of Expression of GlsR17 and miR2
3.6. Identification of miR2 Targets in VSP Genes
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lebwohl, B.; Deckelbaum, R.J.; Green, P.H.R. Giardiasis. Gastrointest. Endosc. 2003, 57, 906–913. [Google Scholar] [CrossRef]
- Müller, N.; von Allmen, N. Recent insights into the mucosal reactions associated with Giardia lamblia infections. Int. J. Parasitol. 2005, 35, 1339–1347. [Google Scholar] [CrossRef] [PubMed]
- Plutzer, J.; Ongerth, J.; Karanis, P. Giardia taxonomy, phylogeny and epidemiology: Facts and open questions. Int. J. Hyg. Environ. Health 2010, 213, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Cai, X.; Bradley, J.E. microRNAs in parasites and parasite infection. RNA Biol. 2013, 10, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef] [PubMed]
- Saraiya, A.A.; Wang, C.C. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008, 4, e1000224. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Saraiya, A.A.; Wang, C.C. The profile of snoRNA-derived microRNAs that regulate expression of variant surface proteins in Giardia lamblia. Cell. Microbiol. 2012, 14, 1455–1473. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Hayashizaki, Y. Exploration of Small RNAs. PLoS Genet. 2008, 4, e22. [Google Scholar] [CrossRef] [PubMed]
- Bratkovič, T.; Rogelj, B. The many faces of small nucleolar RNAs. Biochim. Biophys. Acta 2014, 1839, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Eliaz, D.; Doniger, T.; Tkacz, I.D.; Biswas, V.K.; Gupta, S.K.; Kolev, N.G.; Unger, R.; Ullu, E.; Tschudi, C.; Michaeli, S. Genome-wide analysis of small nucleolar RNAs of Leishmania major reveals a rich repertoire of RNAs involved in modification and processing of rRNA. RNA Biol. 2015, 12, 1222–1255. [Google Scholar] [CrossRef] [PubMed]
- Kolev, N.G.; Tschudi, C.; Ullu, E. RNA interference in protozoan parasites: Achievements and challenges. Eukariot. Cell 2011, 10, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Davis-Dusenbery, B.N.; Hata, A. Mechanisms of control of microRNA biogenesis. J. Biochem. 2010, 48, 381–392. [Google Scholar]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and function. Thromb. Haemost. 2012, 107, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Saraiya, A.A.; Li, W.; Wang, C.C. A microRNA derived from an apparent canonical biogenesis pathway regulates variant surface protein gene expression in Giardia lamblia. RNA 2011, 17, 2152–2164. [Google Scholar] [CrossRef] [PubMed]
- Saraiya, A.A.; Li, W.; Wu, J.; Chang, C.H.; Wang, C.C. The microRNAs in an ancient protist repress the variant-specific surface protein expression by targeting the entire coding sequence. PLoS Pathog. 2014, 10, e1003791. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zheng, W.; Wang, Y.; Li, Y.; Lu, S.; Feng, X. Coexistence of sense and anti-sense mRNAs of variant surface protein in Giardia lamblia trophozoites. Biochem. Biophys. Res. Commun. 2014, 444, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008, 453, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Prucca, C.G.; Slavin, I.; Quiroga, R.; Elías, E.V.; Rivero, F.D.; Saura, A.; Carranza, P.G.; Luján, H.D. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature 2008, 456, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Prucca, C.G.; Rivero, F.D.; Luján, H.D. Regulation of antigenic variation in Giardia lamblia. Annu. Rev. Microbiol. 2011, 65, 611–630. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.Y.; Guo, Y.H.; Zheng, L.L.; Li, Y.; Xu, W.L.; Zhang, Y.C.; Zhou, H.; Lun, Z.R.; Ayala, F.J.; Qu, L.H. Both endo-siRNAs and tRNA-derived small RNAs are involved in the differentiation of primitive eukaryote Giardia lamblia. Proc. Natl. Acad. Sci. USA 2014, 111, 14159–14164. [Google Scholar] [CrossRef] [PubMed]
- Kolev, N.G.; Ullu, E. snoRNAs in Giardia lamblia: A novel role in RNA silencing? Trends Parasitol. 2009, 25, 348–350. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Várallyay, E.; Burgyán, J.; Havelda, Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008, 3, 190–196. [Google Scholar]
