Et tu, Brute? Not Even Intracellular Mutualistic Symbionts Escape Horizontal Gene Transfer
Abstract
:1. Introduction
1.1. Horizontal Gene Transfer: Molecular Signatures and Mechanisms
1.2. Horizontal Gene Transfer as an Evolutionary Force
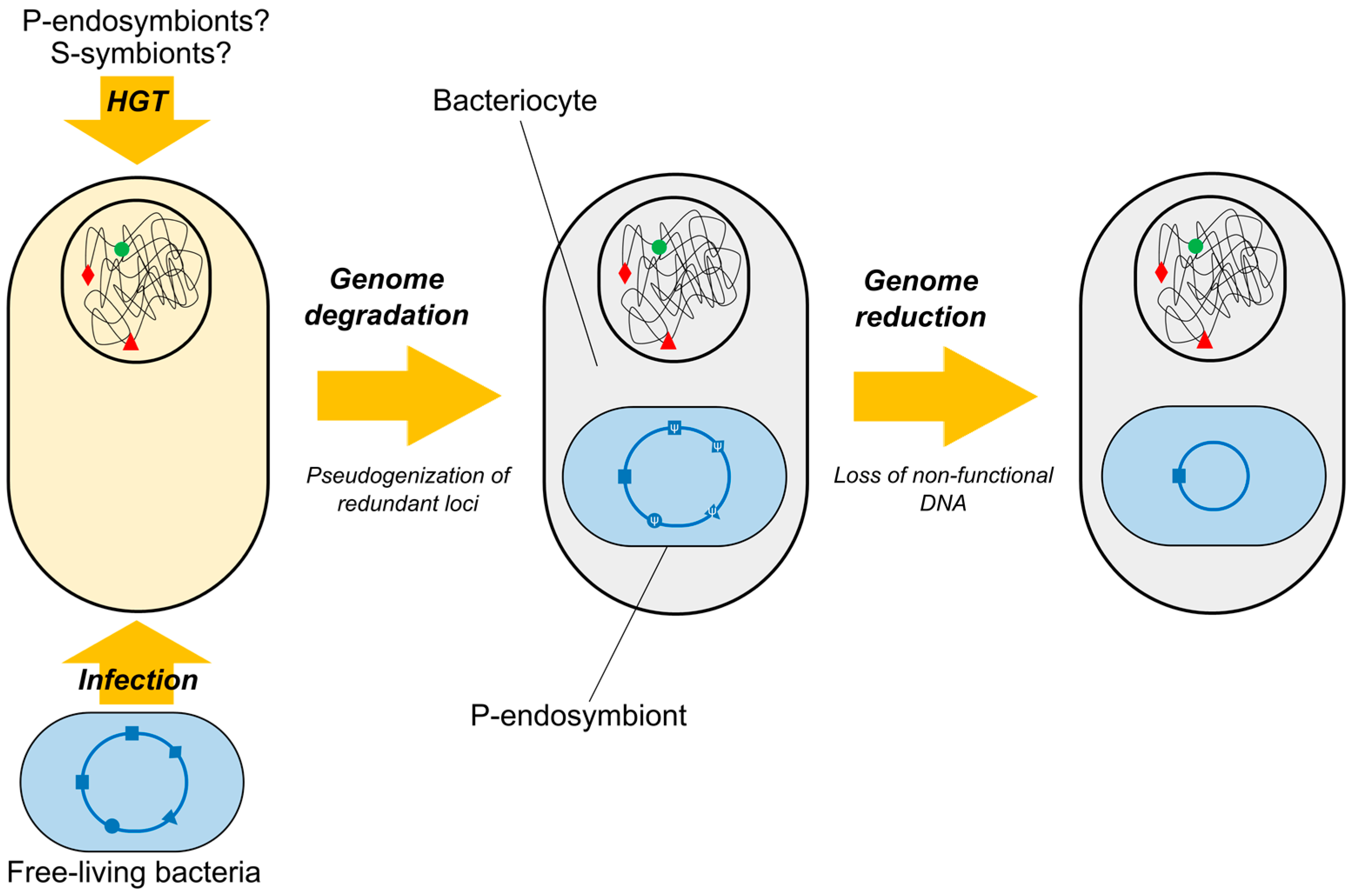
2. Insects-Bacteria Endosymbioses
3. Horizontal Gene Transfer in Insects-Bacteria Endosymbiotic Systems
3.1. Bacteria to Insects and Vice Versa
3.2. Bacteria to Bacteria
3.2.1. Reasons behind Horizontal Gene Transfer Scarcity among Endosymbionts
3.2.2. From Genetic Transfer to Genomic Fusion
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ku, C.; Nelson-Sathi, S.; Roettger, M.; Sousa, F.L.; Lockhart, P.J.; Bryant, D.; Hazkani-Covo, E.; McInerney, J.O.; Landan, G.; Martin, W.F. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 2015, 524, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Acuña, R.; Padilla, B.E.; Flórez-Ramos, C.P.; Rubio, J.D.; Herrera, J.C.; Benavides, P.; Lee, S.J.; Yeats, T.H.; Egan, A.N.; Doyle, J.J.; et al. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl. Acad. Sci. USA 2012, 109, 4197–4202. [Google Scholar] [CrossRef] [PubMed]
- Paganini, J.; Campan-Fournier, A.; Da Rocha, M.; Gouret, P.; Pontarotti, P.; Wajnberg, E.; Abad, P.; Danchin, E.G. Contribution of lateral gene transfers to the genome composition and parasitic ability of root-knot nematodes. PLoS ONE 2012, 7, e50875. [Google Scholar] [CrossRef] [PubMed]
- Flot, J.-F.; Hespeels, B.; Li, X.; Noel, B.; Arkhipova, I.; Danchin, E.G.J.; Hejnol, A.; Henrissat, B.; Koszul, R.; Aury, J.M.; et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 2013, 500, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Pauchet, Y.; Heckel, D.G. The genome of the mustard leaf beetle encodes two active xylanases originally acquired from bacteria through horizontal gene transfer. Proc. R. Soc. B Biol. Sci. 2013, 280. [Google Scholar] [CrossRef] [PubMed]
- Gasmi, L.; Boulain, H.; Gauthier, J.; Hua-Van, A.; Musset, K.; Jakubowska, A.K.; Aury, J.M.; Volkoff, A.N.; Huguet, E.; Herrero, S.; et al. Recurrent domestication by Lepidoptera of genes from their parasites mediated by bracoviruses. PLoS Genet. 2015, 11, e1005470. [Google Scholar] [CrossRef] [PubMed]
- Klasson, L.; Kambris, Z.; Cook, P.E.; Walker, T.; Sinkins, S.P. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genom. 2009, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Schönknecht, G.; Chen, W.-H.; Ternes, C.M.; Barbier, G.G.; Shrestha, R.P.; Stanke, M.; Bräutigam, A.; Baker, B.J.; Banfield, J.F.; Garavito, R.M.; et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 2013, 339, 1207–1210. [Google Scholar] [CrossRef]
- Mashburn-Warren, L.M.; Whiteley, M. Special delivery: Vesicle trafficking in prokaryotes. Mol. Microbiol. 2006, 61, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, B.; Citovsky, V. Transfer of DNA from bacteria to eukaryotes. Am. Soc. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.-H.; Chiu, H.-C.; Delaney, N.F.; Segre, D.; Marx, C.J. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 2011, 332, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Zhang, J. High expression hampers horizontal gene transfer. Genome Biol. Evol. 2012, 4, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Engelberg-Kulka, H.; Glaser, G. Addiction modules and programmed cell death and antideath in bacterial cultures. Ann. Rev. Microbiol. 1999, 53, 43–70. [Google Scholar] [CrossRef] [PubMed]
- Hayes, F. Transposon-based strategies for microbial functional genomics and proteomics. Ann. Rev. Genet. 2003, 37, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, L.; Sigal, N.; Borovok, I.; Nir-Paz, R.; Herskovits, A.A. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 2012, 150, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Gingold, H.; Pilpel, Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 2011, 7, 481. [Google Scholar] [CrossRef] [PubMed]
