Abstract
Rhizobium leguminosarum bv. trifolii is a soil bacterium capable of establishing a symbiotic relationship with clover (Trifolium spp.). Previously, the rosR gene, encoding a global regulatory protein involved in motility, synthesis of cell-surface components, and other cellular processes was identified and characterized in this bacterium. This gene possesses a long upstream region that contains several regulatory motifs, including inverted repeats (IRs) of different lengths. So far, the role of these motifs in the regulation of rosR transcription has not been elucidated in detail. In this study, we performed a functional analysis of these motifs using a set of transcriptional rosR-lacZ fusions that contain mutations in these regions. The levels of rosR transcription for different mutant variants were evaluated in R. leguminosarum using both quantitative real-time PCR and β-galactosidase activity assays. Moreover, the stability of wild type rosR transcripts and those with mutations in the regulatory motifs was determined using an RNA decay assay and plasmids with mutations in different IRs located in the 5′-untranslated region of the gene. The results show that transcription of rosR undergoes complex regulation, in which several regulatory elements located in the upstream region and some regulatory proteins are engaged. These include an upstream regulatory element, an extension of the -10 element containing three nucleotides TGn (TGn-extended -10 element), several IRs, and PraR repressor related to quorum sensing.
1. Introduction
Rhizobium leguminosarum bv. trifolii is a soil bacterium that establishes nitrogen-fixing symbiosis with clover (Trifolium spp.). However, in the absence of compatible host plants, this micro-organism must often exist for a long period of time in the soil, where it is exposed to various environmental conditions. To adapt to these conditions, rhizobia have developed a wide range of strategies which allow them to survive in the soil. Among these adaptations, composition of the bacterial envelope, synthesis of surface polysaccharides, cell motility, and quorum sensing play the most important roles [1,2].
Recently, it was established that a regulatory protein encoded by the rosR gene is essential for the adaptation of R. leguminosarum to stress conditions, affecting the expression of many genes related to the synthesis of surface polysaccharides and cell-surface components, secretion of extracellular proteins, motility, and other cellular processes [3,4,5]. This gene encodes a 15.7 kDa global transcriptional regulator, which contains a Cys2-His2-type zinc finger motif responsible for binding to RosR-box motifs located in the promoter regions of the regulated genes [4,6]. The amino acid sequence of this motif revealed similarity to the Cys2-His2-type zinc finger motif, which is primarily found in DNA-binding proteins of eukaryotic origin [7,8,9]. In contrast to eukaryotic transcription factors (TFs), which typically contain tandem arrays of such Cys2-His2 zinc fingers, bacterial proteins, including the rhizobial RosR/MucR family, possess only one Cys2-His2-type zinc finger motif located in their C-terminal region.
RosR of R. leguminosarum shares significant similarity with Ros of Rhizobium etli and Agrobacterium tumefaciens, and with MucR of Sinorhizobium meliloti and Sinorhizobium fredii [10,11,12,13,14]. Mutations in rosR and mucR affect the function of both free-living rhizobia and during symbiosis with their host plants. In S. meliloti, MucR regulates the expression of genes involved in the synthesis of extracellular polysaccharides (EPS) (succinoglycan and galactoglucan), which are required for effective symbiosis with its macrosymbiont, alfalfa [15]. Thus, in this bacterium, inactivation of mucR abolishes the synthesis of succinoglycan and reduces cell motility, that in consequence, lead to disturbances in symbiosis [12,16,17]. Also in S. fredii, which shows a broad host range for nodulation, mutation in mucR1 leads to a severe decrease in EPS biosynthesis, increased cell aggregation, and a drastic reduction in the nitrogen fixation capacity on Glycine max and Lotus burttii [13,18,19]. At least in this species, MucR1 is also required for the transcriptional activation of ion transporters engaged in bacterial nitrogen fixation in soybean nodules [14]. Similarly, the Rhizobium etli Ros protein is essential for EPS production, as well as for colonization and infection of its host plant, Phaseolus spp. [10]. In addition, a rosR mutant strain of R. leguminosarum produces a significantly smaller amount of EPS than the wild type, is more sensitive to surface-active compounds, exhibits decreased motility, and induces nodules with a high delay on clover roots, which are ineffective in nitrogen fixation [4,5].
Previously, we have established that the R. leguminosarum rosR open reading frame (ORF) possesses a 403 bp long upstream region containing several regulatory motifs [20,21]. Transcription of this gene was found to be driven by two promoters of different strengths: a distal strong promoter P1 and a much weaker proximal, P2 [6]. It was reported that P1 functions as the main promoter, and in addition to the −35 and −10 hexamers recognized by the RNA polymerase (RNAP) with σ70 subunit, it contains two other regulatory elements, an Upstream Promoter (UP) element [22,23] and a 3–4 bp long extension of the −10 element (TGn-extended −10 element), which in other bacteria are known to play a significant role in the initiation of transcription. We have confirmed that the AT-rich UP element located upstream of the −35 hexamer ensures a high level of expression of this gene from P1 [20]. The UP elements are recognized by the α subunits of the RNAP, whereas the 3–4 bp long extended −10 elements, which usually contain two nucleotides TG, located immediately upstream of the −10 hexamer are recognized by the σ subunit of this enzyme [23,24,25,26,27]. The expression of rosR driven by P2 is ~10-fold lower than that from P1. Downstream of the TGA stop codon of rosR, a rho-independent transcriptional terminator is located, which contains 12 nt long inverted repeats (IR) and forms a very stable stem structure.
It was reported that RosR recognizes and binds to a 22 bp RosR-box motif located downstream of rosR promoters and negatively regulates transcription of its own gene [6]. Moreover, binding sites for cAMP receptor protein CRP (cAMP-CRP) and Pho-boxes recognized by an activator PhoB, engaged in a positive regulation of rosR expression under carbon source and phosphate limitation, respectively, and a LysR motif (T-N11-A)3 recognized by proteins of the LysR family, were identified in the rosR upstream region [20,21]. In addition, a few motifs containing inverted repeats of different lengths (named IR1–IR6) were found in this region. Among them, IR5 contained the longest, 12 bp, inverted repeats. Up to now, only the role of IR4 in rosR expression was experimentally confirmed [21]. This motif was engaged in the negative regulation of rosR transcription, since its deletion or mutations resulted in considerably increased expression of the gene.
In this work, we examined the significance of several motifs identified in the upstream region of R. leguminosarum bv. trifolii rosR in the regulation of its own transcription. For this purpose, a set of rosR-lacZ transcriptional fusions, which contained mutations in these motifs was constructed. The levels of rosR transcription in different mutant variants in R. leguminosarum were established using both quantitative real-time PCR and β-galactosidase activity assays. The stability of wild type rosR transcripts and those containing mutations in IR motifs located in the 5′-untranslated region of this gene was determined using an RNA decay assay. In addition, the influence of chosen rhizobial regulatory proteins (a histidine kinase ChvG of a two-component regulatory system, and an activator CinR, and a represor PraR, both involved in quorum sensing) on the expression of rosR was studied.
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1.

Table 1.
Bacterial strains and plasmids used in this study.
R. leguminosarum strains were cultured in 79CA medium with 1% glycerol as a carbon source at 28 °C with agitation (160 rpm) [38]. DF20 and VF39SM strains were grown in 79CA and Vincent’s minimal medium (VMM) supplemented with micronutrients and vitamin stock solution (2 mL L−1) [39] according to [40]. Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37 °C [34]. When required, the media were supplemented with the appropriate antibiotics used at the following final concentrations: kanamycin, 40 μg mL−1, ampicillin, 100 μg mL−1, tetracycline, 10 μg mL−1, and nalidixic acid, 40 μg mL−1.
2.2. DNA Methods and Sequence Analysis
Standard molecular techniques, such as genomic and plasmid DNA isolation, restriction enzyme digestion, electrophoresis, cloning, and transformation were performed according to [34]. For PCR reactions, Pfu DNA Polymerase (Promega, Madison, WI, USA) or Ready-to-use RED-Taq DNA polymerase mix (Sigma-Aldrich, St. Louis, MO, USA) were used. Primers used in this work are listed in Table 2.

Table 2.
Oligonucleotide primers used in this study.
Sequencing was performed using the BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and the ABI Prism 310 apparatus (Applied Biosystems). Database searchers were done with the FASTA and BLAST programs at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/, Bethesda, MD, USA) and the European Bioinformatic Institute (https://www.ebi.ac.uk/, Hinxton, UK). Identification of the IR motifs in the rosR upstream region was performed using Malign and Fuzznuc programs [41,42,43]. RNA secondary structures and their stability were predicted using the Mfold program, version 2.3, and the default settings [44,45].
