B Chromosomes in Grasshoppers: Different Origins and Pathways to the Modern Bs
Abstract
:1. Introduction
2. Material and Methods
2.1. Chromosome Preparation
2.2. DNA Probes Generation
2.3. C-Banding and Fluorescence In Situ Hybridization (FISH) Technique
2.4. Microscopy
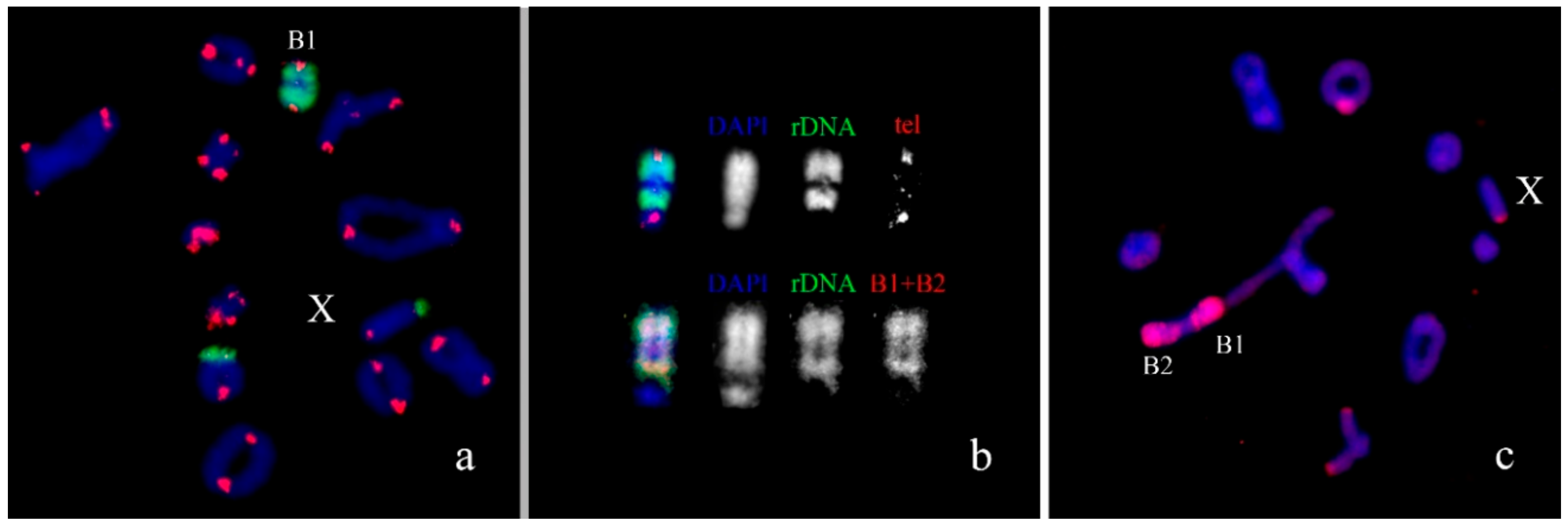
3. Results
B Chromosome Morphology in Studied Grasshopper Species
4. Discussion
4.1. Origin of Dot-Like Bs
4.2. From Dot-Like Bs to the Large Ones
4.3. The Bs with C-Negative Regions
4.4. DNA Content of C-Negative Regions in Bs
4.5. Mechanisms of C-Negative B Chromosome Evolution
4.6. Lesson from neo-Y Chromosomes in Pamphagidae Grasshoppers
4.7. Many Ways to the Bs
4.8. The Maintenance of Bs in Natural Populations
5. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilson, B.E. The supernumerary chromosomes of Hemiptera. Sci. N. Y. 1907, 26, 870–871. [Google Scholar]
- Jones, N. New species with B chromosomes discovered since 1980. Nucleus 2017, 60, 263–281. [Google Scholar] [CrossRef]
- D’Ambrosio, U.; Alonso-Lifante, M.P.; Barros, K.; Kovařík, A.; Mas de Xaxars, G.; Garcia, S. B-chrom: A database on B-chromosomes of plants, animals and fungi. New Phytol. 2017, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Palestis, B.G.; Cabrero, J.; Trivers, R.; Camacho, J.P.M. Prevalence of B chromosomes in Orthoptera is associated with shape and number of A chromosomes. Genetica 2010, 138, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.M.; Sharbel, T.F.; Beukeboom, L.W. B-chromosome evolution. Philos. Trans. R. Soc. B Biol. Sci. 2000, 355, 163–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, J.P.M. Chapter 4—B Chromosomes. In The Evolution of the Genome; Gregory, T., Ed.; Academic Press: Burlington, NJ, USA, 2005; pp. 223–286. ISBN 978-0-12-301463-4. [Google Scholar]
- Blagojević, J.; Vujošević, M. B chromosomes and developmental homeostasis in the yellow-necked mouse, Apodemus flavicollis (Rodentia, Mammalia): Effects on nonmetric traits. Heredity 2004. [Google Scholar] [CrossRef] [PubMed]
- Jojić, V.; Blagojević, J.; Vujošević, M. B chromosomes and cranial variability in yellow-necked field mice (Apodemus flavicollis). J. Mammal. 2011. [Google Scholar] [CrossRef]
- Hewitt, G.M. The integration of supernumerary chromosomes into the orthopteran genome. Cold Spring Harb. Symp. Quant. Biol. 1974, 38, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.M.; Schmid, M.; Cabrero, J. B chromosomes and sex in animals. Sex. Dev. 2011, 5, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Nat. Publ. Gr. 2016, 6, 128333. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Dias, G.B.; de Lima, L.G.; Kuhn, G.C.E.S.S.; Ramos, É.; Martins, C.; Cabral-de-Mello, D.C. High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis. Sci. Rep. 2017, 7, 6422. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; Castillo-Martínez, J.; Cabrero, J.; Gómez, R.; Camacho, J.P.M.; López-León, M.D. High-throughput analysis of satellite DNA in the grasshopper Pyrgomorpha conica reveals abundance of homologous and heterologous higher-order repeats. Chromosoma 2018, 127, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Cabrero, J.; Camacho, J.P.M. Cytogenetic studies in gomphocerine grasshoppers. I. Comparative analysis of chromosome C-banding pattern. Heredity 1986, 56, 365–372. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.L.; Arana, P.; Giráldez, R.; Girfildez, R.; Giráldez, R.; Girfildez, R. Chromosome C-banding patterns in Spanish Acridoidea. Genetica 1983, 61, 65–74. [Google Scholar] [CrossRef]