- Bragg, J.G.; Wagner, A. Protein material costs: Single atoms can make an evolutionary difference. Trends Genet. 2009, 25, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Narra, H.P.; Cordes, M.H.J.; Ochman, H. Structural features and the persistence of acquired proteins. Proteomics 2008, 8, 4772–4781. [Google Scholar] [CrossRef] [PubMed]
- Geiler-Samerotte, K.A.; Dion, M.F.; Budnik, B.A.; Wang, S.M.; Hartl, D.L.; Drummond, D.A. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Gout, J.-F.; Kahn, D.; Duret, L. Paramecium post-genomics consortium. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 2010, 6, e1000944. [Google Scholar] [CrossRef]
- Pál, C.; Papp, B.; Lercher, M.J. Horizontal gene transfer depends on gene content of the host. Bioinformatics 2005, 21. [Google Scholar] [CrossRef] [PubMed]
- Wellner, A.; Gophna, U. Neutrality of foreign complex subunits in an experimental model of lateral gene transfer. Mol. Biol. Evol. 2008, 25, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Golding, I.; Sawai, S.; Guo, L.; Cox, E.C. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005, 3, e229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, G.A.L.; Bower, D.M.; Prazeres, D.M.F.; Monteiro, G.A.; Prather, K.L.J. Rational engineering of Escherichia coli strains for plasmid biopharmaceutical manufacturing. Biotechnol. J. 2012, 7, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Aravind, L. Horizontal gene transfer in prokaryotes: Quantification and classification. Ann. Rev. Microbiol. 2001, 55, 709–742. [Google Scholar] [CrossRef] [PubMed]
- Beiko, R.G.; Harlow, T.J.; Ragan, M.A. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 14332–14337. [Google Scholar] [CrossRef] [PubMed]
- Martiny, A.C.; Huang, Y.; Li, W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ. Microbiol. 2009, 11, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Monchy, S.; Taghavi, S.; Zhu, W.; Ramos, J.; van der Lelie, D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. 2011, 35, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Walk, S.T.; Gordon, D.M.; Feldgarden, M.; Tiedje, J.M.; Konstantinidis, K.T. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl. Acad. Sci. USA 2011, 108, 7200–7205. [Google Scholar] [CrossRef] [PubMed]
- Gogarten, J.P.; Doolittle, W.F.; Lawrence, J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002, 19, 2226–2238. [Google Scholar] [CrossRef] [PubMed]
- Wybouw, N.; Pauchet, Y.; Heckel, D.G.; Van Leeuwen, T. Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol. Evol. 2016, 8, 1785–1801. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Palmer, J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 2015, 25, R911–921. [Google Scholar] [CrossRef] [PubMed]
- Schönknecht, G.; Weber, A.P.M.; Lercher, M.J. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. BioEssays 2014, 36, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Brachat, S.; Dietrich, F.S. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1102–1115. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.; Veronese, P.; Gold, S.E.; Thomma, B.P.H.J.; Chen, Z.; Henrissat, B.; Lee, Y.H.; Park, J.; et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaramillo, V.D.A.; Sukno, S.A.; Thon, M.R. Identification of horizontally transferred genes in the genus Colletotrichum reveals a steady tempo of bacterial to fungal gene transfer. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Hu, X.; Sun, H.; Yang, Y.; Huang, J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Whitaker, N.; González-Rivera, C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 2014, 1843, 1578–1591. [Google Scholar] [CrossRef] [PubMed]