2.3. Construction of rosR-lacZ Transcriptional Fusions Containing Changed Sequences of Regulatory Motifs
The broad-host-range vector pMP220 carrying a promoter-less lacZ gene was used to construct plasmid fusions containing the rosR upstream region with mutated regulatory sequences [36]. For this purpose, plasmid pB31 containing the entire rosR gene and primers harboring changed sequences within the regulatory motifs (UP element, TGn-extended −10 element, IR1, IR2, IR3, IR5, IR6, and the RosR-box) and specific sequences for restriction enzymes were used (Table 2). Using primer pairs EP1/EB1, EP1/EB2, EP1/EB3, EP1/EB4, EP1/EB5, EP1/EB6, EP1/EB7, EP1/EB8, EP1/EB9, EP1/EB10, EP1/EB11, and EP1/EB12, 12 PCR products were obtained (PCR1 to PCR12), which encompassed the 5′-part of the rosR regulatory region. 12 DNA fragments encompassing the 3′-part of the rosR regulatory region were obtained using primer pairs BX13/RR1 (PCR13), BX14/RR1 (PCR14), BX15/RR1 (PCR15), BX16/RR1 (PCR16), BX17/RR1 (PCR17), BX18/RR1 (PCR18), BX19/RR1 (PCR19), BX20/RR1 (PCR20), BX21/RR1 (PCR21), BX22/RR1 (PCR22), BX23/RR1 (PCR23), and BX24/RR1 (PCR24). The first set of PCR products (PCR1–PCR8 and PCR11–PCR12) was digested with EcoRI and BamHI enzymes, whereas the second set of PCR products (PCR13–PCR20, PCR23, and PCR24) was digested with BamHI and XbaI. The exceptions were PCR9, PCR10, PCR21, and PCR22 fragments which were digested with PstI instead of BamHI. Next, PCR fragments (PCR1 and PCR13, PCR2 and PCR14, PCR3 and PCR15, PCR4 and PCR16, PCR5 and PCR17, PCR6 and PCR18, PCR7 and PCR19, PCR8 and PCR20, PCR9 and PCR21, PCR10 and PCR22, PCR11 and PCR23, and PCR12 and PCR24) were ligated together, and with pUC19 digested with EcoRI and XbaI, and then used to transform E. coli DH5α. The correctness of cloning was verified by PCR amplification, plasmid DNA isolation, restriction enzyme digestion, and sequencing. Subsequently, the resulting plasmids pPUC1–pPUC12 and the pMP220 vector were digested with EcoRI and XbaI, ligated, and introduced into DH5α strain. The correctness of the resulting plasmid constructs (named pM1 to pM12) was verified by colony PCR and enzyme digestions. The plasmids were then used to transform E. coli S17-1; the obtained derivatives were used as donor strains in bi-parental conjugation with R. leguminosarum bv. trifolii strain Rt24.2 as a recipient. The resulting Rt24.2 derivatives containing different rosR-lacZ transcriptional fusions were used for the determination of the amounts of different rosR transcript variants, as well as for the determination of β-galactosidase activities.
2.4. Construction of Plasmids Containing the Entire rosR Gene and Mutations in the Regulatory Motifs
To establish the stability of different rosR transcript variants, a set of plasmids containing the entire rosR gene and mutated sequences within the regulatory motifs was constructed using the expression vector pQE-31. For this purpose, pPUC7–pPUC12 plasmids and primers EP3/RR1 were used as templates for the amplification of the 5′-parts of the rosR upstream region (with mutations of IR5, IR6, and the RosR-box); pB31 and primers rosR-P/rosR-H were used for the amplification of the 3′-part of rosR (Table 2). A set of obtained PCR products (PCR25 to PCR30) was digested with EcoRI and PstI, whereas the PCR product obtained with primers rosR-P/rosR-H (PCR31) was digested with PstI and HindIII. Next, PCR fragments were ligated together (PCR25 and PCR31, PCR26 and PCR31, PCR27 and PCR31, PCR28 and PCR31, PCR29 and PCR31, and PCR30 and PCR31, accordingly) and the resulting products were subcloned into pQE-31 digested with EcoRI and HindIII. The different plasmids obtained (pQM7 to pQM12) were used to transform E. coli JM101. The control plasmid pQM1 was constructed using a 0.8 bp PCR fragment obtained with primers EP3 and rosR-H. The correctness of the resultant plasmids (pQM1 and pQM7–pQM12) was verified by PCR, enzyme digestions, and sequencing. E. coli JM101 derivatives containing these plasmids were used for the determinations of the life-time of different variants of rosR mRNAs.
2.5. RNA Isolation
To isolate total RNA from R. leguminosarum and E. coli strains, 5 mL of 24 h cultures in 79CA and LB, respectively, supplemented with the appropriate antibiotics were used. Bacterial pellets obtained after centrifugation of the cultures (12,000 × g, 10 min) were suspended in 1 mL of TRI-zol Reagent (Zymo Research, Irvine, CA, USA), vigorously mixed, and left for 5 min at room temperature. Next, the samples were centrifuged, and the supernatants were added to 1 mL of 95% ethanol, mixed, and subsequently applied to Zymo-Spin IIC columns from the Direct-Zol RNA MiniPrep kit (Zymo Research). Further steps of RNA purification were performed according to the manufacturer’s instructions. The concentration and purity of the isolated RNAs were determined spectrophotometrically using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Traces of DNA present in the RNA preparations were removed using the TURBO DNA-free kit containing DNase (Life Technologies, Waltham, MA, USA). The effectiveness of DNA elimination from RNA samples was checked by PCR using Ready-to-use RED-Taq DNA polymerase mix (Sigma-Aldrich) and primer pairs specific to rhizobial genes, pssA encoding a glucosyltransferase involved in the first step of EPS synthesis (pssAG1f/pssA2r) and pssY encoding a putative glycosyltransferase (pssY5f/pssY5r), or primers specific to the E. coli 16S ribosomal RNA (rRNA) gene (16Sec-F1/16Sec-R1).
2.6. Reverse Transcription and Quantitative Real-Time PCR
Real Time quantitative PCR (RT-qPCR) was used to determine the quantities and decay rates of the different variants of rosR transcripts. Complementary DNA (cDNA) was synthesized using 1 μg of total RNA and random hexamer primers (High-Capacity cDNA Reverse Transcription Kit, Life Technologies) as described earlier [46]. The following temperature profile was used: 25 °C (10 min), 37 °C (120 min), and 85 °C (5 min). Real Time quantitative PCRs were done using a Step One Plus PCR System and a Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) in a 10-μL reaction volume, which contained gene-specific primers (Table 2). To determine the ratio of rosR cDNA to recA cDNA (encoding a protein required for homologous recombination) in R. leguminosarum samples, primers rosR4-Fw/rosR4-Rv specific to the 5′-region of the rosR ORF and primers recA2-Fw/recA2-Rv specific to the recA coding region were used. In the case of cDNAs obtained using RNAs isolated from E. coli strains, the same primers (rosR4-Fw and rosR4-Rv) for rosR, and primers 16SEc-F1/16SEc-R1 specific to 16S rRNA were employed. The temperature profile was as follows: denaturation at 95 °C for 1 min; followed by 40 cycles at 95 °C for 15 s, 58 °C for 30 s, and 60 °C for 30 s. Quantification cycle (Cq) values for rosR-specific transcripts were normalized to recA, a housekeeping gene involved in homologous recombination in the R. leguminosarum background, or to the 16S rRNA gene in the E. coli background. The data shown are averages for triplicate biological replicates.
2.7. Determination of RNA Decay
The determination of rosR mRNA stability in E. coli cells was performed according to a method described by Pratte and Thiel [47,48], with minor modifications. For these analyses, E. coli JM101 derivatives containing plasmids pQM1 and pQM7–pQM12 were grown in 50 mL of LB supplemented with ampicillin at 37 °C (160 rpm) up to OD600 = 0.4 (optical density at 600 nm). Next, isopropyl-β-d-1-thiogalactopyranoside was added to a final concentration of 1 mM, and the cultures were grown for 1.5 h. After this time, 5 mL aliquots were collected from each culture (the 0 h time-point), 100 μg mL−1 rifampin was added to the cultures to inhibit new transcription, and the growth was continued for 1 h. Then, 10, 20, 30, 45, and 60 min after the addition of rifampin, 5 mL aliquots of the cultures were collected, quickly chilled on ice, harvested, and frozen. For each strain and time-point tested, three biological replicates were used. RNAs were isolated from these bacterial pellets using TRI-Reagent as described above. Reverse transcription and RT-qPCR were performed as described above (Section 2.6).