- Cissell, K.A.; Deo, S.K. Trends in microRNA detection. Anal. Bioanal. Chem. 2009, 394, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. Sequence requirements for micro RNA processing and function in human cells. RNA 2003, 9, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ruan, K. MicroRNA detection by microarray. Anal. Bioanal. Chem. 2009, 394, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Cirera, S.; Busk, P.K. Quantification of miRNAs by a simple and specific qPCR method. Methods Mol. Biol. 2014, 1182, 73–81. [Google Scholar]
- Kramer, M.F. Stem-loop RT-qPCR for miRNAS. Curr. Protoc. Mol. Biol. 2011. [Google Scholar] [CrossRef]
- Czimmerer, Z.; Hulvely, J.; Simandi, Z.; Varallyay, E.; Havelda, Z.; Szabo, E.; Varga, A.; Dezso, B.; Balogh, M.; Horvath, A.; et al. A Versatile Method to Design Stem-Loop Primer-Based Quantitative PCR Assays for Detecting Small Regulatory RNA Molecules. PLoS ONE 2013, 8, e55168. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.H.; Wang, S.L.; Tang, L.L.; Liu, B.; Ye, W.L.; Wang, L.L.; Wang, Z.Y.; Zhou, M.T.; Chen, B.C. Universal Stem-Loop Primer Method for Screening and Quantification of MicroRNA. PLoS ONE 2014, 9, e115293. [Google Scholar] [CrossRef] [PubMed]
- Gautam, V.; Singh, A.; Singh, S.; Sarkar, A.K. An Efficient LCM-Based Method for Tissue Specific Expression Analysis of Genes and miRNAs. Sci. Rep. 2016, 6, 21577. [Google Scholar] [CrossRef] [PubMed]
- Balcells, I.; Cirera, S.; Busk, P.K. Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol. 2011, 11, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busk, P.K. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinform. 2014, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.; Brennecke, J.; Czech, B.; Aravin, A.; Hannon, G.J. Preparation of Small RNA Libraries for High-Throughput Sequencing. Cold Spring Harb. Protoc. 2012, 2012, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.W. Gel Purification of RNA. Cold Spring Harb. Protoc. 2013, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Aurrecoechea, C.; Brestelli, J.; Brunk, B.P.; Carlton, J.M.; Dommer, J.; Fischer, S.; Gajria, B.; Gao, X.; Gingle, A.; Grant, G.; et al. GiardiaDB and TrichDB: Integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009, 37, D526–D530. [Google Scholar] [CrossRef] [PubMed]
- Martínez, I.; Nogueda, B.; Martínez-Hernández, F.; Espinoza, B. Microsatellite and mini-exon analysis of Mexican human DTU I Trypanosoma cruzi strains and their susceptibility to nifurtimox and benznidazole. Vector Borne Zoonotic Dis. 2013, 13, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Wilkening, S.; Bader, A. Quantitative real-time polymerase chain reaction: Methodical analysis and mathematical model. J. Biomol. Tech. 2004, 15, 107–111. [Google Scholar] [PubMed]
- Pfaffl, M.W. Quantification Strategies in Real-Time PCR. In The Real-Time PCR Encyclopedia, A–Z of Quantitative PCR; Bustin, S.A., Ed.; International University Line: La Jolla, CA, USA, 2004; pp. 87–120. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Marcial-Quino, J.; Fierro, F.; De la Mora-De la Mora, I.; Enríquez-Flores, S.; Gómez-Manzo, S.; Vanoye-Carlo, A.; Garcia-Torres, I.; Sierra-Palacios, E.; Reyes-Vivas, H. Validation of housekeeping genes as an internal control for gene expression studies in Giardia lamblia using quantitative real-time PCR. Gene 2016, 581, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Reichow, S.L.; Hamma, T.; Ferre-D’Amare, A.R.; Varani, G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007, 35, 1452–1464. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Aparicio, M.; Huang, K.; Wafula, E.K.; Honaas, L.A.; Wickett, N.J.; Timko, M.P.; dePamphilis, C.W.; Yoder, J.; Westwood, J.H. Application of qRT-PCR and RNA-Seq analysis for the identification of housekeeping genes useful for normalization of gene expression values during Striga hermonthica development. Mol. Biol. Rep. 2013, 40, 3395–3407. [Google Scholar] [CrossRef] [PubMed]