- Boto, L. Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc. Biol. Sci. 2014, 281. [Google Scholar] [CrossRef] [PubMed]
- Dubilier, N.; Bergin, C.; Lott, C. Symbiotic diversity in marine animals: The art of harnessing chemosynthesis. Nat. Rev. Microbiol. 2008, 6, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Moya, A.; Peretó, J.; Gil, R.; Latorre, A. Learning how to live together: Genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 2008, 9, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Novelli, J.; Jiang, D.; Dailey, H.A.; Landmann, F.; Ford, L.; Taylor, M.J.; Carlow, C.K.; Kumar, S.; Foster, J.M.; et al. Interdomain lateral gene transfer of an essential ferrochelatase gene in human parasitic nematodes. Proc. Natl. Acad. Sci. USA 2013, 110, 7748–7753. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, S.; Thézé, J.; Chebbi, M.A.; Giraud, I.; Moumen, B.; Ernenwein, L.; Grève, P.; Gilbert, C.; Cordaux, R. Birth of a W sex chromosome by horizontal transfer of Wolbachia bacterial symbiont genome. Proc. Natl. Acad. Sci. USA 2016, 113, 15036–15041. [Google Scholar] [CrossRef] [PubMed]
- Danchin, E.G.J.; Rosso, M.-N.; Vieira, P.; de Almeida-Engler, J.; Coutinho, P.M.; Henrissat, B.; Abad, P. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc. Natl. Acad. Sci. USA 2010, 107, 17651–17656. [Google Scholar] [CrossRef] [PubMed]
- Boschetti, C.; Pouchkina-Stantcheva, N.; Hoffmann, P.; Tunnacliffe, A. Foreign genes and novel hydrophilic protein genes participate in the desiccation response of the bdelloid rotifer Adineta ricciae. J. Exp. Biol. 2011, 214, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H. Insect symbiosis: An Introduction. In Insect Symbiosis, 1st ed.; Bourtzis, K., Miller, T.A., Eds.; CRC Press: Boca Raton, FL, USA, 2003; Volume 1, pp. 1–16. ISBN 9780849312861. [Google Scholar]
- McCutcheon, J.P.; McDonald, B.R.; Moran, N.A. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. USA 2009, 106, 15394–15399. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, J.P.; Moran, N.A. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2010, 2, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. The nutritional quality of phloem sap utilized by natural aphid populations. Ecol. Entomol. 1993, 18, 31–38. [Google Scholar] [CrossRef]
- Sandström, J.; Pettersson, J. Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J. Insect Physiol. 1994, 40, 947–955. [Google Scholar] [CrossRef]
- Dinant, S.; Bonnemain, J.-L.; Girousse, C.; Kehr, J. Phloem sap intricacy and interplay with aphid feeding. C. R. Biol. 2010, 333, 504–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prosser, W.A.; Douglas, A.E. A test of the hypotheses that nitrogen is upgraded and recycled in an aphid (Acyrthosiphon pisum) symbiosis. J. Insect Physiol. 1992, 38, 93–99. [Google Scholar] [CrossRef]
- López-Madrigal, S.; Beltrà, A.; Resurrección, S.; Soto, A.; Latorre, A.; Moya, A.; Gil, R. Molecular evidence for ongoing complementarity and horizontal gene transfer in endosymbiotic systems of mealybugs. Front. Microbiol. 2014, 5, 449. [Google Scholar] [CrossRef] [PubMed]
- Houk, E.J.; Griffiths, G.W. Intracellular symbiotes of the Homoptera. Ann. Rev. Entomol. 1980, 25, 161–187. [Google Scholar] [CrossRef]
- Baumann, P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Ann. Rev. Microbiol. 2005, 59, 155–189. [Google Scholar] [CrossRef] [PubMed]
- Munson, M.A.; Baumann, P.; Moran, N.A. Phylogenetic relationships of the endosymbionts of mealybugs (Homoptera: Pseudococcidae) based on 16S rDNA sequences. Mol. Phylogenet. Evol. 1992, 1, 26–30. [Google Scholar] [CrossRef]
- Sauer, C.; Stackebrandt, E.; Gadau, J.; Hölldobler, B.; Gross, R. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: Proposal of the new taxon Candidatus Blochmannia gen. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 1877–1886. [Google Scholar] [CrossRef] [PubMed]