2.8. β-Galactosidase Assay
For this assay, R. leguminosarum and E. coli derivatives carrying plasmids with different transcriptional rosR-lacZ fusions were grown in a medium (79CA for R. leguminosarum, and LB for E. coli) supplemented with tetracycline for 24 h. The β-galactosidase activity was determined as described earlier using orto-nitrophenyl-β-d-galactopiranoside as a substrate [4].
2.9. Statistical Analysis
The statistical analyses of data were performed using the Student’s t-test and significant differences between the analyzed samples were established at p < 0.05.
3. Results
3.1. Directed-Mutagenesis and Functional Analysis of Regulatory Motifs Located in the rosR Upstream Region
Our previous studies showed that transcription of rosR is driven by two promoters, P1 and P2, each of them containing sequences highly similar to the eubacterial −35 and −10 sigma factor (σ70) binding sites (Figure 1) [6,20]. Among these, P1, containing two additional elements (UP and TGn-extended −10 elements), is the primary promoter responsible for ~90% of the rosR transcriptional activity. Moreover, rosR possesses a very long upstream region (403 bp), in which several putative regulatory motifs containing inverted repeats of different lengths (named IR1 to IR6) were identified. Among these, IR5 is the longest, containing 12 bp inverted repeats (Figure 1) [20,21]. Such long upstream regions are often described as target sites for the regulation of gene expression [49,50].
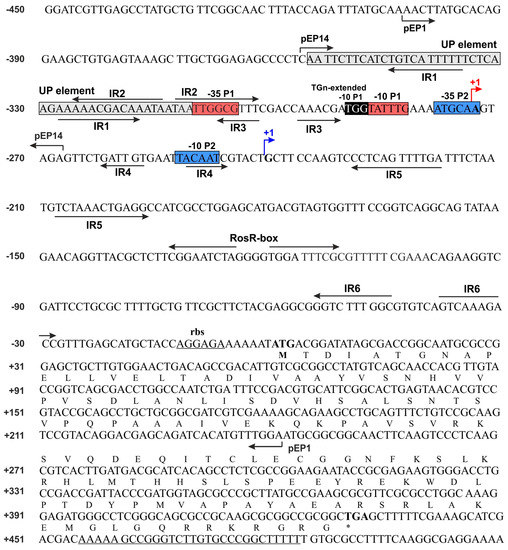
Figure 1.
Nucleotide sequence of Rhizobium leguminosarum bv. trifolii Rt24.2 rosR, including its upstream region. The amino acid sequence of RosR is presented in the single letter code. P1 and P2 are promoters, whereas TS1 and TS2 are transcription start sites. The −35 and −10 hexamers of P1 and P2 promoters are marked by red and blue boxes, and TS1 and TS2 by red and blue arrows. The upstream promoter (UP) element and 3-4 bp long extension of the −10 hexamer (TGn-extended -10 element) are marked by grey and black boxes, respectively. The inverted repeats IR1 to IR6 and a palindromic sequence of the RosR-box are marked by inverted arrows. Over-line short arrows indicate the upstream and downstream endpoints of PCR fragments in the individual plasmid fusions (pEP1 and pEP14), respectively. pEP1 contains the rosR upstream region from −403 to +243 bp, whereas pEP14 harbors a promoter region from −357 to −268 bp. The ribosome-binding site (rbs) and a palindromic sequence of the rho-independent terminator are underlined.
In this study, we proceeded to establish the significance of individual regulatory motifs identified in the rosR upstream region for the expression of this gene. For this purpose, a set of transcriptional fusions between mutated versions of the rosR promoter (prosR) and a promoter-less lacZ gene was constructed on the low-copy plasmid pMP220. Using oligonucleotide primers with changed sequences within the regulatory motifs, we obtained 12 plasmid constructs, named pM1 to pM12 (Figure 2). These prosR-lacZ fusions harbored mutations within the UP element (pM1), 5′- and 3′-parts of the IR1 motif (pM2 and pM3), 3′-part of IR2 (pM4), 3′-part of IR3 (pM5), TGn-extended −10 element (pM6), 5′- and 3′-parts of IR5 (pM7 and pM8), 5′- and 3′-parts of IR6 (pM9 and pM10), and 5′- and 3′-parts of the RosR-box (pM11 and pM12) (Figure 2). Plasmid pEP1 containing the wild type prosR-lacZ fusion was used as a control.
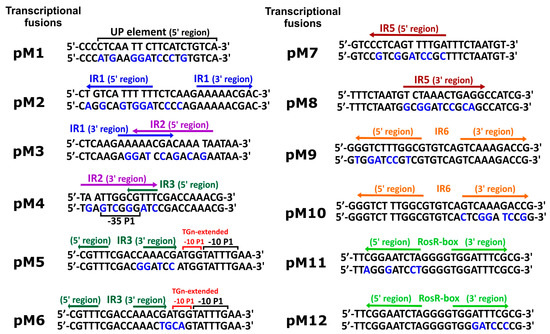
Figure 2.
Nucleotide sequences of regulatory motifs identified in the upstream region of R. leguminosarum bv. trifolii Rt24.2 rosR and mutations introduced in these motifs (UP element, TGn-extended −10 element, and inverted repeats IR1, IR2, IR3, IR5, and IR6). Changed nucleotides in the sequences of the regulatory motifs on individual plasmids (pM1 to pM12) are designated by blue letters. UP and TGn-extended -10 elements are marked by black and red over-line lines, RosR-box is marked by light green arrows, whereas inverted repeats IR1, IR2, IR3, IR5, and IR6 are designated by blue, purple, dark green, dark red, and orange arrows, respectively.
These plasmids were transferred by conjugation into R. leguminosarum bv. trifolii strain Rt24.2 in which subsequently the influence of mutations in the regulatory motifs on the rosR expression was studied. For this purpose, total RNA was isolated from the Rt24.2 derivatives containing pEP1 and pM1–pM12 plasmids, and used in RT-qPCR experiments (Figure 3). Comparative analysis of abundance of rosR transcripts in the strains containing different pM plasmids indicated that almost all changes introduced into the sequences of the analyzed regulatory motifs (with the exception of pM6) affected the level of rosR expression. Among these, the mutations present in pM2, pM5, pM9, pM11, and pM12 resulted in increased amounts of rosR transcripts in relation to that in the strain containing pEP1 (1.76-, 1.78-, 1.94-, 1.97-, and 30.29-fold for pM12, pM2, pM9, pM11, and pM5, respectively). This suggested that the motifs changed in pM2 (IR1), pM5 (IR3), and pM9 (IR6) were engaged in the negative regulation of rosR expression, and confirmed the role of the RosR-box (pM11 and pM12) in this type of regulation, as previously established [6].
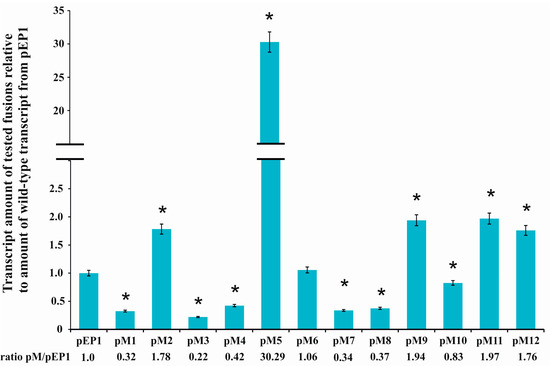
Figure 3.
Quantitative Real Time PCR (RT-qPCR) analysis of rosR transcript levels using total RNA isolated from the R. leguminosarum bv. trifolii Rt24.2 derivatives containing pEP1 and pM1–pM12. Expression of rosR was normalized to the expression of recA ± standard deviation (SD). The ratio of the amount of transcripts carrying mutations within regulatory motifs to the amount of wild type transcript (pEP1) is given below the graph. Significant differences in the expression of rosR in the Rt24.2(pM) strains in relation to its expression in the control strain Rt24.2(pEP1) are indicated by an asterisk (* p < 0.05).
On the other hand, significantly reduced amounts of rosR mRNAs were detected in Rt24.2 carrying the remaining plasmids (pM1, pM3, pM4, pM7, pM8, and pM10) in relation to the control Rt24.2(pEP1) (the ratio pM/pEP1 varied from 0.22 to 0.83, depending on the plasmid tested) (Figure 3). These data suggested that the UP element, and several IR motifs (IR2, IR5, and the 3′-part of IR6) participate in the positive regulation of the expression of this gene. Among the changes introduced in the examined motifs, the greatest effect was observed for the mutation in the 3′-part of IR3 located just upstream of the −10 P1 hexamer (~30-fold increase in rosR transcript amount in pM5 in relation to that of the control pEP1) (Figure 3).
In addition, the effect of mutations in the regulatory motifs on the level of rosR expression was examined using the β-galactosidase activity assay. These analyses were performed in both R. leguminosarum bv. trifolii and E. coli derivatives carrying plasmids pEP1 and pM1–pM12 (Figure 4).