- Abdelaziz, M.; Teruel, M.; Chobanov, D.; Camacho, J.P.M.; Cabrero, J. Physical mapping of rDNA and satDNA in A and B chromosomes of the grasshopper Eyprepocnemis plorans from a Greek population. Cytogenet. Genome Res. 2007, 119, 143–146. [Google Scholar] [CrossRef] [PubMed]
- López-León, M.D.; Neves, N.; Schwarzacher, T.; Heslop-Harrison, J.S.; Hewitt, G.M.; Camacho, J.P.M. Possible origin of a B chromosome deduced from its DNA composition using double FISH technique. Chromosom. Res. 1994, 2, 87–92. [Google Scholar] [CrossRef]
- Teruel, M.; Ruíz-Ruano, F.J.; Marchal, J.A.; Sánchez, A.; Cabrero, J.; Camacho, J.P.M.; Perfectti, F. Disparate molecular evolution of two types of repetitive DNAs in the genome of the grasshopper Eyprepocnemis plorans. Heredity 2014, 112, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Makunin, A.I.; Kichigin, I.G.; Larkin, D.M.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Yang, F.; Proskuryakova, A.A.; Vorobieva, N.V.; Chernyaeva, E.N.; O’Brien, S.J.; et al. Contrasting origin of B chromosomes in two cervids (Siberian roe deer and grey brocket deer) unravelled by chromosome- specific DNA sequencing. BMC Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Makunin, A.I.; Rajičić, M.; Karamysheva, T.V.; Romanenko, S.A.; Druzhkova, A.S.; Blagojević, J.; Vujošević, M.; Rubtsov, N.B.; Graphodatsky, A.S.; Trifonov, V.A. Low-pass single-chromosome sequencing of human small supernumerary marker chromosomes (sSMCs) and Apodemus B chromosomes. Chromosoma 2018, 127, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Makunin, A.; Romanenko, S.; Beklemisheva, V.; Perelman, P.; Druzhkova, A.; Petrova, K.; Prokopov, D.; Chernyaeva, E.; Johnson, J.; Kukekova, A.; et al. Sequencing of Supernumerary Chromosomes of Red Fox and Raccoon Dog Confirms a Non-Random Gene Acquisition by B Chromosomes. Genes 2018, 9, 405. [Google Scholar] [CrossRef] [PubMed]
- Loreto, V.; Cabrero, J.; López-León, M.D.; Camacho, J.P.M.; Souza, M.J. Possible autosomal origin of macro B chromosomes in two grasshopper species. Chromosome Res. 2008, 16, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Darlington, C.D.; la Cour, L.F. The Handling of Chromosomes, 5th ed.; George Allen And Unwin Ltd.: London, UK, 1969; ISBN 0045750130. [Google Scholar]
- Bugrov, A.G.; Karamysheva, T.V.; Rubtsov, D.N.; Andreenkova, O.V. Comparative FISH analysis of distribution of B chromosome repetitive DNA in A and B chromosomes in two subspecies of Podisma sapporensis (Orthoptera, Acrididae). Cytogenet. Genome Res. 2004, 106, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Telenius, H.; Carter, N.; Bebb, C.E.; Nordensjold, M.; Ponder, B.A.; Tunnacliffe, A. Degenerate oligonucleotide-primed PCR: General amplification of targeted DNA by single degenerate primer. Genomics 1992, 13, 718–725. [Google Scholar] [CrossRef]