- Thao, M.L.; Baumann, P. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl. Environ. Microbiol. 2004, 70, 3401–3406. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Sayavedra, L.; Sámano-Sánchez, H.; Roth, A.; Martínez-Romero, E. Evolutionary relationships of flavobacterial and enterobacterial endosymbionts with their scale insect hosts (Hemiptera: Coccoidea). J. Evol. Biol. 2012, 25, 2357–2368. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 2873–2878. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Kikuchi, Y.; Nikoh, N.; Shimada, M.; Fukatsu, T. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 2006, 4, e337. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Prieto, M.; Latorre, A.; Moya, A. Scanty microbes, the “symbionelle” concept. Environ. Microbiol. 2014, 16, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Yamashita, A.; Toh, H.; Ishikawa, H.; Dunbar, H.E.; Moran, N.A.; Hattori, M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 2006, 314, 267. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, J.P.; von Dohlen, C.D. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 2011, 21, 1366–1372. [Google Scholar] [CrossRef] [PubMed]
- López-Madrigal, S.; Latorre, A.; Porcar, M.; Moya, A.; Gil, R. Complete genome sequence of “Candidatus Tremblaya princeps” strain PCVAL, an intriguing translational machine below the living-cell status. J. Bacteriol. 2011, 193, 5587–5588. [Google Scholar] [CrossRef] [PubMed]
- Bennett, G.M.; Moran, N.A. Small, smaller, smallest: The origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol. Evol. 2013, 5, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Price, D.R.G.; Wilson, A.C.C. A substrate ambiguous enzyme facilitates genome reduction in an intracellular symbiont. BMC Biol. 2014, 12, 110. [Google Scholar] [CrossRef] [PubMed]
- Moya, A.; Gil, R.; Latorre, A. The evolutionary history of symbiotic associations among bacteria and their animal hosts: A model. Clin. Microbiol. Infect. 2009, 15, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Sudakaran, S.; Kost, C.; Kaltenpoth, M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017, 25, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.C.; Duncan, R.P. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc. Natl. Acad. Sci. USA 2015, 112, 201423305. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.A.; Latorre, A.; Sabater-Muñoz, B.; Moya, A.; Moran, N.A. Side-stepping secondary symbionts: Widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 2003, 12, 1061–1075. [Google Scholar] [CrossRef] [PubMed]
- Nováková, E.; Hypsa, V.; Moran, N.A. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 2009, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Dale, C.; Welburn, S.C. The endosymbionts of tsetse flies: Manipulating host-parasite interactions. Int. J. Parasitol. 2001, 31, 628–631. [Google Scholar] [CrossRef]
- Moran, N.A.; Russell, J.A.; Koga, R.; Fukatsu, T. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 2005, 71, 3302–3310. [Google Scholar] [CrossRef] [PubMed]
- Brumin, M.; Kontsedalov, S.; Ghanim, M. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci. 2011, 18, 57–66. [Google Scholar] [CrossRef]
- Duron, O.; Bouchon, D.; Boutin, S.; Bellamy, L.; Zhou, L.; Engelstädter, J.; Hurst, G.D. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008, 6. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Feldhaar, H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 2011, 36, 533–543. [Google Scholar] [CrossRef]