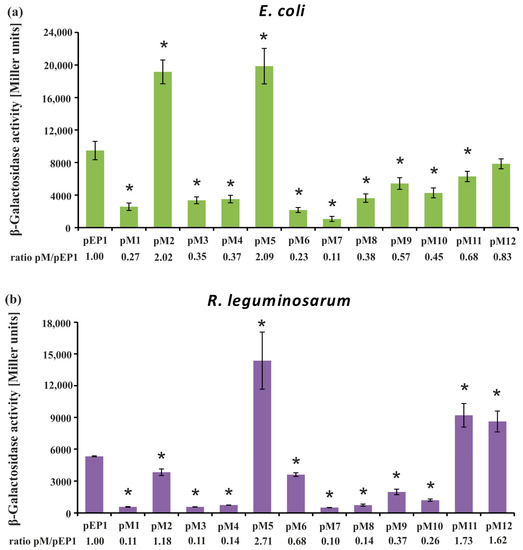
Figure 4.
Determination of rosR expression in Escherichia coli DH5α (a) and R. leguminosarum bv. trifolii Rt24.2 (b) containing pEP1 and pM1–pM12 plasmids using the β-galactosidase activity assay. The ratio of β-galactosidase activity in the strains carrying pM plasmids to that in the strain carrying the control plasmid pEP1 is given below each graph. β-Galactosidase activity for pMP220 in E. coli was 12.5 ± 2.7, and in R. leguminosarum bv. trifolii it was 22.4 ± 3.6 Miller units. Significant differences in the expression level of rosR in the Rt24.2(pM) strains in relation to its expression in the control strain Rt24.2(pEP1), and in the E. coli DH5α(pM) in relation to E. coli DH5α(pEP1), are indicated by an asterisk (* p < 0.05).
Using this approach, we observed a tendency in rosR expression profiles in the two tested bacterial backgrounds that was similar to the RT-qPCR data, although values of β-galactosidase activity obtained in E. coli, which does not possess a RosR ortholog, were significantly higher than in R. leguminosarum (i.e., ~2-fold higher for the wild type pEP1 fusion). In general, β-galactosidase activity values obtained in both these strains carrying transcriptional fusions pM1, pM3, pM4, and pM6–pM10 were significantly lower than pEP1, indicating that the motifs, whose mutated sequences were harbored by these plasmids, are involved in the positive regulation of rosR expression. As noticed for pM1 in both tested backgrounds, mutation of the 5′-end of the UP element strongly reduced the transcriptional activity of this fusion, which confirmed the essential role of this promoter element in its activity. Moreover, the significance of IR2, which is located just upstream of the −35 P1 hexamer, in rosR expression was confirmed, since mutations within both parts of this motif highly reduced the level of rosR transcription (see pM3 and pM4) (Figure 3 and Figure 4). In this assay, a negative effect of the mutation in the TGn-extended −10 element on rosR expression (pM6) was observed; this effect was more pronounced in E. coli than in Rt24.2. Further, mutations in the motifs located downstream of transcription start sites TS1 and TS2 (IR5 and IR6) resulted in reduced rosR expression (plasmids pM7 to pM10), indicating their contribution to the determination of the optimal level of rosR expression. In contrast, mutations in both 5′- and 3′-parts of the RosR-box (pM11 and pM12) resulted, as expected, in elevated rosR transcription in the R. leguminosarum background. In these experiments, the strong positive effect of the mutation located within the 3′-end of IR3 (plasmid pM5) on the expression of this gene was also confirmed (Figure 4). This suggested that among the studied regulatory motifs, the IR3 sequence plays the most essential role in the determination of the rosR transcript levels. To establish whether the effects observed for pM5 and pM2 were caused by a generation of new promoter sequences as a result of the changes introduced into the motifs, in silico sequence analyses were performed. However, they did not reveal any novel promoter sequences (data not shown).
All these data indicate that the analyzed motifs are involved in either positive or negative regulation of rosR transcription, suggesting that their collective activity might ensure the optimal level of its expression.
3.2. Determination of Secondary Structures of rosR Transcripts and Their Stability
Next, we performed additional analyses to establish whether the IR motifs located downstream of TS1 and TS2 (i.e., IR5, IR6, and the RosR-box) play a role in the generation of secondary structures in rosR mRNAs and their stability. Previously, we reported that two types of rosR transcripts of different lengths (766 nt and 733 nt long) are synthesized [6,21]. These transcripts contain 273 nt and 240 nt 5′-untranslated regions, respectively, in which IR5, the RosR-box, and IR6 are located. In the current study, we performed in silico sequence analysis of both the longer and shorter wild type rosR transcripts and their mutated variants with sequence changes within these motifs. We found that the shortening of the 766 nt transcript at the 5′-end did not dramatically influence its secondary structure and stability, which was confirmed by high and similar free energy values (ΔG) (−348.89 for the 766 nt long and −334.87 kcal/mol for the 733 nt long transcript, respectively) (Table 3).

Table 3.
Minimal free energy (ΔG) of secondary structures of the wild type rosR transcripts and their derivatives containing mutations in IR motifs.
In silico predictions revealed that the tested motifs were involved in the formation of secondary structures in rosR transcripts (data not shown). However, although mutations introduced into IR5, the RosR-box, and IR6 affected the formation of secondary structures in these transcripts, they only slightly decreased transcript stability. In fact, the greatest changes in the absolute ΔG values were from −348.89 and −334.87 kcal/mol, for the wild type transcripts, to −334.05 and −330.29 kcal/mol, in the case of mutations in the 3′-part of IR5 (Table 3). Alterations of the 3′-part of the RosR-box did not affect ΔG values of the rosR transcripts.
To experimentally confirm the role of individual IR motifs in the stability of rosR transcripts, a set of plasmids containing the entire rosR ORF with either the wild-type or altered (by mutations in IR5, IR6, or the RosR-box) upstream region was constructed using the pQE-31 vector (pQM1 and pQM7–pQM12). To avoid the influence of other rhizobial regulators than RosR on rosR expression, we examined the stability of these transcripts in an E. coli background. The analysis was performed by quantifying mRNA after the addition of rifampin, which inhibits the initiation but not elongation of transcription involving RNAP [51]. In this experiment, the strains containing the complete wild type rosR sequence (plasmid pQM1) or mutations in the 5′- and 3′-parts of IR5 (plasmids pQM7 and pQM8), 5′- and 3′-parts of IR6 (pQM9 and pQM10), and 5′- and 3′-parts of the RosR-box (pQM11 and pQM12) were used. The abundance of different variants of rosR mRNAs was measured by RT-qPCR at various time-points after the addition of rifampin (from 0 to 60 min). The data presented in Figure 5 were normalized to the amount of the wild type transcript synthesized in E. coli (pQM1) at time 0 min.
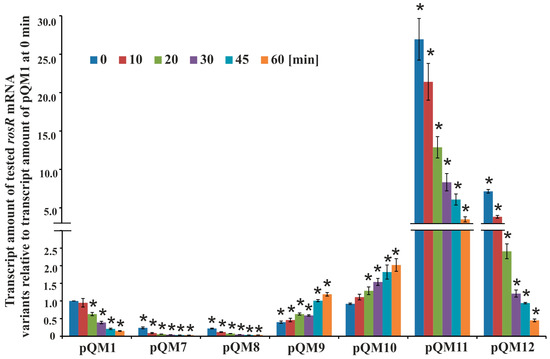
Figure 5.
The stability of different variants of rosR transcripts. Transcript stability was determined by measuring transcript amount by RT-qPCR, using RNAs isolated from E. coli derivatives carrying pQM1 and pQM7–pQM12 plasmids with mutations in the IR5, IR6, and RosR-box motifs. Bacteria were collected 0, 10, 20, 30, 45, and 60 min after the addition of rifampin. The data shown are averages from three biological replicates ± SD. The statistically significant differences between the amount of different variants of rosR transcripts in relation to the amount of the wild type transcript in the control strain E. coli(pQM1) at the same time-point are indicated by an asterisk (* p < 0.05).
