Origin and Evolution of the Neo-Sex Chromosomes in Pamphagidae Grasshoppers through Chromosome Fusion and Following Heteromorphization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples of Pamphagidae Grasshoppers
2.2. Chromosome Preparation
2.3. Microdissected DNA Library and DNA Probe Preparation
2.4. Fluorescence In Situ Hybridization
2.5. Microscopy
2.6. Image Analysis by the Visualization of the Specific Signal In Silico Software
2.7. Chromosome Nomenclature
3. Results
3.1. Microdissected DNA Probes Derived from Neo-Sex Chromosomes
- FISH signal of background intensity was usually observed in C-positive regions of some autosomes containing no interspersed repeats. It indicated to the absence of homology between DNA of chromosome region and DNA probe (Figures S5 and S6);
- FISH signal produced by interspersed repeats was usually observed in C-negative regions of the autosomes. The intensity of the signal was slightly but distinctive higher than background level. It was decreased by the VISSIS software analysis (Figures S5 and S6);
- Specific FISH signal that was stronger than signal produced by interspersed repeats. The signal was observed in C-negative regions of dissected chromosome or chromosome region after reverse painting. It was produced by hybridization of both unique DNA sequences from region of dissection and interspersed repeats (Figures S5 and S6);
- Strong specific FISH signal was observed in C-positive regions of dissected chromosomes after reverse painting. We should note that strong FISH signal was observed also in C-positive regions of some autosomes. It was registered in region containing clustered repeats homologous to repeats of microdissected chromosome (Figures S5 and S6).
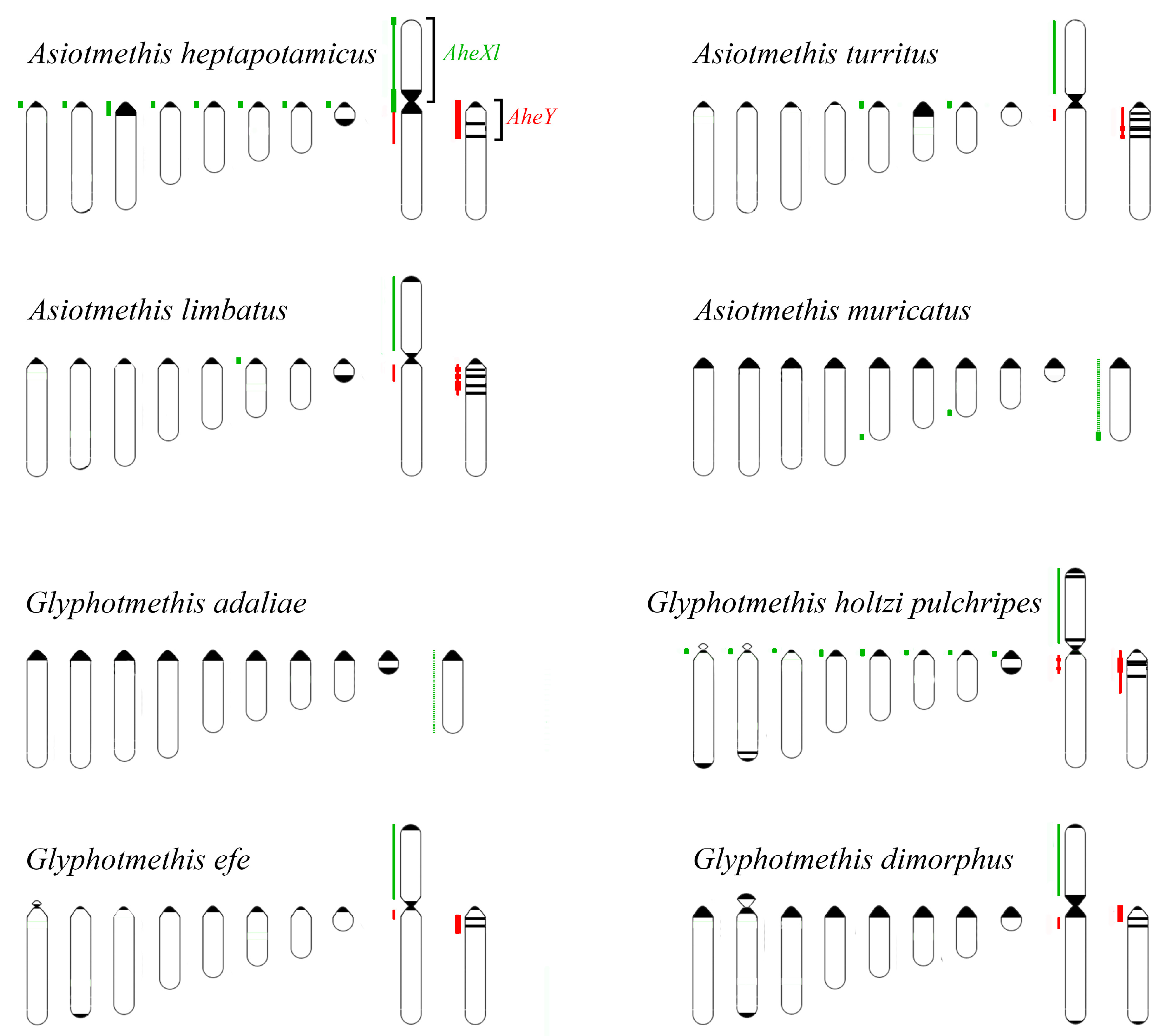
3.2. Chromosome Painting in Pamphagidae Grasshoppers
4. Discussion
4.1. Particularities of Comparative Cytogenetics in Grasshoppers
4.2. Comparative Cytogenetics of C-Negative Regions in the Pamphagidae Neo-Sex Chromosomes
4.3. Comparative Cytogenetics of C-Positive Regions of Chromosomes in Pamphagidae Grasshoppers
4.4. Conjugation of the Neo-Sex Chromosomes in Pamphagidae Species
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Manyard-Smith, J. The Evolution of Sex; Cambridge University Press: Cambidge, UK, 1978. [Google Scholar]
- Hartfield, M.; Keighteley, P.D.; Keightley, P.D.; Keighteley, P.D. Current hypotheses for the evolution of sex and recombination. Integr. Zool. 2012, 7, 192–209. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. Genetic Variability, Twin Hybrids and Constant Hybrids, in a Case of Balanced Lethal Factors. Genetics 1918, 3, 422–499. [Google Scholar] [PubMed]