- Werren, J.H.; Windsor, D.M. Wolbachia infection frequencies in insects: Evidence of a global equilibrium? Proc. Biol. Sci. 2000, 267, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Hilgenboecker, K.; Hammerstein, P.; Schlattmann, P.; Telschow, A.; Werren, J.H. How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol. Lett. 2008, 281, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Kondo, N.; Nikoh, N.; Ijichi, N.; Shimada, M.; Fukatsu, T. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc. Natl. Acad. Sci. USA 2002, 99, 14280–14285. [Google Scholar] [CrossRef] [PubMed]
- Dunning-Hotopp, J.C.; Clark, M.E.; Oliveira, D.C.S.G.; Foster, J.M.; Fischer, P.; Muñoz-Torres, M.C.; Giebel, J.D.; Kumar, N.; Ishmael, N.; Wang, S.; et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 2007, 317, 1753–1756. [Google Scholar] [CrossRef] [PubMed]
- Woolfit, M.; Iturbe-Ormaetxe, I.; McGraw, E.A.; O’Neill, S.L. An ancient horizontal gene transfer between mosquito and the endosymbiotic bacterium Wolbachia pipientis. Mol. Biol. Evol. 2009, 26, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Blaxter, M. Symbiont genes in host genomes: Fragments with a future? Cell Host Microbe 2007, 2, 211–213. [Google Scholar] [CrossRef] [PubMed]
- Nikoh, N.; Tanaka, K.; Shibata, F.; Kondo, N.; Hizume, M.; Shimada, M.; Fukatsu, T. Wolbachia genome integrated in an insect chromosome: Evolution and fate of laterally transferred endosymbiont genes. Genome Res. 2008, 18, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Nikoh, N.; Nakabachi, A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009, 7. [Google Scholar] [CrossRef] [PubMed]
- Nikoh, N.; McCutcheon, J.P.; Kudo, T.; Miyagishima, S.; Moran, N.A.; Nakabachi, A. Bacterial genes in the aphid genome: Absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010, 6, e1000827. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; Nikoh, N.; Koga, R.; Ross, L.; Duncan, R.P.; Fujie, M.; Tanaka, M.; Satoh, N.; Bachtrog, D.; Wilson, A.C.; et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 2013, 153, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Nakabachi, A.; Richards, S.; Qu, J.; Murali, S.C.; Gibbs, R.A.; Moran, N.A. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol. Biol. Evol. 2014, 31, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.-B.; Chen, W.; Hasegawa, D.K.; Simmons, A.M.; Wintermantel, W.M.; Ling, K.-S.; Fei, Z.; Liu, S.S.; Douglas, A.E. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol. Evol. 2015, 7, 2635–2647. [Google Scholar] [CrossRef] [PubMed]
- Bastolla, U.; Moya, A.; Viguera, E.; van Ham, R.C.H.J. Genomic determinants of protein folding thermodynamics in prokaryotic organisms. J. Mol. Biol. 2004, 343, 1451–1466. [Google Scholar] [CrossRef] [PubMed]
- Kupper, M.; Gupta, S.K.; Feldhaar, H.; Gross, R. Versatile roles of the chaperonin GroEL in microorganism-insect interactions. FEMS Microbiol. Lett. 2014, 353, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Shigenobu, S.; Watanabe, H.; Hattori, M.; Sakaki, Y.; Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 2000, 407, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Park, J.T. An anhydro-N-acetylmuramyl-l-alanine amidase with broad specificity tethered to the outer membrane of Escherichia coli. J. Bacteriol. 2007, 189, 5634–5641. [Google Scholar] [CrossRef] [PubMed]
- Jollès, P. Lysozymes: Model Enzymes in Biochemistry and Biology, 1st ed.; Birkhäuser: Basel, Switzerland, 1996; p. 449. [Google Scholar]
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; de Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldón, T.; Ghanim, M.; et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010, 11, R21. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Valero, L.; Soriano-Navarro, M.; Pérez-Brocal, V.; Heddi, A.; Moya, A.; García-Verdugo, J.M.; Latorre, A. Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J. Bacteriol. 2004, 186, 6626–6633. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, M.; Koga, R.; Tsuchida, T.; Meng, X.-Y.; Fukatsu, T. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: Novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 2005, 71, 4069–4075. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Ishida, K.; Hongoh, Y.; Ohkuma, M.; Miyagishima, S. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Curr. Biol. 2014, 24, R640–641. [Google Scholar] [CrossRef] [PubMed]