In general, the wild type rosR transcript proved to be very stable and its degradation was very slow, which confirmed its long half-life (25.2 ± 4.2 min); 95% of its amount at time 0 min was detected after 10 min, and 15% after 60 min from the transcription inhibition. However, the patterns of degradation of the mutated versions of rosR transcripts were essentially different from that of the wild type. Mutations in both 5′- and 3′-parts of IR5 and in the 5′-part of IR6 negatively affected rosR transcription, as suggested by the very low transcript amounts found in E. coli(pQM7), E. coli(pQM8), and E. coli(pQM9) strains at 0 min in relation to control strain E. coli(pQM1) (Figure 5). This difference was about 4-fold for transcripts from E. coli(pQM7) and E. coli(pQM8), and 2.5-fold for that from E. coli(pQM9). Surprisingly, the transcripts containing mutations in IR5 differed in their stability with respect to those having mutations in IR6. The first ones decayed very quickly and their half-life was 8.2 ± 1.7 min (pQM7) and 11.3 ± 3.2 min (pQM8), respectively. In contrast, the transcripts with mutated 5′- or 3′-parts of IR6 (pQM9 and pQM10) behaved similarly and were very stable. They did not degrade or even, their amounts slightly increased during the time of the experiment, which could be explained in part by the fact that RNA polymerase molecules that were already transcribing at the time of rifampin addition would complete their transcripts, as it has been described previously for the ATPase operon of Prochlorococcus MED4 [52].
In contrast, we observed another effect in the case of mutations within the RosR-box (Figure 5). Significantly higher amounts of rosR transcripts in E. coli(pQM11) and E. coli(pQM12) strains were found in relation to the control E. coli(pQM1) at 0 min. This indicated that the introduced changes positively affected the level of rosR transcription in this genetic background. Moreover, this finding suggested that the R. leguminosarum RosR protein was effectively synthesized in E. coli from pQM plasmids and bound to the wild type sequence of the RosR-box, strongly repressing rosR transcription. This was confirmed by pronounced differences in the abundance of rosR transcripts in E. coli(pQM11) and E. coli(pQM12) in comparison with E. coli(pQM1). However, mutations within the RosR-box did not affect the stability of the synthesized transcripts as pronouncedly as mutations in IR5, as indicated by the half-lives (for transcripts containing changed 5′- and 3′-regions of the RosR-box these were 17.8 ± 3.0 min (pQM11) and 12.5 ± 4.1 min (pQM12), respectively).
In summary, the presented data confirmed that the IR5 and IR6 motifs play an important role in the determination of the rosR transcription level and/or stabilization of the synthesized transcript. These motifs are engaged in the formation of secondary structures of rosR RNA and their stability, that in consequence, affects transcript life-time in bacterial cells.
3.3. The Influence of CinR, PraR, and ChvG on the Expression of rosR
In addition, we decided to study whether some rhizobial regulators could affect the expression of rosR. For this experiment, we selected proteins that play an important role in quorum sensing (transcriptional regulators CinR and PraR) and rhizobial signaling (histidine kinase ChvG of a two-component regulatory system), since mutations in genes encoding these proteins lead to effects that are similar to those observed for the rosR mutant (i.e., changes in biofilm formation, cell-surface properties, and the synthesis of surface polysaccharides) [4,29,31,33,53].
First, plasmid fusions pEP1 and pEP14 (Figure 6a) were transferred to cinR, praR, and chvG mutants and their wild type strains, and β-galactosidase activities were assayed (Figure 6b). The pEP1 plasmid contained the entire rosR upstream region (from −403 to +243 bp), whereas pEP14 harbored a shorter promoter region encompassing only the UP element and the P1 promoter (from −357 to −268 bp) (Figure 1 and Figure 6a). The levels of rosR expression in the tested strains carrying pEP1 and pEP14 were high and similar to those described previously for the R. leguminosarum strain Rt24.2 [20]. In the three wild type strains tested, β-galactosidase activities provided by pEP14 were higher than those obtained in the presence of pEP1. When the individual plasmid fusions were analyzed, no differences were observed in the β-galactosidase activity levels in the chvG mutant DF20 and its wild type strain VF39SM, and in the cinR mutant A552 and its wild type strain 8401 (Figure 6b). This indicated that CinR or ChvG do not affect rosR expression. In contrast, a significant difference in β-galactosidase activities was noted for the pEP1 fusion in the praR mutant A963 and its wild type strain 3841 (Figure 6b), suggesting that PraR could affect rosR expression. Frederix and others [29] have previously characterized a CAAC-N5-GTTG consensus recognized by PraR. We identified a sequence motif CAAGTAGAGTTC in the rosR upstream region (from −275 to −263 nt), which showed a similarity to PraR-binding site (nucleotides identical to those in the consensus are underlined). This sequence, present in pEP1, was truncated in pEP14.
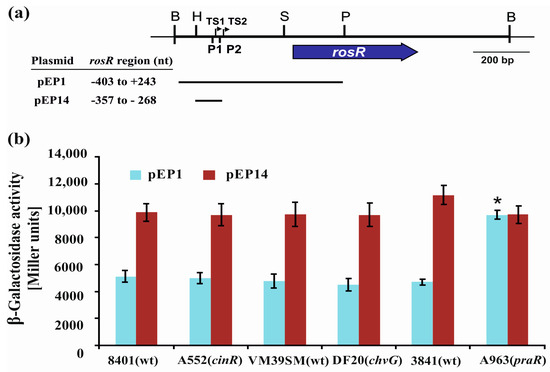
Figure 6.
(a) A physical and genetic map of plasmid pB31 carrying R. leguminosarum rosR. The blue arrow below the map shows the direction of rosR transcription. P1 and P2 are promoter sequences, whereas TS1 and TS2 are transcription start sites. Lengths of the rosR upstream region fragments in pEP1 and pEP14 plasmids are shown as horizontal lines. (b) Transcriptional activity of rosR assayed in wild type R. leguminosarum strains (8401, VM39SM, and 3841) and their derivatives with mutations in cinR, encoding a positive regulator involved in quorum sensing (A552), chvG, encoding a histidine kinase ChvG of a two-component regulatory system (DF20), and praR, encoding a repressor related to quorum sensing (A963), that carried pEP1 and pEP14 plasmids with different fragments of the rosR upstream region fused with a promoter-less lacZ gene. β-Galactosidase activities (presented as Miller units) for the tested plasmid pEP fusions are given as the averages from three independent experiments ± SD. Significant differences in rosR expression between 3841 and A963 (praR) for the individual fusions tested are indicated by an asterisk (* p < 0.05). B: BamHI; H: HindIII; P: PstI; S: SphI.
4. Discussion
In this study, we showed that transcription of the R. leguminosarum rosR gene undergoes a complex regulation, in which several cis-regulatory elements and a previously unidentified trans-acting factor are engaged. Mutational analysis of regulatory motifs identified in the rosR upstream region confirmed a significant role of some of these elements in the modulation of transcription and/or transcript stability of this gene. In general, transcription of rosR was high (see transcriptional activity of rosR in the wild type fusion pEP1 in Figure 4 and Figure 6), and the main promoter P1 was responsible for this effect (see transcriptional activity of rosR provided by UP and P1 in pEP14 in Figure 6).
According to the definition of Gottesman, RosR belongs to global regulators, on the basis of its pleiotropic phenotype and ability to regulate operons associated with different metabolic pathways [54]. In fact, RosR directly or indirectly affects a large group of R. leguminosarum genes (1106), with the majority of them negatively regulated, indicating that RosR functions mainly as a repressor. These genes are associated with the synthesis of cell-surface components, envelope biogenesis, motility, transport and metabolism of carbohydrates and nitrogen sources, and other cellular processes, such as signal transduction and transcription regulation [4,55]. The RosR-box motifs identified in the promoter regions of genes directly regulated by RosR shared a low similarity with the RosR-box consensus. It is well-known that the regulatory effect of individual TFs depends on their concentration and affinity to binding sites; to function, weak sites (i.e., sites with a low sequence similarity to the consensus) require high concentrations of TFs, whereas strong binding sites work with a lower amount of TFs [56]. In contrast to local TFs that tend to have high-affinity sites, global TFs are less specific, bind to a larger collection of sites, and therefore, must be expressed at higher levels [57,58]. This is in agreement with our observation of a high level of rosR transcription. This effect was associated with the activity of the P1 promoter, which in addition to the two core elements (−35 and −10 hexamers) recognized by RNAP with σ70 subunit, contains two cis-regulatory elements (UP and TGn-extended −10) that are present only in a small number of bacterial promoters [59]. Three domains of RNAP σ70 are responsible for recognizing and binding the −10 hexamer (domain 2), −10 extension (domain 3), and −35 hexamer (domain 4), whereas C-terminal domains of the two RNAP α subunits can interact with the UP element located upstream of the −35 region. UP elements characterized in E. coli promoters are ~30 bp A/T-rich sequences that contain two distal and proximal regions, and their presence can stimulate transcription up to 300-fold, depending on the gene studied [23,24,25,60]. Several studies reported that UP elements enhance the transcription of downstream genes, although so far they have been described only for a small number of bacterial promoters, mainly in E. coli (such as P1 of the rRNA rrnB and the guaB promoter required for the de novo synthesis of GMP) but also in other bacteria, such as Bacillus subtilis [23,24,25,61,62,63,64]. Moreover, the presence of UP in promoters served by RNAP containing σ70 might reduce their dependence on the consensus of the −10 or −35 elements [27]. To the best of our knowledge, R. leguminosarum rosR P1 is the first described example of a rhizobial promoter containing UP and TGn-extended −10 elements [23,24,25,26,27].