- Rubtsov, N.B.; Karamisheva, T.V.; Astakhova, N.M.; Liehr, T.; Claussen, U.; Zhdanova, N.S. Zoo-FISH with region-specific paints for mink chromosome 5q: Delineation of inter-and intrachromosomal rearrangements in human, pig, and fox. Cytogenet. Genome Res. 2000, 90, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Jetybayev, I.Y.; Bugrov, A.G.; Ünal, M.; Buleu, O.G.; Rubtsov, N.B. Molecular cytogenetic analysis reveals the existence of two independent neo-XY sex chromosome systems in Anatolian Pamphagidae grasshoppers. BMC Evol. Biol. 2017, 17, 20. [Google Scholar] [CrossRef] [PubMed]
- Jetybayev, I.E.; Bugrov, A.G.; Karamysheva, T.V.; Camacho, J.P.M.; Rubtsov, N.B. Chromosomal localization of ribosomal and telomeric DNA provides new insights on the evolution of Gomphocerinae grasshoppers. Cytogenet. Genome Res. 2012, 138, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Jetybayev, I.Y.; Bugrov, A.G.; Buleu, O.G.; Bogomolov, A.G.; Rubtsov, N.B. Origin and evolution of the neo-sex chromosomes in Pamphagidae grasshoppers through chromosome fusion and following heteromorphization. Genes 2017, 8, 323. [Google Scholar] [CrossRef] [PubMed]
- Warchałowska-Śliwa, E.; Bugrov, A.G.; Tatsuta, H.; Akimoto, S.I. B chromosomes, translocation between B and autosomes, and C-heterochromatin polymorphism of the grasshopper Podisma sapporensis Shir. (Orthoptera, Acrididae) in Hokkaido, northern Japan. Folia Biol. 2001, 49, 63–76. [Google Scholar]
- Bugrov, A.G.; Karamysheva, T.V.; Pyatkova, M.S.; Rubtsov, D.N.; Andreenkova, O.V.; Elzbieta, W.-S.; Rubtsov, N.B. B chromosomes of the Podisma sapporensis Shir. (Orthoptera, Acrididae) analysed by chromosome microdissection and FISH. Folia Biol. 2003, 51, 1–12. [Google Scholar]
- Dzyubenko, V.V.; Karagyan, G.A.; Çiplak, B.; Rubtsov, N.B.; Bugrov, A.G. The B chromosome polymorphism in Armenian and Turkey populations of the grasshopper Eyprepocnemis plorans plorans (Charpentier) (Orthoptera, Acrididae, Eyprepocnemidinae). Tsitologiya 2006, 48, 1016–1022. [Google Scholar]
- Liehr, T.; Claussen, U.; Starke, H. Small supernumerary marker chromosomes (sSMC) in humans. Cytogenet. Genome Res. 2004, 107, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Bugrov, A.G.; Karamysheva, T.V.; Perepelov, E.A.; Elisaphenko, E.A.; Rubtsov, D.N.; Warchałowska-Śliwa, E.E.; Tatsuta, H.; Rubtsov, N.B. DNA content of the B chromosomes in grasshopper Podisma kanoi Storozh. (Orthoptera, Acrididae). Chromosom. Res. 2007, 15, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Montiel, E.E.; Cabrero, J. Gypsy, RTE and Mariner transposable elements populate Eyprepocnemis plorans genome. Genetica 2012, 140, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Bueno, D.; Cabral-de-Mello, D.C. Chromosomal mapping of two Mariner-like elements in the grasshopper Abracris flavolineata (Orthoptera: Acrididae) reveals enrichment in euchromatin. Eur. J. Entomol. 2014, 111, 329–334. [Google Scholar] [CrossRef]