- Charlesworth, B. The Evolution of Sex Chromosomes. Science 1991, 251, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- White, M.J.D. Animal Cytology and Evolution; Cambridge University Press: Cambridge, UK, 1973. [Google Scholar]
- Steinemann, M.; Steinemann, S. Enigma of Y chromosome degeneration: neo-Y and neo-X chromosomes of Drosophila miranda a model for sex chromosome evolution. Genetica 1998, 102–103, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Charlesworth, D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B 2000, 355, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Evolution of the Y Sex Chromosome in Animals. Bioscience 1996, 46, 331–334. [Google Scholar] [CrossRef]
- Schartl, M.; Schmid, M.; Nanda, I. Dynamics of vertebrate sex chromosome evolution: From equal size to giants and dwarfs. Chromosoma 2016, 125, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.d.B.; Yano, C.F.; Sember, A.; Bertollo, L.A.C. Chromosomal Evolution in Lower Vertebrates: Sex Chromosomes in Neotropical Fishes. Genes 2017, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T. A review of sex determining mechanisms in geckos (Gekkota: Squamata). Sex. Dev. 2010, 4, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Steinlein, C. Sex chromosomes, sex-linked genes and sex determination in the vertebrate class amphibia. EXS 2001, 143–176. [Google Scholar] [CrossRef]
- Kumar, S.; Kumari, R.; Sharma, V. Genetics of dioecy and causal sex chromosomes in plants. J. Genet. 2014, 93, 241–277. [Google Scholar] [CrossRef] [PubMed]
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, V.B.; Bachtrog, D. Evolution of sex chromosomes in insects. Annu. Rev. Genet. 2010, 44, 91–112. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.B.; Clark, A.G. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 2005, 307, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Bernardo Carvalho, A.; Koerich, L.B.; Clark, A.G. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009, 25, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.C.; Lin, F.J.; Chang, H.Y. Stepwise chromosome evolution in Drosophila albomicans. Heredity 1999, 83, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.H.; Chang, C.H.; Chang, H.Y. Early-stage evolution of the neo-Y chromosome in Drosophila albomicans. Zool. Stud. 2011, 50, 338–349. [Google Scholar]
- Castillo, E.R.; Marti, D.A.; Bidau, C.J. Sex and neo-sex chromosomes in Orthoptera: A review. J. Orthoptera Res. 2010, 19, 213–231. [Google Scholar] [CrossRef]
- Castillo, E.R.D.; Bidau, C.J.; Martí, D.A. Neo-sex chromosome diversity in Neotropical melanopline grasshoppers (Melanoplinae, Acrididae). Genetica 2010, 138, 775–786. [Google Scholar] [CrossRef] [PubMed]
- Saez, F. Gradient of the heterochromatinization in the evolution of the sexual system neo-X neo-Y. Port. Acta Biol. A 1963, 7, 111–138. [Google Scholar]