- Gruwell, M.E.; Hardy, N.B.; Gullan, P.J.; Dittmar, K. Evolutionary relationships among primary endosymbionts of the mealybug subfamily Phenacoccinae (Hemiptera: Coccoidea: Pseudococcidae). Appl. Environ. Microbiol. 2010, 76, 7521–7525. [Google Scholar] [CrossRef] [PubMed]
- Von Dohlen, C.D.; Kohler, S.; Alsop, S.T.; McManus, W.R. Mealybug beta-proteobacterial endosymbionts contain gamma-proteobacterial symbionts. Nature 2001, 412, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Thao, M.; Gullan, P.; Baumann, P. Secondary (γ-Proteobacteria.) endosymbionts infect the primary (β-Proteobacteria.) endosymbionts of mealybugs multiple times and coevolve with their hosts. Appl. Environ. Microbiol. 2002, 68, 3190–3197. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; McCutcheon, J.P. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc. Natl. Acad. Sci. USA 2016, 113, E5416–5424. [Google Scholar] [CrossRef] [PubMed]
- López-Madrigal, S.; Balmand, S.; Latorre, A.; Heddi, A.; Moya, A.; Gil, R. How does Tremblaya princeps get essential proteins from its nested partner Moranella endobia in the mealybug Planoccocus citri? PLoS ONE 2013, 8, e77307. [Google Scholar] [CrossRef] [PubMed]
- Thao, M.L.; Moran, N.A.; Abbot, P.; Brennan, E.B.; Burckhardt, D.H.; Baumann, P. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl. Environ. Microbiol. 2000, 66, 2898–2905. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N.J.; van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 2013, 23, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Santos-Garcia, D.; Farnier, P.-A.; Beitia, F.; Zchori-Fein, E.; Vavre, F.; Mouton, L.; Moya, A.; Latorre, A.; Silva, F.J. Complete genome sequence of “Candidatus Portiera aleyrodidarum” BT-QVLC, an obligate symbiont that supplies amino acids and carotenoids to Bemisia tabaci. J. Bacteriol. 2012, 194, 6654–6655. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Moran, N.A. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 2012, 8, 986–989. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.-F.; Xia, F.; Johnson, K.W.; Brown, C.D.; Bartom, E.; Tuteja, J.H.; Stevens, R.; Grossman, R.L.; Brumin, M.; White, K.P.; et al. Comparison of the genome sequences of “Candidatus Portiera aleyrodidarum” primary endosymbionts of the whitefly Bemisia tabaci B and Q biotypes. Appl. Environ. Microbiol. 2013, 79, 1757–1759. [Google Scholar] [CrossRef] [PubMed]
- Popa, O.; Hazkani-Covo, E.; Landan, G.; Martin, W.; Dagan, T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011, 21, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Andersson, S.G.E.; Battin, T.J.; Prosser, J.I.; Schimel, J.P.; Whitman, W.B.; Hallin, S. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 2010, 8, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Andreani, N.A.; Hesse, E.; Vos, M. Prokaryote genome fluidity is dependent on effective population size. ISME J. 2017, 11, 1719–1721. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.R.; Bergstrom, C.T. Bacteria are different: Observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc. Natl. Acad. Sci. USA 2000, 97, 6981–6985. [Google Scholar] [CrossRef] [PubMed]
- Maisnier-Patin, S.; Paulander, W.; Pennhag, A.; Andersson, D.I. Compensatory evolution reveals functional interactions between ribosomal proteins S12, L14 and L19. J. Mol. Biol. 2007, 366, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Davids, W.; Zhang, Z. The impact of horizontal gene transfer in shaping operons and protein interaction networks—Direct evidence of preferential attachment. BMC Evol. Biol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Huang, H.V. Homologous recombination in Escherichia coli: Dependence on substrate length and homology. Genetics 1986, 112, 441–457. [Google Scholar] [PubMed]