- López-León, M.D.; Cabrero, J.; Dzyubenko, V.V.; Bugrov, A.G.; Karamysheva, T.V.; Rubtsov, N.B.; Camacho, J.P.M. Differences in ribosomal DNA distribution on A and B chromosomes between eastern and western populations of the grasshopper Eyprepocnemis plorans plorans. Cytogenet. Genome Res. 2008, 121, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Estévez, M.; Badisco, L.; Broeck, J.V.; Perfectti, F.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. B chromosomes showing active ribosomal RNA genes contribute insignificant amounts of rRNA in the grasshopper Eyprepocnemis plorans. Mol. Genet. Genom. 2014, 289, 1209–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-León, M.D.; Cabrero, J.; Camacho, J.P.M. Unusually high amount of inactive ribosomal DNA in the grasshopper Stauroderus scalaris. Chromosom. Res. 1999, 7, 83–88. [Google Scholar] [CrossRef]
- Bugrov, A.G.; Jetybayev, I.E.; Karagyan, G.H.; Rubtsov, N.B. Sex chromosome diversity in Armenian toad grasshoppers (Orthoptera, Acridoidea, Pamphagidae). Comp. Cytogenet. 2016, 10, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Meštrović, N.; Mravinac, B. Centromere identity from the DNA point of view. Chromosoma 2014, 123, 313–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Ruano, F.J.; Cabrero, J.; López-León, M.D.; Sánchez, A.; Camacho, J.P.M. Quantitative sequence characterization for repetitive DNA content in the supernumerary chromosome of the migratory locust. Chromosoma 2018, 127, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Cabrero, J.; López-León, M.D.; Bakkali, M.; Camacho, J.P.M. Common origin of B chromosome variants in the grasshopper Eyprepocnemis plorans. Heredity 1999, 83, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T.; Buleu, O.; Karamysheva, T.; Bugrov, A.; Rubtsov, N. New Insights into Phasmatodea Chromosomes. Genes 2017, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T. Small Supernumerary Marker Chromosomes (sSMC); Springer: Berlin/Heidelberg, Germany, 2012; ISBN 978-3-642-20765-5. [Google Scholar]
- Eichler, E.E. Masquerading Repeats: Paralogous Pitfalls of the Human Genome. Genome Res. 1998, 8, 758–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichler, E.E.; Archidiacono, N.; Rocchi, M. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 1999, 9, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Horvath, J.E.; Bailey, J.A.; Locke, D.P.; Eichler, E.E. Lessons from the human genome: Transitions between euchromatin and heterochromatin. Hum. Mol. Genet. 2001, 10, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Dorer, D.R.; Henikoff, S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 1994, 77, 993–1002. [Google Scholar] [CrossRef]
- Torres, E.M.; Williams, B.R.; Amon, A. Aneuploidy: cells losing their balance. Genetics 2008, 179, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Zadesenets, K.S.; Vizoso, D.B.; Schlatter, A.; Konopatskaia, I.D.; Berezikov, E.; Schärer, L.; Rubtsov, N.B. Evidence for karyotype polymorphism in the free-living flatworm, Macrostomum lignano, a model organism for evolutionary and developmental biology. PLoS ONE 2016, 11, e0164915. [Google Scholar] [CrossRef] [PubMed]