In this study, we performed mutational analyses of 12 sequence motifs located in the rosR upstream region, including the UP and TGn-extended −10 elements, to examine their role in the transcription of this gene. rosR expression is driven by two promoters, the strong P1 promoter and the very weak P2 promoter. The fact that the majority of the motifs studied are located immediately upstream or even inside P1 most probably means that their mutations mainly affect P1-driven rosR expression. Based on the results obtained for different variants of this region (plasmids pM1–pM12), we observed that the UP present in the rosR P1 promoter was longer (46 nt) than those characterized in E. coli (30 nt), and that it was functional in both R. leguminosarum and E. coli backgrounds, since mutations of both the 5′- and 3′-regions of this element (pM1 and pM3, respectively) considerably reduced rosR transcription in both bacteria (3-fold in E. coli and 10-fold in R. leguminosarum) (Figure 4). This confirmed that the UP element plays an essential role in the stimulation of rosR expression from the P1 promoter. However, alteration of the 5′-region of the IR1 element located in UP (pM2) had an opposite effect, resulting in a ~2-fold increase of rosR transcription in both tested bacteria (Figure 3 and Figure 4). To elucidate whether the changes introduced in the sequence of this motif might have resulted in the appearance of an additional promoter sequence, bioinformatics analyses of the rosR upstream region containing the changed IR1 5′-end were performed; the analyses did not reveal any such new sequences. Therefore, we propose that the alteration of this sequence may have contributed to a stronger interaction of RNAP α subunits with this regulatory region or may be connected with the loss of either a repressor-binding site or a silencing region, which would normally attenuate rosR expression from P1. In contrast, mutation of IR2 (pM3 and pM4) had a negative effect on rosR transcription. Taken together, these results indicated that not only the A/T-rich composition of UP, but also its local structural organization and sequence might be important for transcription initiation, and that IR1 and IR2 exert a negative and positive effect on rosR expression, respectively. Surprisingly, we did not observe a pronounced effect of the mutation in the TGn-extended −10 element (plasmid pM6) (Figure 3 and Figure 4). Based on the approach used, only a weak negative effect was observed in R. leguminosarum using β-galactosidase assay and no effect in RT-qPCR. The effect of this mutation was more pronounced in E. coli (a 4-fold decrease of rosR transcription in pM6 in relation to the wild type pEP1). These data indicated that the 3 bp long −10 extension does not play such an important role in the transcription of this gene in R. leguminosarum, as described for some E. coli genes [58,59,60]. However, a mutation in the IR3 3′-region located just upstream of the −10 extension (pM5) resulted in a strong positive effect in both genetic backgrounds tested, confirming that IR3 also plays a negative role in rosR transcription. Further, in this case the performed in silico analyses excluded the occurrence of a new, additional promoter in this region. Thus, one of the possible explanations of this phenomenon might be the loss of a site recognized by a repressor, which would be conserved between α- and γ-Proteobacteria. Moreover, the possibility of the loss of a silencing region that normally attenuates rosR expression from P1 to allow regulation via the downstream promoter P2 cannot be excluded. Another possibility is that the observed elevated rosR transcription from pM5 is associated with a putative interaction of RNAP σ70 domains with the IR3 3′-region, which might be stronger in the case of its mutated version than in the wild type sequence. Our results suggest that, apart from the −10 extension, a short sequence (7 bp) adjacent to this motif might be engaged in the RNAP σ70–rosR promoter binding, and that at least in some cases, e.g., rosR, the −10 extension might be longer than 3 bp. The fact that the mutations in pM5 and pM2 similarly affected rosR expression in both R. leguminosarum and E. coli suggests that such element(s) may be conserved in both bacteria. Moreover, mutations introduced in IR1–IR3 most probably do not affect the rosR mRNA stability since these motifs are located upstream of the transcription start sites. Similarly, a highly complex mechanism of action was detected for other bacterial global regulatory proteins. For example, fumarate-nitrate reduction regulator Fnr was found to play a dual role in the regulation of arcA, which encodes an aerobic respiratory control protein in E. coli, depending on the growth conditions tested (anaerobiosis/aerobiosis) [65,66]. This gene possesses a long non-coding upstream region (530 bp) containing five promoters recognized by RNAPσ70 and Fnr can function either as an activator from a distal arcAp1 promoter or as a repressor from arcAp3 promoter by binding to the same Fnr-box sequence in this region (−284 bp).
In this study, we also established the role of the IR5 and IR6 motifs, and the RosR-box, located downstream of the transcription start sites TS1 and TS2, in the formation and stabilization of rosR RNA secondary structures. In silico sequence analysis of the wild type and mutated versions of rosR transcripts indicated that these sequence motifs impacted the RNA secondary structure. Based on the results obtained using plasmids pM7–pM12, we experimentally confirmed that mutations of both 5′- and 3′-parts of IR5 and IR6 negatively affected rosR transcription in R. leguminosarum and E. coli (Figure 3 and Figure 4). The greatest reduction was observed for mutations within IR5 (in the rhizobial background). RNA decay analysis performed in E. coli using plasmids pQM1 and pQM7–pQM12 confirmed that wild type rosR transcripts were very stable in bacterial cells, and that IR5 located at the 5′-end of the rosR mRNAs plays the most essential role in their synthesis and protection against degradation (Figure 5). In contrast, the IR6 motif decreased the stability of the transcript since its inactivation increased the half-life of rosR mRNA. Thus, our data indicated that IR5 and IR6 play opposite roles in the stability of rosR transcripts. Moreover, considerably higher amounts of rosR transcripts were detected in the case of plasmids pQM11 and pQM12, which harbor mutations within the RosR-box, than in the wild type pQM1. This suggested that (i) RosR was effectively synthesized in E. coli, and (ii) this rhizobial protein was functional in E. coli (i.e., recognized the wild type RosR-box and negatively regulated the transcription of its own gene). However, this motif did not play such an important role in the stability of rosR transcripts as the IR5 and IR6 motifs.
As reported by Pratte and Thiel, for several genes in nif cluster (nifB1, nifS1, nifH1, nifE1, nifD1, nifU1, nifK1, nifN1, nifX1, hesA1, and fdxH1) encoding proteins involved in nitrogen fixation in the cyanobacterium Anabaena variabilis [47], the half-lives of individual nif mRNAs are very different; from as high as 33 min (for nifH1) and ~20 min (for nifD1, hesA1 required for efficient nitrogen fixation, and fdxH1 encoding the [2Fe-2S] ferredoxin that is an electron donor to nitrogenase, a key enzyme in nitrogen fixation), to as low as ~8 min (nifE1 and nifU1). In comparison to these data, R. leguminosarum rosR transcript belongs to those characterized by high life-time. These authors also showed that the degradation patterns of these mRNAs were strictly different, confirming that this is a specific property of individual gene’s mRNA. The structural organization of promoters of these nif genes and their transcript stability proved to be important for their abundance and life-time in the cell. Similarly to our findings for IR5, they reported that the stem-loop structure upstream of nifH1 controlled the abundance of nifH1 mRNA through transcript processing and stabilization [48]. Stem-loops stabilize transcripts when they are at the extreme 5′-end of the transcript, since these double-stranded structures prevent mRNA recognition by 5′-exonuclease [67,68].
In this work, we also examined whether some regulatory proteins which play an important role in processes such as quorum sensing (CinR and PraR) or signaling (ChvG) in rhizobia, affect rosR transcription [29,31,33,53]. Based on the results obtained for the wild type R. leguminosarum and its chvG, cinR, and praR mutant strains, we confirmed that PraR may act as a trans-regulatory factor able to repress rosR expression (Figure 6). In fact, a sequence with a high similarity to the PraR-binding site was identified downstream of the P1 -10 motif. However, further studies are required to effectively prove a direct effect of PraR on the R. leguminosarum rosR expression. Inhibition of rosR expression by PraR in E. coli, which lacks a PraR ortholog, might provide evidence for the direct interaction. We plan to perform these types of studies in the near future. Recently, Frederix and others [53] characterized a consensus sequence recognized by PraR, and reported that this TF effectively binds to the promoters of rapA2, rapB, and rapC (which encode adhesins), plyB, rosR, and its own promoter, and negatively regulates transcription of these genes. PraR is important for biofilm formation both in vitro and on plant roots, i.e., during a step that precedes the initiation of rhizobial infection of legume roots. Mutation in praR enhanced root biofilms and improved nodulation competitiveness of the bacterium, most probably by increasing the expression of genes coding for proteins involved in bacterial attachment to host root surfaces. All these data indicate that RosR and PraR are important elements of the rhizobial regulatory network, which is required by the bacteria to constantly monitor the extracellular physiological conditions and to respond by modifying gene expression pattern to adjust their growth [69,70].