- Ansell, B.R.E.; McConville, M.J.; Baker, L.; Korhonen, P.K.; Young, N.D.; Hall, R.S.; Rojas, C.A.A.; Svärd, S.G.; Gasser, R.B.; Jex, A.R. Time-Dependent Transcriptional Changes in Axenic Giardia duodenalis Trophozoites. PLoS Negl. Trop. Dis. 2015, 9, e0004261. [Google Scholar] [CrossRef] [PubMed]
- Thomson, T.; Lin, H. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu. Rev. Cell Dev. Biol. 2009, 25, 355–376. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, J.; Zhou, X. A review: microRNA detection methods. Org. Biomol. Chem. 2015, 13, 2226–2238. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; King, K.S.; Donahue, C.P.; Khrapko, K.; Kosik, K.S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003, 9, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.G.; Calin, G.A.; Meloon, B.; Gamliel, N.; Sevignani, C.; Ferracin, M.; Dumitru, C.D.; Shimizu, M.; Zupo, S.; Dono, M.; et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 9740–9744. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tan, R.; Wong, L.; Fekete, R.; Halsey, J. Quantitation of microRNAs by real-time RT-qPCR. Methods Mol. Biol. 2011, 687, 113–134. [Google Scholar]
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Gandelma, O.; Jackson, R.; Kiddle, G.; Tisi, L. Loop-Mediated Amplification Accelerated by Stem Primers. Int. J. Mol. Sci. 2011, 12, 9108–9124. [Google Scholar] [CrossRef] [PubMed]
- Bissels, U.; Wild, S.; Tomiuk, S.; Holste, A.; Hafner, M.; Tuschl, T.; Bosio, A. Absolute quantification of microRNAs by using a universal reference. RNA 2009, 15, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.C.; Li, S.C.; Lin, W.C.; Shin, J.W.; Hu, S.N.; Yu, X.M.; Huang, T.Y.; Chen, S.C.; Chen, H.C.; Chen, S.J.; et al. Identification of microRNA in the protist Trichomonas vaginalis. Genomics 2009, 93, 487–493. [Google Scholar] [CrossRef] [PubMed]



| Primer | Sequence (5′–3′) |
|---|---|
| Stem-loop-GlsR17 | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGACTAT |
| GlsR17-Forward | TGTTTTGTTTCTAGACCTCCTGG |
| qGlsR17-Forward | CAGGACACAGGCGGAG |
| miR2-Forward | TGCAGCCTAATCACCGC |
| UniLoop-Reverse | GTGCAGGGTCCGAGGT |
| pJET1.2 Forward | CGACTCACTATAGGGAGAGCGGC |
| pJET1.2 Reverse | AAGAACATCGATTTTCCATGGCAG |
| pJET2-Forward | CAATTAGTAGCATCACGC |
| Ald-Forward | GAGTCCGTGAAGATGGCGA |
| Ald-Reverse | GTCCCAAGTTCAGCCTCCAC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcial-Quino, J.; Gómez-Manzo, S.; Fierro, F.; Vanoye-Carlo, A.; Rufino-González, Y.; Sierra-Palacios, E.; Castillo-Villanueva, A.; Castillo-Rodríguez, R.A.; Rodríguez-Bustamante, E.; Arreguin-Espinosa, R.; et al. Stem-Loop RT-qPCR as an Efficient Tool for the Detection and Quantification of Small RNAs in Giardia lamblia. Genes 2016, 7, 131. https://doi.org/10.3390/genes7120131
Marcial-Quino J, Gómez-Manzo S, Fierro F, Vanoye-Carlo A, Rufino-González Y, Sierra-Palacios E, Castillo-Villanueva A, Castillo-Rodríguez RA, Rodríguez-Bustamante E, Arreguin-Espinosa R, et al. Stem-Loop RT-qPCR as an Efficient Tool for the Detection and Quantification of Small RNAs in Giardia lamblia. Genes. 2016; 7(12):131. https://doi.org/10.3390/genes7120131
Chicago/Turabian StyleMarcial-Quino, Jaime, Saúl Gómez-Manzo, Francisco Fierro, America Vanoye-Carlo, Yadira Rufino-González, Edgar Sierra-Palacios, Adriana Castillo-Villanueva, Rosa Angélica Castillo-Rodríguez, Eduardo Rodríguez-Bustamante, Roberto Arreguin-Espinosa, and et al. 2016. "Stem-Loop RT-qPCR as an Efficient Tool for the Detection and Quantification of Small RNAs in Giardia lamblia" Genes 7, no. 12: 131. https://doi.org/10.3390/genes7120131
APA StyleMarcial-Quino, J., Gómez-Manzo, S., Fierro, F., Vanoye-Carlo, A., Rufino-González, Y., Sierra-Palacios, E., Castillo-Villanueva, A., Castillo-Rodríguez, R. A., Rodríguez-Bustamante, E., Arreguin-Espinosa, R., & Reyes-Vivas, H. (2016). Stem-Loop RT-qPCR as an Efficient Tool for the Detection and Quantification of Small RNAs in Giardia lamblia. Genes, 7(12), 131. https://doi.org/10.3390/genes7120131