- Lorenz, M.G.; Wackernagel, W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 1994, 58, 563–602. [Google Scholar] [PubMed]
- Majewski, J.; Cohan, F.M. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics 1999, 153, 1525–1533. [Google Scholar] [PubMed]
- McCutcheon, J.P.; McDonald, B.R.; Moran, N.A. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009, 5, e1000565. [Google Scholar] [CrossRef] [PubMed]
- Singer, G.A.C.; Hickey, D.A. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol. Biol. Evol. 2000, 17, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Boël, G.; Letso, R.; Neely, H.; Price, W.N.; Wong, K.-H.; Su, M.; Luff, J.; Valecha, M.; Everett, J.K.; Acton, T.B.; et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 2016, 529, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, J.; Gill, R.T. Amino acid content of recombinant proteins influences the metabolic burden response. Biotechnol. Bioeng. 2005, 90, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.M.; Kemper, A.; Gruber-Vodicka, H.; Cardini, U.; van der Geest, M.; Kleiner, M.; Bulgheresi, S.; Mußmann, M.; Herbold, C.; Seah, B.K.; et al. Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat. Microbiol. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Dietrich, C.; Brune, A. Genome analysis of Endomicrobium proavitum suggests loss and gain of relevant functions during the evolution of intracellular symbionts. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.F.; Scharf, M.E. Lower termite associations with microbes: Synergy, protection, and interplay. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Nikoh, N.; Hosokawa, T.; Moriyama, M.; Oshima, K.; Hattori, M.; Fukatsu, T. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA 2014, 111, 10257–10262. [Google Scholar] [CrossRef] [PubMed]
- Gil, R.; Vargas, C.; López-Madrigal, S.; Santos-García, D.; Latorre, A.; Moya, A. Tremblaya phenacola PPER: An evolutionary beta-gammaproteobacterium collage. ISME J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.-Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2010, 107, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Penz, T.; Schmitz-Esser, S.; Kelly, S.E.; Cass, B.N.; Müller, A.; Woyke, T.; Malfatti, S.A.; Hunter, M.S.; Horn, M. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 2012, 8, e1003012. [Google Scholar] [CrossRef] [PubMed]
- Sait, M.; Aitchison, K.; Wheelhouse, N.; Wilson, K.; Lainson, F.A.; Longbottom, D.; Smith, D.G. Genome sequence of Lawsonia intracellularis strain N343, isolated from a sow with hemorrhagic proliferative enteropathy. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Joardar, V.; Williams, K.P.; Driscoll, T.; Hostetler, J.B.; Nordberg, E.; Shukla, M.; Walenz, B.; Hill, C.A.; Nene, V.M.; et al. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J. Bacteriol. 2012, 194, 376–394. [Google Scholar] [CrossRef] [PubMed]
- Sjödin, A.; Svensson, K.; Ohrman, C.; Ahlinder, J.; Lindgren, P.; Duodu, S.; Johansson, A.; Colquhoun, D.J.; Larsson, P.; Forsman, M. Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC Genomics 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Kellner, R.L.L.; Dettner, K. Differential efficacy of toxic pederin in deterring potential arthropod predators of Paederus (Coleoptera: Staphylinidae) offspring. Oecologia 1996, 107, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Kellner, R.L.L. Molecular identification of an endosymbiotic bacterium associated with pederin biosynthesis in Paederus sabaeus (Coleoptera: Staphylinidae). Insect Biochem. Mol. Biol. 2002, 32, 389–395. [Google Scholar] [CrossRef]
