Abstract
Copper (Cu) is involved in fundamental biological processes for plant growth and development. However, Cu excess is harmful to plants. Thus, Cu in plant tissues must be tightly regulated. In this study, we found that the peanut Yellow Stripe-Like family gene AhYSL3.1 is involved in Cu transport. Among five AhYSL genes, AhYSL3.1 and AhYSL3.2 were upregulated by Cu deficiency in peanut roots and expressed mainly in young leaves. A yeast complementation assay suggested that the plasma membrane-localized AhYSL3.1 was a Cu-nicotianamine complex transporter. High expression of AhYSL3.1 in tobacco and rice plants with excess Cu resulted in a low concentration of Cu in young leaves. These transgenic plants were resistant to excess Cu. The above results suggest that AhYSL3.1 is responsible for the internal transport of Cu in peanut.
1. Introduction
Copper (Cu) is an essential micronutrient that plays key roles in multiple plant processes, including photosynthesis, respiration, carbon and nitrogen metabolism, oxidative stress, cell wall synthesis, and hormone perception [1,2]. Cu deficiency in plants often leads to stunted growth, chlorosis in young leaves, and defects in development [3]. In contrast, excess Cu can also be toxic to plants due to the production of highly toxic hydroxyl radicals by redox cycling between Cu(I) and Cu(II). Cu toxicity usually results in reduction of plant biomass, inhibition of root growth, chlorosis, bronzing, and necrosis [4]. Moreover, excess Cu in the environment exerts harmful effects on all living organisms [5]. Therefore, Cu homeostasis must be tightly regulated in plants, and indeed all organisms [6].
For maintenance of appropriate Cu levels in plant tissues, a sophisticated homeostatic network is controlled by various types of transporter proteins. The conserved Cu transporter (CTR) family proteins or CTR-like transporters (COPT) have been well characterized. In Arabidopsis (Arabidopsis thaliana), six CTR family members have been identified [7]. Among them, COPT1 and COPT2 restored the growth of a mutant yeast strain, while COPT3 and COPT5 partially rescued the defective growth of mutant yeast, as determined by a functional complementation assay [8]. The high-affinity copper transporter COPT1 plays an important role in Cu uptake and pollen development [9,10]. However, COPT2 participates in the cross-talk between iron-deficiency responses and low-phosphate signaling [11]. The tonoplast-localized COPT5 functions in inter-organ reallocation of copper ions from the root to reproductive organs [3] and is required for photosynthetic electron transport under Cu scarcity [12]. The plasma membrane transporter AtCOPT6 is responsible for Cu distribution and homeostasis during Cu limitation and excess [13,14]. The rice (Oryza sativa) CTR family consists of seven members. It was suggested that rice COPTs function alone or cooperatively to mediate Cu transport in various tissues [13,15]. OsCOPT expression is affected at the transcriptional level by multiple factors, including the levels of Cu and other bivalent cations [13].
The P-type heavy-metal ATPase (HMA) family proteins have also been implicated in the transport of Cu in plants. Arabidopsis HMA1, HMA6/PAA1, and HMA8/PAA2 are required for Cu delivery in chloroplasts [16,17,18,19]. Moreover, AtHMA5 is involved in Cu detoxification and tolerance in Arabidopsis [20,21]. In rice, the transcript level of the Cu+ transporter OsHMA5 was upregulated by excess Cu, but not by deprivation of Cu or other metals. Mutation of OsHMA5 resulted in a decreased Cu concentration in the xylem sap and shoots, but increased Cu concentration in the roots, suggesting its role in Cu loading of xylem in roots [22]. In contrast, the OsHMA4 was proved to function to sequester Cu into root vacuoles [23]. The expression of OsHMA9 was induced by a high concentration of Cu, Zn, and Cd, and it plays a role in the efflux of these metals from cells [24]. It was reported that the cucumber HMA5-like ATPases may contribute to vacuolar sequestration of Cu in vacuoles under Cu excess [25].