- Zadesenets, K.S.; Schärer, L.; Rubtsov, N.B. New insights into the karyotype evolution of the free-living flatworm Macrostomum lignano (Platyhelminthes, Turbellaria). Sci. Rep. 2017, 7, 6066. [Google Scholar] [CrossRef] [PubMed]
- Zadesenets, K.; Ershov, N.; Berezikov, E.; Rubtsov, N.; Zadesenets, K.S.; Ershov, N.I.; Berezikov, E.; Rubtsov, N.B. Chromosome Evolution in the Free-Living Flatworms: First Evidence of Intrachromosomal Rearrangements in Karyotype Evolution of Macrostomum lignano (Platyhelminthes, Macrostomida). Genes 2017, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Hamid Al-Rikabi, A.B.; Pekova, S.; Fan, X.; Jancuskova, T.; Liehr, T. Small Supernumerary Marker Chromosome May Provide Information on Dosage-insensitive Pericentric Regions in Human. Curr. Genom. 2018, 19, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M.; John, B. Parallel Polymorphism for Supernumerary Segments in Chorthippus parallelus (ZETTERSTEDT) I. British Populations. Chromosoma 1968, 25, 319–342. [Google Scholar] [CrossRef] [PubMed]
- Gosálvez, J.; López-Fernández, C. Extra heterochromatin in natural populations of Gomphocerus Sibiricus (Orthoptera: Acrididae). Genetica 1981, 56, 197–204. [Google Scholar] [CrossRef]
- López-Fernández, C.; de la Vega García, C.; Gosálvez, J. Unstable B-Chromosomes in Gomphocerus Sibiricus (Orthoptera). Caryologia 1986, 39, 185–192. [Google Scholar] [CrossRef]
- Gusachenko, A.M.; Vysotskaya, L.V.; Bugrov, A.G. Cytogeneic analysis of Siberian Grasshopper (Orthoptera, Acrididae) from Altay. Dokl. Biol. Sci. 1993, 328, 250–252. [Google Scholar]
- Cella, D.M.; Ferreira, A. The Cytogenetics of Abracris flavolineata (Orthoptera, Caelifera, Ommatolampinae, Abracrini). Rev. Bras. Genet. 1991, 14, 315–329. [Google Scholar]
- Henriques-Gil, N.; Santos, J.L.; Arana, P. Evolution of a complex B-chromosome polymorphism in the grasshopper Eyprepocnemis plorans. Chromosoma 1984, 89, 290–293. [Google Scholar] [CrossRef]
- Bakkali, M.; Camacho, J.P.M. The B chromosome polymorphism of the grasshopper Eyprepocnemis plorans in North Africa: III. Mutation rate of B chromosomes. Heredity 2004, 92, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Camacho, J.P.M.; Carballo, A.R.; Cabrero, J. The B-chromosome system of the grasshopper Eyprepocnemis plorans subsp. plorans (Charpentier). Chromosoma 1980, 80, 163–176. [Google Scholar] [CrossRef]
- Gallagher, A.; Hewitt, G.; Gibson, I. Differential Giemsa staining of heterochromatic B-chromosomes in Myrmeleotettix maculatus (Thunb.) (Orthoptera: Acrididae). Chromosoma 1972, 40, 167–172. [Google Scholar] [CrossRef]
- Cabrero, J.; Viseras, E.; Camacho, J.P.M. The B-chromosomes of Locusta migratoria I. Detection of negative correlation between mean chiasma frequency and the rate of accumulation of the B’s; a reanalysis of the available data about the transmission of these B-chromosomes. Genetica 1984, 64, 155–164. [Google Scholar] [CrossRef]
- Bensasson, D.; Petrov, D.A.; Zhang, D.X.; Hartl, D.L.; Hewitt, G.M. Genomic gigantism: DNA loss is slow in mountain grasshoppers. Mol. Biol. Evol. 2001, 18, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.R. Animal Genome Size Database. Available online: http://www.genomesize.com (accessed on 18 October 2018).