5. Conclusions
Bacterial TFs play an important role in the genetic regulation of transcription in response to external and internal cellular stimuli. In R. leguminosarum, the TF encoded by rosR is a global regulator that plays an essential role in this regulatory network. Here, we reported that transcription of rosR undergoes a complex regulation, with the involvement of several cis-acting regulatory elements identified in its upstream region and the trans-acting repressor PraR. Among them, apart from the −35 and −10 boxes of the distal P1 promoter, the most essential elements are the A/T-rich UP element located upstream of the P1 −35 hexamer, the IR3 3′-region located upstream of the −10 extension, as well as the inverted repeats IR5 and IR6. These elements are involved in the modulation of the level of rosR transcription and/or the formation of secondary structures and the stability of rosR transcripts, which, consequently affect their life-time in the cell. Most probably, such a complex arrangement of both negative and positive cis-acting elements (even overlapping) in the upstream region of this gene is linked to the requirement for fine-tuning of the regulation of the expression of global regulatory proteins, such as RosR.
Acknowledgments
We thank J. Allan Downie (John Innes Centre, Norwich, United Kingdom) for providing strains 3841, 8401, A963 and A552, and Christopher Yost (University of Regina, Canada) for providing strains VF39SM and DF20. This work was supported by the Polish National Science Centre (NCN) (grant no. DEC-2012/07/B/NZ1/00099).
Author Contributions
M.J. and I.W. conceived and designed the experiments; K.R., M.J., P.L., and I.W. performed the experiments; M.J., J.-M.V., and I.W. analyzed the data; K.R., M.J., P.L., and I.W. contributed reagents/materials/analysis tools; M.J. and J.-M.V. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Downie, J.A. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 2010, 34, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Pini, F.; Galardini, M.; Bazzicalupo, M.; Mengoni, A. Plant-bacteria association and symbiosis: Are there common genomic traits in Alphaproteobacteria? Genes 2011, 2, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Jaszek, M.; Janczarek, M.; Kuczyński, K.; Piersiak, T.; Grzywnowicz, K. The response of the Rhizobium leguminosarum bv. trifolii wild-type and exopolysaccharide-deficient mutants to oxidative stress. Plant Soil 2014, 376, 75–94. [Google Scholar] [CrossRef]
- Rachwał, K.; Matczyńska, E.; Janczarek, M. Transcriptome profiling of a Rhizobium leguminosarum bv. trifolii rosR mutant reveals the role of the transcriptional regulator RosR in motility, synthesis of cell-surface components, and other cellular processes. BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Rachwał, K.; Boguszewska, A.; Kopcińska, J.; Karaś, M.; Tchórzewski, M.; Janczarek, M. The Regulatory protein RosR affects Rhizobium leguminosarum bv. trifolii protein profiles, cell surface properties, and symbiosis with clover. Front. Microbiol. 2016, 7, 1–21. [Google Scholar] [CrossRef]
- Janczarek, M.; Skorupska, A. The Rhizobium leguminosarum bv. trifolii RosR: Transcriptional regulator involved in exopolysaccharide production. Mol. Plant Microbe Interact. 2007, 20, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Shi, Y. The galvanization of biology: A growing appreciation for the roles of zinc. Science 1996, 271, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Cooley, M.B.; D’Souza, M.R.; Kado, C.I. The virC and virD operons of the Agrobacterium Ti plasmid are regulated by the ros chromosomal gene: Analysis of the cloned ros gene. J. Bacteriol. 1991, 173, 2608–2616. [Google Scholar] [CrossRef] [PubMed]
- Malgieri, G.; Russo, L.; Esposito, S.; Baglivo, I.; Zaccaro, L.; Pedone, E.M.; Di Blasio, B.; Isernia, C.; Pedone, P.V.; Fattorusso, R. The prokaryotic Cys2His2 zinc-finger adopts a novel fold as revealed by the NMR structure of Agrobacterium tumefaciens Ros DNA-binding domain. Proc. Natl. Acad. Sci. USA 2007, 104, 17341–17346. [Google Scholar] [CrossRef] [PubMed]
- Bittinger, M.A.; Milner, J.L.; Saville, B.J.; Handelsman, J. rosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol. Plant Microbe Interact. 1997, 10, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Chou, A.Y.; Archdeacon, J.; Kado, C.I. Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. Proc. Natl. Acad. Sci. USA 1998, 95, 5293–5298. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.; Roxlau, A.; Weng, W.M.; Schmidt, M.; Quandt, J.; Niehaus, K.; Jording, D.; Arnold, W.; Pühler, A. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant Microbe Interact. 1995, 8, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Jurado, S.; Alias-Villegas, C.; Navarro-Gómez, P.; Zehner, S.; Murdoch, P.D.; Rodríguez-Carvajal, M.A.; Soto, M.J.; Ollero, F.-J.; Ruiz-Sainz, J.E.; Göttfert, M.; et al. The Sinorhizobium fredii HH103 MucR1 global regulator is connected with the nod regulon and is required for efficient symbiosis with Lotus burttii and Glycine max cv. Williams. Mol. Plant Microbe Interact. 2016, 29, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Wu, L.J.; Zhang, B.; Hu, Y.; Li, Y.; Zhang, X.X.; Guo, H.J.; Liu, L.X.; Chen, W.X.; Zhang, Z.; et al. MucR is required for transcriptional activation of conserved ion transporters to support nitrogen fixation of Sinorhizobium fredii in soybean nodules. Mol. Plant Microbe Interact. 2016, 29, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Ruberg, S.; Kuster, H.; Roxlau, A.A.; Keller, M.; Ivashina, T.; Cheng, H.P.; Walker, G.C.; Puhler, A. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: Genetic organization and properties of the encoded gene products. J. Bacteriol. 1997, 179, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Bertram-Drogatz, P.A.; Rüberg, S.; Becker, A.; Pühler, A. The regulatory protein MucR binds to a short DNA region located upstream of the mucR coding region in Rhizobium meliloti. Mol. Gen. Genet. 1997, 254, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Bertram-Drogatz, P.A.; Quester, I.; Becker, A.; Pühler, A. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 1998, 257, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Jurado, S.; Rodríguez-Navarro, D.N.; Kawaharada, Y.; Perea, J.F.; Gil-Serrano, A.; Jin, H.; An, Q.; Rodríguez-Carvajal, M.A.; Andersen, S.U.; Sandal, N.; et al. Sinorhizobium fredii HH103 Invades Lotus burttii by crack entry in a nod factor-and surface polysaccharide-dependent manner. Mol. Plant Microbe Interact. 2016, 29, 925–937. [Google Scholar] [CrossRef] [PubMed]
- López-Baena, F.J.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.A.; Vinardell, J.-M. Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 2016, 17, 755. [Google Scholar] [CrossRef] [PubMed]
- Janczarek, M.; Skorupska, A. Rhizobium leguminosarum bv. trifolii rosR gene expression is regulated by catabolic repression. FEMS Microbiol. Lett. 2009, 291, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Janczarek, M.; Skorupska, A. Modulation of rosR expression and exopolysaccharide production in Rhizobium leguminosarum bv. trifolii by phosphate and clover root exudates. Int. J. Mol. Sci. 2011, 12, 4132–4155. [Google Scholar] [CrossRef] [PubMed]
- Ross, W.; Gosink, K.K.; Salomon, J.; Igarashi, K.; Zou, C.; Ishihama, A.; Severinov, K.; Gourse, R.L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 1993, 262, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Estrem, S.T.; Gaal, T.; Ross, W.; Gourse, R.L. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 1998, 95, 9761–9766. [Google Scholar] [CrossRef] [PubMed]
- Estrem, S.T.; Ross, W.; Gall, T.; Chen, Z.W.; Niu, W.; Ebright, R.H.; Gourse, R.L. Bacterial promoter architecture: Subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase α subunit. Genes Dev. 1999, 13, 2134–2147. [Google Scholar] [CrossRef] [PubMed]
- Ross, W.; Aiyar, S.E.; Salomon, J.; Gourse, R.L. Escherichia coli promoters with UP elements of different strengths: Modular structure of bacterial promoters. J. Bacteriol. 1998, 18, 5375–5383. [Google Scholar]
- Gourse, R.L.; Ross, W.; Gaal, T. UPs and downs in bacterial transcription initiation: The role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 2000, 37, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Busby, S.J. More pieces in the promoter jigsaw: Recognition of -10 regions by alternative sigma factors. Mol. Microbiol. 2009, 72, 809–811. [Google Scholar] [CrossRef] [PubMed]
- Johnston, A.W.; Beringer, J.E. Identification of the rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 1975, 87, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Frederix, M.; Edwards, A.; McAnulla, C.; Downie, J.A. Co-ordination of quorum-sensing regulation in Rhizobium leguminosarum by induction of an anti-repressor. Mol. Microbiol. 2011, 81, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.W.; Hombrecher, G.; Johnston, A.B. Plasmid determined nodulation and nitrogen fixation abilities in Rhizobium phaseoli. Mol. Gen. Genet. 1982, 186, 449–452. [Google Scholar] [CrossRef]
- Lithgow, J.K.; Wilkinson, A.; Hardman, A.; Rodelas, B.; Wisniewski-Dyé, F.; Williams, P.; Downie, J.A. The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol. Microbiol. 2000, 37, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Priefer, U.B. Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum biovar viciae VF39. J. Bacteriol. 1989, 171, 6161–6168. [Google Scholar] [CrossRef] [PubMed]
- Foreman, D.L.; Vanderlinde, E.M.; Bay, D.C.; Yost, C.K. Characterization of a gene family of outer membrane proteins (ropB) in Rhizobium leguminosarum bv. viciae VF39SM and the role of the sensor kinase ChvG in their regulation. J. Bacteriol. 2010, 192, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989; pp. 11–26, 94–144, 162–186. ISBN 1936113422. [Google Scholar]
- Simon, R.; Quandt, J.; Klipp, W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene 1989, 80, 161–169. [Google Scholar] [CrossRef]
- Spaink, H.P.; Okker, R.J.; Wijffelman, C.A.; Pees, E.; Lugtenberg, B.J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 1987, 9, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Qiagen. Available online: https://www.qiagen.com/us/ (accessed on 16 October 2017).