- Manley, G.V. Paederus fuscipes [Col.: Staphylinidae]: A predator of rice fields in west Malaysia. Entomophaga 1977, 22, 47–59. [Google Scholar] [CrossRef]
- Jain, R.; Rivera, M.C.; Lake, J.A. Horizontal gene transfer among genomes: The complexity hypothesis. Proc. Natl. Acad. Sci. USA 1999, 96, 3801–3806. [Google Scholar] [CrossRef] [PubMed]
- Cohen, O.; Gophna, U.; Pupko, T. The complexity hypothesis revisited: Connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 2011, 28, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Szabó, G.; Schulz, F.; Toenshoff, E.R.; Volland, J.-M.; Finkel, O.M.; Belkin, S.; Horn, M. Convergent patterns in the evolution of mealybug symbioses involving different intrabacterial symbionts. ISME J. 2017, 11, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Comas, I.; Moya, A.; Azad, R.K.; Lawrence, J.G.; Gonzalez-Candelas, F. The evolutionary origin of Xanthomonadales genomes and the nature of the horizontal gene transfer process. Mol. Biol. Evol. 2006, 23, 2049–2057. [Google Scholar] [CrossRef] [PubMed]
- López-Madrigal, S.; Latorre, A.; Porcar, M.; Moya, A.; Gil, R. Mealybugs nested endosymbiosis: Going into the “matryoshka” system in Planococcus citri in depth. BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- López-Madrigal, S.; Latorre, A.; Moya, A.; Gil, R. The link between independent acquisition of intracellular gamma-endosymbionts and concerted evolution in Tremblaya princeps. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]

| Host | Endosymbiont | Gene Number | Function | Source | Ref. |
|---|---|---|---|---|---|
| Acyrthosiphon pisum | Buchnera aphidicola | 12 | peptidoglycan metabolism (defensive, control) | Rickettsia, Wolbachia | [88,89] |
| Planococcus citri | “Ca. Tremblaya princeps” “Ca. Moranella endobia” | 22 | Lys, Met, riboflavin and biotin biosynthesis (nutritional) peptidoglycan metabolism (control) | Arsenophonus, Cardinium, Rickettsia, Serratia, Sodalis, Wolbachia | [90] |
| Pachypsylla venusta | “Ca. Carsonella ruddii” | 10 | Phe, Arg, riboflavin biosynthesis (nutritional) DNA mismatch repair (informational) | Carsonella, Rickettsia, Wolbachia | [91] |
| Bemisia tabaci | “Ca. Portiera aleyrodidarum” “Ca. Hamiltonella defensa” | 10 | Arg, Lys, Phe, thiamine biosynthesis/ urea degradation (nutritional) | Pantoea & stinkbugs gut symbionts, Rickettsiales, Niastella, Isosphaera | [92] |
| Host | Endosymbiont | Gene Number | Function | Source | Ref. |
|---|---|---|---|---|---|
| Diaphorina citri | “Ca. Carsonella ruddii” “Ca. Profftella armatura” | 20 | diaphorin biosynthesis (defensive) | Paederus-associated Pseudomonas | [108] |
| Cimex lectularius | Wolbachia pipientis | 9 | biotin and thiamine biosynthesis (nutritional) | Cardinium, Rickettsia | [128] |
| Phenacoccus peruvianus | “Ca. Tremblaya phenacola” | 80 | nutritional informational | Sodalis-allied clade | [129] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Madrigal, S.; Gil, R. Et tu, Brute? Not Even Intracellular Mutualistic Symbionts Escape Horizontal Gene Transfer. Genes 2017, 8, 247. https://doi.org/10.3390/genes8100247
López-Madrigal S, Gil R. Et tu, Brute? Not Even Intracellular Mutualistic Symbionts Escape Horizontal Gene Transfer. Genes. 2017; 8(10):247. https://doi.org/10.3390/genes8100247
Chicago/Turabian StyleLópez-Madrigal, Sergio, and Rosario Gil. 2017. "Et tu, Brute? Not Even Intracellular Mutualistic Symbionts Escape Horizontal Gene Transfer" Genes 8, no. 10: 247. https://doi.org/10.3390/genes8100247
APA StyleLópez-Madrigal, S., & Gil, R. (2017). Et tu, Brute? Not Even Intracellular Mutualistic Symbionts Escape Horizontal Gene Transfer. Genes, 8(10), 247. https://doi.org/10.3390/genes8100247