- Bugrov, A.G.; Jetybayev, I.E.; Karagyan, G.H.; Rubtsov, N.B. Cytogenetics Sex chromosome diversity in Armenian toad grasshoppers (Orthoptera, Acridoidea, Pamphagidae). Comp. Cytogenet. 2016, 10, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Jetybayev, I.Y.; Bugrov, A.G.; Ünal, M.; Buleu, O.G.; Rubtsov, N.B. Molecular cytogenetic analysis reveals the existence of two independent neo-XY sex chromosome systems in Anatolian Pamphagidae grasshoppers. BMC Evol. Biol. 2017, 17, 20. [Google Scholar] [CrossRef] [PubMed]
- Ünal, M. Pamphagidae (Orthoptera: Acridoidea) from the Palaearctic Region: Taxonomy, classification, keys to genera and a review of the tribe Nocarodeini I.Bolívar. Zootaxa 2016, 4206, 1–223. [Google Scholar] [CrossRef] [PubMed]
- Bugrov, A.G.; Karamysheva, T.V.; Rubtsov, D.N.; Andreenkova, O.V. Comparative FISH analysis of distribution of B chromosome repetitive DNA in A and B chromosomes in two subspecies of Podisma sapporensis (Orthoptera, Acrididae). Cytogenet. Genome Res. 2004, 106, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, N.B.; Karamisheva, T.V.; Astakhova, N.M.; Liehr, T.; Claussen, U.; Zhdanova, N.S. Zoo-FISH with region-specific paints for mink chromosome 5q: Delineation of inter-and intrachromosomal rearrangements in human, pig and fox. Cytogenet. Genome Res. 2000, 90, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, N.; Senger, G.; Kuzcera, H.; Neumann, A.; Kelbova, C.; Junker, K.; Beensen, V.; Claussen, U. Interstitial deletion of chromosome 6q: Precise definition of the breakpoints by microdissection, DNA amplification and reverse painting. Hum. Genet. 1996, 97, 705–709. [Google Scholar] [CrossRef] [PubMed]
- Pinkel, D.; Straume, T.; Gray, J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA 1986, 83, 2934–2938. [Google Scholar] [CrossRef] [PubMed]
- Bogomolov, A.G.; Zadesenets, K.S.; Karamysheva, T.V.; Podkolodnyi, N.L.; Rubtsov, N.B. Visualization of Chromosome-Specific DNA Sequences by Fluorescence In Situ Hybridization of Microdissection DNA Probes With Metaphase Chromosomes. Russ. J. Genet. 2012, 2, 413–420. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Cabrero, J.; Viseras, E. C-heterochromatin variation in the genus Eumigus (Orthoptera: Pamphagoidea). Genetica 1981, 56, 185–188. [Google Scholar] [CrossRef]
- Bugrov, A.G.; Buleu, O.G.; Jetybayev, I.E. Chromosome polymorphism in populations of Asiotmethis heptapotamicus (Zub.) (Pamphagidae, Thrinchinae) from Kazakhstan. Eurasian Entomol. J. 2016, 15, 545–549. [Google Scholar]
- Buleu, O.G.; Jetybaev, I.E.; Chobanov, D.P.; Bugrov, A.G. Cytogenetic features of some Pamphagidae grasshoppers from Morocco. Eurasian Entomol. J. 2015, 14, 555–560. [Google Scholar]
- Gregory, T.R. Animal Genome Size Database. Available online: http://www.genomesize.com (accessed on 10 November 2017).