- Vincent, J.M. A manual for the practical study of root nodule bacteria. In International Biological Program Handbook no. 15; Blackwell Scientific Publications, Ltd.: Oxford, UK; pp. 153–159. ISBN 0632064102.
- Brown, C.M.; Dilworth, M.J. Ammonia assimilation by rhizobium cultures and bacteroids. J. Gen. Microbiol. 1975, 86, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Vanderlinde, E.M.; Yost, C.K. Mutation of the sensor kinase chvG in Rhizobium leguminosarum negatively impacts cellular metabolism, outer membrane stability, and symbiosis. J. Bacteriol. 2012, 194, 768–777. [Google Scholar] [CrossRef] [PubMed]
- AliBee-Multiple Alignment Program. Available online: http://www.genebee.msu.su/services/malign_reduced.html (accessed on 16 October 2017).
- Fuzznuc Program. Available online: http://emboss.ch.embnet.org?Pise (accessed on 16 October 2017).
- Zucker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- The UNAFold Web Server. Available online: http://www.bioinfo.rpi.edu/applications/mfold (accessed on 16 October 2017).
- Newbury, S.F.; Smith, N.H.; Robinson, E.C.; Hiles, I.D.; Higgins, C.F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell 1987, 48, 297–310. [Google Scholar] [CrossRef]
- Wojda, I.; Taszłow, P. Heat shock affects host-pathogen interaction in Galleria mellonella infected with Bacillus thuringiensis. J. Insect Physiol. 2013, 59, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Pratte, B.S.; Thiel, T. Regulation of nitrogenase gene expression by transcript stability in the cyanobacterium Anabaena variabilis. J. Bacteriol. 2014, 196, 3609–3621. [Google Scholar] [CrossRef] [PubMed]
- Pratte, B.S.; Ungerer, J.; Thiel, T. Role of RNA secondary structure and processing in stability of the nifH1 transcript in the cyanobacterium Anabaena variabilis. J. Bacteriol. 2015, 197, 1408–1422. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.V.; Pontel, L.B.; Vescovi, E.G.; Soncini, F.C. Regulation of magnesium homeostasis in Salmonella: Mg2+ targets the mgtA transcript for degradation by RNase E. FEMS Microbiol. Lett. 2008, 280, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Gastebois, A.; Mathieu-Demazière, C.; Sorroche, F.; Masson-Boivin, C.; Batut, J.; Garnerone, A.M. transcriptomic insight in the control of legume root secondary infection by the Sinorhizobium meliloti transcriptional regulator Clr. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sippel, A.; Hartmann, G. Mode of action of rifampicin on the RNA polymerase reaction. Biochim. Biophys. Acta 1968, 157, 218–219. [Google Scholar] [CrossRef]
- Steglich, C.; Lindell, D.; Futschik, M.; Rector, T.; Steen, R.; Chisholm, S.W. Short RNA half-lives in the slow-growing marine cyanobacterium Prochlorococcus. Genome Biol. 2010, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Frederix, M.; Edwards, A.; Swiderska, A.; Stanger, A.; Karunakaran, R.; Williams, A.; Abbruscato, P.; Sanchez-Contreras, M.; Poole, P.S.; Downie, J.A. Mutation of praR in Rhizobium leguminosarum enhances root biofilms, improving nodulation competitiveness by increased expression of attachment proteins. Mol. Microbiol. 2014, 93, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S. Bacterial regulation: Global regulatory networks. Annu. Rev. Genet. 1984, 18, 415–444. [Google Scholar] [CrossRef] [PubMed]
- Janczarek, M.; Rachwał, K.; Kopcińska, J. Genetic characterization of the Pss region and the role of PssS in exopolysaccharide production and symbiosis of Rhizobium leguminosarum bv. trifolii with clover. Plant Soil 2015, 396, 257–275. [Google Scholar] [CrossRef]
- Alon, U. Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 2007, 8, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Lozada-Chavez, I.; Angarica, V.E.; Collado-Vides, J.; Contreras-Moreira, B. The role of DNA-binding specificity in the evolution of bacterial regulatory networks. J. Mol. Biol. 2008, 379, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Balleza, E.; Lopez-Bojorquez, L.N.; Martinez-Antonio, A.; Resendis-Antonio, O.; Lozada-Chavez, I.; Balderas-Martinez, Y.I.; Encarnacion, S.; Collado-Vides, J. Regulation by transcription factors in bacteria: Beyond description. FEMS Microbiol. Rev. 2009, 33, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.F.; Busby, S.J. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004, 2, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Aiyar, S.E.; Gourse, R.L.; Ross, W. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase alpha subunit. Proc. Natl. Acad. Sci. USA 1998, 95, 14652–14657. [Google Scholar] [CrossRef] [PubMed]
- Leirmo, S.; Gourse, R.L. Factor-independent activation of Escherichia coli rRNA transcription. I. Kinetic analysis of the roles of the upstream activator region and supercoiling on transcription of the rrnB P1 promoter in vitro. J. Mol. Biol. 1991, 220, 555–568. [Google Scholar] [CrossRef]
- Husnain, S.I.; Thomas, M.S. The UP element is necessary but not sufficient for growth rate-dependent control of the Escherichia coli guaB promoter. J. Bacteriol. 2008, 190, 2450–2457. [Google Scholar] [CrossRef] [PubMed]
- Banner, C.D.; Moran, C.P., Jr.; Losick, R. Deletion analysis of a complex promoter for a developmentally regulated gene from Bacillus subtilis. J. Mol. Biol. 1983, 168, 351–365. [Google Scholar] [CrossRef]
- Busby, S.; Spassky, A.; Chan, B. RNA polymerase makes important contacts upstream from base pair -49 at the Escherichia coli galactose operon P1 promoter. Gene 1987, 53, 145–152. [Google Scholar] [CrossRef]
- Compan, I.; Touati, D. Anaerobic activation of arcA transcription in Escherichia coli: Roles of Fnr and ArcA. Mol. Microbiol. 1994, 11, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Shimizu, K. Transcriptional regulation of main metabolic pathways of cyoA, cydB, fnr, and fur gene knockout Escherichia coli in C-limited and N-limited aerobic continuous cultures. Microb. Cell Fact. 2011, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Emory, S.A.; Bouvet, P.; Belasco, J.G. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992, 6, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Bensing, B.A.; Meyer, B.J.; Dunny, G.M. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 1996, 93, 7794–7799. [Google Scholar] [CrossRef] [PubMed]
- Maltsev, N.; Marland, E.; Yu, G.X.; Bhatnagar, S.; Lusk, R. Sentra, a database of signal transduction proteins. Nucl. Acids Res. 2002, 30, 349–350. [Google Scholar] [CrossRef] [PubMed]
- Rui, S.; Tse-Dinh, Y.C. Topoisomerase function during bacterial responses to environmental challenge. Front. Biosci. 2003, 8, 256–263. [Google Scholar]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).