- Li, X.-J.; Zhang, D.-C.; Wang, W.-Q.; Zheng, J.-Y. The chromosomal C-banding karyotypes of two pamphagid species from China. Chin. Bull. Entomol. 2008, 45, 549–553. [Google Scholar]
- Camacho, J.P.M.; Ruiz-Ruano, F.J.; Martín-Blázquez, R.; López-León, M.D.; Cabrero, J.; Lorite, P.; Cabral-de-Mello, D.C.; Bakkali, M. A step to the gigantic genome of the desert locust: Chromosome sizes and repeated DNAs. Chromosoma 2015, 124, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Jetybayev, I.; Karamysheva, T.; Bugrov, A.; Rubtsov, N. Cross-hybridization of repetitive sequences from the pericentric heterochromatic region of Chorthippus apricarius (L.) with the chromosomes of grasshoppers from the Gomphocerini tribe. Eurasian Entomol. J. 2010, 9, 433–436. [Google Scholar]
- Eckert, W.A.; Plass, C.; Weith, A.; Traut, W.; Winking, H. Transcripts from amplified sequences of an inherited homogeneously staining region in chromosome 1 of the house mouse (Mus musculus). Mol. Cell. Biol. 1991, 11, 2229–2235. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Domínguez, B.; Ruiz-Ruano, F.J.; Cabrero, J.; Corral, J.M.; López-León, M.D.; Sharbel, T.F.; Camacho, J.P.M. Protein-coding genes in B chromosomes of the grasshopper Eyprepocnemis plorans. Sci. Rep. 2017, 7, 45200. [Google Scholar] [CrossRef] [PubMed]
- Yudkin, D.V.; Trifonov, V.A.; Kukekova, A.V.; Vorobieva, N.V.; Rubtsova, N.V.; Yang, F.; Acland, G.M.; Ferguson-Smith, M.A.; Graphodatsky, A.S. Mapping of KIT adjacent sequences on canid autosomes and B chromosomes. Cytogenet. Genome Res. 2007, 116, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Kichigin, I.G.; Lisachov, A.P.; Giovannotti, M.; Makunin, A.I.; Kabilov, M.R.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Graphodatsky, A.S.; Trifonov, V.A. First report on B chromosome content in a reptilian species: The case of Anolis carolinensis. Mol. Genet. Genom. 2018. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Conte, M.A.; Fantinatti, B.E.A.A.; Cabral-De-Mello, D.C.; Carvalho, R.F.; Vicari, M.R.; Kocher, T.D.; Martins, C. Origin and evolution of B chromosomes in the cichlid fish Astatotilapia latifasciata based on integrated genomic analyses. Mol. Biol. Evol. 2014, 31, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.E.; Mitchell, S.E.; O’Neill, R.J. Pericentric and centromeric transcription: A perfect balance required. Chromosom. Res. 2012, 20, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Simboeck, E.; Déjardin, J. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 2015, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bugrov, A.G.; Grozeva, S. Neo-XY chromosome sex determination in four species of the pamphagid grasshoppers (Orthoptera, Acridoidea, Pamphagidae) from Bulgaria. Caryologia 1998, 51, 115–121. [Google Scholar] [CrossRef] [Green Version]
- Bugrov, A.G.; Warchałowska-Śliwa, E. Chromosome numbers and C-banding patterns in some Pamphagidae grasshoppers (Orthoptera, Acridoidea) from the Caucasus, Central Asia, and Transbaikalia. Folia Biol. 1997, 45, 133–138. [Google Scholar]
- Bugrov, A.G.; Buleu, O.G.; Jetybayev, I.E. Chromosome polymorphism in populations of Asiotmethis heptapotamicus (Zub.) (Pamphagidae, Thrinchinae) from Kazakhstan. Eurasian Entomol. J. 2016, 15, 545–549. [Google Scholar]