- Palacios-Gimenez, O.M.; Castillo, E.R.; Martí, D.A.; Cabral-de-Mello, D.C. Tracking the evolution of sex chromosome systems in Melanoplinae grasshoppers through chromosomal mapping of repetitive DNA sequences. BMC Evol. Biol. 2013, 13, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacios-Gimenez, O.M.; Marti, D.A.; Cabral-de-Mello, D.C. Neo-sex chromosomes of Ronderosia bergi: Insight into the evolution of sex chromosomes in grasshoppers. Chromosoma 2015, 124, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Nat. Publ. Gr. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed]
- Bugrov, A.G.; Karamysheva, T.V.; Perepelov, E.A.; Elisaphenko, E.A.; Rubtsov, D.N.; Warchałowska-Śliwa, E.E.; Tatsuta, H.; Rubtsov, N.B. DNA content of the B chromosomes in grasshopper Podisma kanoi Storozh. (Orthoptera, Acrididae). Chromosome Res. 2007, 15, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Cabrero, J.; Camacho, J.P.M. Location and expression of ribosomal RNA genes in grasshoppers: Abundance of silent and cryptic loci. Chromosome Res. 2008, 16, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Anjos, A.; Ruiz-Ruano, F.J.; Camacho, J.P.M.; Loreto, V.; Cabrero, J.; de Souza, M.J.; Cabral-de-Mello, D.C. U1 snDNA clusters in grasshoppers: Chromosomal dynamics and genomic organization. Heredity 2015, 114, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Teruel, M.; Cabrero, J.; Montiel, E.E.; Acosta, M.J.; Sánchez, A.; Camacho, J.P.M. Microdissection and chromosome painting of X and B chromosomes in Locusta migratoria. Chromosome Res. 2009, 17, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Cabrero, J.; Bugrov, A.; Warchałowska-Sliwa, E.; López-León, M.D.; Perfectti, F.; Camacho, J.P.M. Comparative FISH analysis in five species of Eyprepocnemidine grasshoppers. Heredity 2003, 90, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Bugrov, A.; Karamysheva, T.V.; Pyatkova, M.S.; Rubtsov, D.N.; Andreenkova, O.V.; Warchalowska-Sliwa, E.; Rubtsov, N.B. B chromosomes of the Podisma sapporensis Shir. (Orthoptera, Acrididae) analyzed by chromosome microdissection and FISH. Folia Biol. 2003, 51, 1–11. [Google Scholar]
- Menezes-de-Carvalho, N.Z.; Palacios-Gimenez, O.M.; Milani, D.; Cabral-de-Mello, D.C. High similarity of U2 snDNA sequence between A and B chromosomes in the grasshopper Abracris flavolineata. Mol. Genet. Genom. 2015, 290, 1787–1792. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fang, X.; Yang, P.; Jiang, X.; Jiang, F.; Zhao, D.; Li, B.; Cui, F.; Wei, J.; Ma, C.; et al. The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 2014, 5, 2957. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.M.; Ruiz-Ruano, F.J.; Martín-Blázquez, R.; López-León, M.D.; Cabrero, J.; Lorite, P.; Cabral-de-Mello, D.C.; Bakkali, M. A step to the gigantic genome of the desert locust: Chromosome sizes and repeated DNAs. Chromosoma 2015, 124, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Ferguson-Smith, M.A.; Trifonov, V. Mammalian karyotype evolution. Nat. Rev. Genet. 2007, 8, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Mrasek, K.; Heller, A.; Rubtsov, N.; Trifonov, V.; Starke, H.; Claussen, U.; Liehr, T. Detailed Hylobates lar karyotype defined by 25-color FISH and multicolor banding. Int. J. Mol. Med. 2003, 12, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Mrasek, K.; Heller, A.; Rubtsov, N.; Trifonov, V.; Starke, H.; Rocchi, M.; Claussen, U.; Liehr, T. Reconstruction of the female Gorilla gorilla karyotype using 25-color FISH and multicolor banding (MCB). Cytogenet. Genome Res. 2001, 93, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Makunin, A.I.; Kichigin, I.G.; Larkin, D.M.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Yang, F.; Proskuryakova, A.A.; Vorobieva, N.V.; Chernyaeva, E.N.; O’Brien, S.J.; et al. Contrasting origin of B chromosomes in two cervids (Siberian roe deer and grey brocket deer) unravelled by chromosome-specific DNA sequencing. BMC Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Kichigin, I.G.; Giovannotti, M.; Makunin, A.I.; Ng, B.L.; Kabilov, M.R.; Tupikin, A.E.; Barucchi, V.C.; Splendiani, A.; Ruggeri, P.; Rens, W.; et al. Evolutionary dynamics of Anolis sex chromosomes revealed by sequencing of flow sorting-derived microchromosome-specific DNA. Mol. Genet. Genom. 2016, 291, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, N.B.; Karamysheva, T.V.; Bogdanov, A.S.; Likhoshvay, T.V.; Kartavtseva, I.V. Comparative FISH analysis of C-positive regions of chromosomes of wood mice (Rodentia, Muridae, Sylvaemus). Russ. J. Genet. 2011, 47, 1096–1110. [Google Scholar] [CrossRef]
- Pita, M.; Zabal-Aguirre, M.; Arroyo, F.; Gosálvez, J.; López-Fernández, C.; De La Torre, J. Arcyptera fusca and Arcyptera tornosi repetitive DNA families: Whole-comparative genomic hybridization (W-CGH) as a novel approach to the study of satellite DNA libraries. J. Evol. Biol. 2008, 21, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Jetybayev, I.; Karamysheva, T.; Bugrov, A.; Rubtsov, N. Cross-hybridization of repetitive sequences from the pericentric heterochromatic region of Chorthippus apricarius (L.) with the chromosomes of grasshoppers from the Gomphocerini tribe. Eurasian Entomol. J. 2010, 9, 433–436. [Google Scholar]
- Fry, K.; Salser, W. Nucleotide sequences of HS-α satellite DNA from kangaroo rat dipodomys ordii and characterization of similar sequences in other rodents. Cell 1977, 12, 1069–1084. [Google Scholar] [CrossRef]
- Ünal, M. Revision of the genus Glyphotmethis Bey-Bienko, 1951 (Orthoptera: Pamphagidae). Zootaxa 2007, 1581, 1–36. [Google Scholar]
- Palacios-Gimenez, O.M.; Dias, G.B.; de Lima, L.G.; Kuhn, G.C.e.S.; Ramos, É.; Martins, C.; Cabral-de-Mello, D.C. High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis. Sci. Rep. 2017, 7, 6422. [Google Scholar] [CrossRef] [PubMed]







| Taxa | Species * | Location | Specimen Number |
|---|---|---|---|
| Thrinchini, Thrinchinae | Asiotmethis muricatus (Pallas, 1771) | Kazakhstan | 12 |
| 50°45.377′ N; 51°37.493′ E | |||
| Asiotmethis turritus (Fischer von Waldheim, 1833) | Armenia | 9 | |
| 40°10.220′ N; 44°.22.458′ E | |||
| Asiotmethis heptapotamicus songoricus (Shumakov, 1949) | Kazakhstan | 19 | |