- Teruel, M.; Cabrero, J.; Perfectti, F.; Camacho, J.P.M. B chromosome ancestry revealed by histone genes in the migratory locust. Chromosoma 2010, 119, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Rubtzov, N.B.; Borissov, Y.M.; Karamysheva, T.V.; Bochkarev, M.N.; Rubtsov, N.B.; Borosov, Y.M.; Karamysheva, T.V.; Bochkarev, M.N.N. The mechanisms of formation and evolution of B chromosomes in Korean field mice Apodemus peninsulae (Mammalia, Rodentia). Russ. J. Genet. 2009, 45, 389–396. [Google Scholar] [CrossRef]
- Rubtsov, N.B.; Kartavtseva, I.V.; Roslik, G.V.; Karamysheva, T.V.; Pavlenko, M.V.; Iwasa, M.A.; Koh, H.S. Features of the B chromosome in Korean field mouse Apodemus peninsulae (Thomas, 1906) from Transbaikalia and the Far East identified by the FISH method. Russ. J. Genet. 2015, 51, 278–288. [Google Scholar] [CrossRef]









| Taxa | Species Name | Location | Year | N |
|---|---|---|---|---|
| Acrididae | ||||
| Gomphocerinae | Chorthippus apricarius (L.) | Foothills of Trans-Ili Alatau mountains near Almaty, Kazakhstan | 2005 | 3 ʘ from 2♀ |
| Chorthippus jacobsoni (Ikonn.) | Foothills of Trans-Ili Alatau mountains near Almaty, Kazakhstan | 2007 | 7 ʘ from 3♀ | |
| Aeropus sibiricus (L.) | Kurai steppe, Altay mountains, Russian Federation | 2007 | 6♂ | |
| Oedipodinae | Bryodema gebleri (F-W.) | Kurai steppe, Altay mountains, Russian Federation | 2017 | 12♂ |
| Melanoplinae | Podisma sapporensis sapporensis Shir. | Hokkaido, Japan | 2000, 2005 | 157♂, 37 ʘ |
| Eyprepocnemidinae | Eyprepocnemis plorans plorans (Charp.) | Megri and Yerevan, Armenia, Antalia, Turkey, Malaga, Spain (kindly provided by JPM Camacho) | 2003, 2007 | 381♂ 551♂ |
| Eyprepocnemis plorans meridionalis Uv. | Springbok, South Africa | 2004 | 33♂ | |
| Pamphagidae | ||||
| Pamphaginae | Nocaracris tardus Ünal, Bugrov & Jetybayev | Sultandag mountains, Turkey | 2014 | 7♂ |
| Nocaracris cyanipes (F-W.) | Prielbrusie National park Kabardino-Balkaria, Russia | 1987 | 5♂ | |
| Trinchinae | Asiotmethis heptapotamicus songoricus Shum. | Ayagoz, East Kazakhstan | 2015 | 18♂ |
| DNA probe | Species | Dissected Chromosome/Region |
|---|---|---|
| WCPNtaB | Nocaracris tardus | Whole B |
| PCPCapBq | Chorthippus apricarius | Long arm of the B |
| PCPCapAc | Chorthippus apricarius | Pericentric region of autosome |
| WCPEplBa4 | Eyprepocnemis plorans | Whole Ba4 |
| PCPPsaB1-B2dist | Podisma sapporensis | Distal parts of B1 and B2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jetybayev, I.Y.; Bugrov, A.G.; Dzuybenko, V.V.; Rubtsov, N.B. B Chromosomes in Grasshoppers: Different Origins and Pathways to the Modern Bs. Genes 2018, 9, 509. https://doi.org/10.3390/genes9100509
Jetybayev IY, Bugrov AG, Dzuybenko VV, Rubtsov NB. B Chromosomes in Grasshoppers: Different Origins and Pathways to the Modern Bs. Genes. 2018; 9(10):509. https://doi.org/10.3390/genes9100509
Chicago/Turabian StyleJetybayev, Ilyas Yerkinovich, Alexander Gennadievich Bugrov, Victoria Vladimirovna Dzuybenko, and Nikolay Borisovich Rubtsov. 2018. "B Chromosomes in Grasshoppers: Different Origins and Pathways to the Modern Bs" Genes 9, no. 10: 509. https://doi.org/10.3390/genes9100509
APA StyleJetybayev, I. Y., Bugrov, A. G., Dzuybenko, V. V., & Rubtsov, N. B. (2018). B Chromosomes in Grasshoppers: Different Origins and Pathways to the Modern Bs. Genes, 9(10), 509. https://doi.org/10.3390/genes9100509