| 47°52.494′ N; 80°6.435′ E | |||
| Asiotmethis limbatus (Charpentier, 1845) | Bulgaria, Harmanli ** | 11 | |
| Glyphotmethis adaliae (Uvarov, 1928) | Turkey | 6 | |
| 37°37.518′ N; 29°13.948′ E | |||
| Glyphotmethis dimorphus dimorphus (Uvarov, 1934) | Turkey | 11 | |
| 38°18.438′ N; 31°43.676′ E | |||
| Glyphotmethis efe (Ünal, 2007) | Turkey | 10 | |
| 39°03.285′ N; 29°26.741′ E | |||
| Glyphotmethis holtzi pulchripes (Uvarov, 1943) | Turkey | 14 | |
| 38°46.688′ N; 34°51.215′ E | |||
| Pamphaginae, Nocarodeini | Nocaracris citripes (syn. Paranocaracris citripes citripes (Uvarov, 1949)) | Turkey | 13 |
| 37°05.779′ N; 28°50.972′ E | |||
| Nocaracris cyanipes (Fischer von Waldheim, 1846) | Armenia | 9 | |
| 40°39.116′ N; 44°58.525′ E | |||
| Nocaracris rubripes (syn. Paranocaracris rubripes (Fischer von Waldheim, 1846)) | Armenia | 3 | |
| 40°23.111′ N; 44°15.324′ E | |||
| Nocaracris furvus furvus (syn. Oronothrotes furvus (Mishchenko, 1951)) | Turkey | 27 | |
| 38°21.258′ N; 28°06.713′ E | |||
| Nocaracris idrisi (syn. Paranocaracris citripes idrisi (Karabağ, 1953)) | Turkey | 9 | |
| 40°35.385′ N; 31°17.293′ E | |||
| Nocaracris sureyana (syn. Paranocaracris sureyana (Ramme, 1951)) | Turkey | 3 | |
| 39°02.353′ N; 29°17.074′ E | |||
| Nocaracris tardus Ünal et al. 2016 (syn. Paranocaracris sp.) | Turkey | 7 | |
| 38°16.672′ N; 31°19.491′ E | |||
| Paranocarodes anatoliensis anatoliensis (syn. Paranocarodes fieberi anatoliensis Demirsoy, 1973) | Turkey | 2 | |
| 37°48.527′ N; 30°45.472′ E | |||
| Paranocarodes karabagi (syn. Pseudosavalania karabagi Demirsoy, 1973) | Turkey | 15 | |
| 39°03.285′ N; 29°26.741′ E | |||
| Paranocarodes tolunayi tolunayi (syn. Paranocarodes fieberi tolunayi Karabag, 1949) | Turkey | 2 | |
| 40°40.937′ N; 31°46.489′ E | |||
| Paranocarodes turkmen (Ünal, 2014) | Turkey | 2 | |
| 39°54.453′ N; 30°41.477′ E |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jetybayev, I.Y.; Bugrov, A.G.; Buleu, O.G.; Bogomolov, A.G.; Rubtsov, N.B. Origin and Evolution of the Neo-Sex Chromosomes in Pamphagidae Grasshoppers through Chromosome Fusion and Following Heteromorphization. Genes 2017, 8, 323. https://doi.org/10.3390/genes8110323
Jetybayev IY, Bugrov AG, Buleu OG, Bogomolov AG, Rubtsov NB. Origin and Evolution of the Neo-Sex Chromosomes in Pamphagidae Grasshoppers through Chromosome Fusion and Following Heteromorphization. Genes. 2017; 8(11):323. https://doi.org/10.3390/genes8110323
Chicago/Turabian StyleJetybayev, Ilyas Yerkinovich, Alexander Gennadievich Bugrov, Olesya Georgievna Buleu, Anton Gennadievich Bogomolov, and Nikolay Borisovich Rubtsov. 2017. "Origin and Evolution of the Neo-Sex Chromosomes in Pamphagidae Grasshoppers through Chromosome Fusion and Following Heteromorphization" Genes 8, no. 11: 323. https://doi.org/10.3390/genes8110323
APA StyleJetybayev, I. Y., Bugrov, A. G., Buleu, O. G., Bogomolov, A. G., & Rubtsov, N. B. (2017). Origin and Evolution of the Neo-Sex Chromosomes in Pamphagidae Grasshoppers through Chromosome Fusion and Following Heteromorphization. Genes, 8(11), 323. https://doi.org/10.3390/genes8110323