Nicotianamine (NA) is a key compound for metal transport [26,27]. It was reported that NA is required for delivery of Cu in the xylem [28,29]. The Yellow Stripe-Like (YSL) family proteins have been found to participate in Cu–NA transport. Arabidopsis YSL2 (AtYSL2) is suggested to transport both Fe(II)–NA and Cu–NA [30], although another report argues against transport of these substrates [31]. The AtYSL2 transcriptional level is regulated by Fe and Cu [30]. Importantly, the Arabidopsis small ubiquitin-like modifier (SUMO) E3 ligase SIZ1, which regulates the expression of AtYSL1 and AtYSL3, functions in excess Cu tolerance in plants [32]. Additionally, rice YSL16 is involved in the delivery of Cu–NA to developing young tissues and seeds through phloem transport. The expression of OsYSL16 was upregulated by Zn and Fe deprivation, but not by Mn and Cu deficiency, in the roots. It was demonstrated by yeast expression assay that OsYSL16 transports Cu–NA, but not ionic Cu, and the Cu-deoxymugineic acid (DMA) complex. Knockout of OsYSL16 resulted in a significant reduction in Cu–NA translocation from older to younger leaves and from the flag leaf to the panicle [33]. The osysl16 mutant showed low pollen germination which could be rescued by addition of Cu [34]. In addition to Cu translocation, OsYSL16 is also involved in Fe allocation in rice plants [35,36] by transporting Fe(III)-DMA, as indicated by a yeast complementation assay [36]. DMA is an Fe(III)-chelating molecule biosynthesized in graminaceous plants, which functions in Fe uptake from the soil and Fe translocation within the plant [37]. In our previous study, we identified five peanut YSL genes. Among these, the expression of only AhYSL1 was induced by Fe deficiency in the roots. Yeast functional complementation indicated that AhYSL3.1 transports not only Fe(III)–DMA, but also Fe(II)–NA, while AhYSL1 specifically transports Fe(III)–DMA. It was implied that AhYSL1 functions in Fe acquisition in the peanut maize intercropping system [38]. However, the functions of other peanut YSL genes remain unknown. In the present study, the peanut AhYSL3.1 gene was characterized. AhYSL3.1 is suggested to be responsible for internal transport of Cu in peanut.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Peanut (Arachis hypogaea L. cv. Luhua 14) plants were grown in 5 L boxes with the following nutrient solution: 0.7 mM K2SO4, 0.1 mM KCl, 0.1 mM KH2PO4, 2.0 mM Ca(NO3)2, 0.5 mM MgSO4, 10 μM H3BO3, 0.5 μM MnSO4, 0.5 μM ZnSO4, 0.2 μM CuSO4, 0.01 μM (NH4)6Mo7O24, and 100 μM Fe(III)-EDTA. For metal treatments, the levels of the corresponding metals in the above nutrient solution were modified. Peanut plants were cultured in a greenhouse with aeration and 30 °C light/25 °C dark cycles under natural light conditions.
2.2. Quantitative Real-Time PCR
For analysis of AhYSL gene expression in response to metal treatments, roots or leaves of peanut plants subjected to Cu deficiency (–Cu), Mn deficiency (–Mn), Zn deficiency (–Zn), or excess Cu (25 μM Cu) in hydroponics for 5 days were harvested. For analysis of tissue expression of AhYSL genes, parts of roots, stems, young leaves, and old leaves from 7-day-old peanut plants were sampled.
The samples were subjected to total RNA extraction and then treated with RNase-free DNase I (Takara, Tokyo, Japan) to remove genomic DNA contamination [39]. First-strand complementary DNA (cDNA) was synthesized using ReverTra Ace reverse transcriptase (Toyobo, Tokyo, Japan) by priming with the d(T)17-adaptor primer. Quantitative real-time PCR was performed using the StepOnePlus™ real-time PCR system (Applied Biosystems, Foster City, CA, USA) and SYBR Premix Ex Taq (Perfect Real Time) reagent (Takara, Tokyo, Japan) using primers reported previously [38]. The PCR products were confirmed by DNA sequencing (3130 Genetic Analyzer, Applied Biosystems, Tokyo, Japan). The peanut ubiquitin gene [40] was used as an internal control.
2.3. Subcellular Localization of AhYSL3.1
The AhYSL3.1 open reading frame (ORF) without a stop codon and containing XbaI and BamHI restriction enzyme sites was amplified by PCR using the primers 5′-TCTAGAATGAATCCAATGGAAATCAAT-3′ and 5′-GGATCCCTTGGATGTAAAGAAGCT-3′. The PCR product was then subcloned into the CaMV35S-sGFP(S65T)-NOS3’ vector (kindly provided by Dr. Yasuo Niwa, University of Shizuoka, Japan) to generate the AhYSL3.1-sGFP construct. The AhYSL3.1-sGFP construct or the control CaMV35S-sGFP, together with 35S-DsRed (Clontech, Mountain View, CA, USA), was then bombarded into onion epidermal cells by DNA particle bombardment as described by Mizuno et al. [41]. After incubation in the dark for 16 h, fluorescent cells were imaged by confocal microscopy (LSM5Pascal; Carl Zeiss, Göttingen, Germany).
2.4. In Situ Hybridization
In situ hybridization was performed as described previously [39]. Peanut roots grown hydroponically were fixed in formalin-acetic acid-alcohol and embedded in paraffin. The root tissues were sectioned into 10 μm slices and mounted on slides. The 3′UTR fragment of AhYSL3.1 was amplified using the primers: AhYSL3.1probe-F, 5′-TTTGCGATAGCAGCCAACTTGGTGAG-3′ and AhYSL3.1probe-R, 5′-AATTGTAGTTGCAAACTAGATACACTGATC-3′. After subcloning into the pCR®-Blunt II-TOPO® vector (Invitrogen, Carlsbad, CA, USA), the plasmid was linearized using BamHI. T7 RNA polymerase was used to generate sense and antisense probes, and the generated probes were labeled with digoxigenin-11-UTP (Roche, Mannheim, Germany). The slides were pretreated with proteinase K and subjected to acetylation and then hybridized for 16 h with the sense or antisense probe at 50 °C. After washing, the sections were incubated with anti-digoxigenin alkaline phosphatase conjugate (Roche) and stained with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche).
2.5. Functional Complementation in Yeast
The AhYSL3.1 ORF with NotI and BamHI restriction sites was amplified by PCR using the primers: YAhYSL3.1-F, 5′-TGCGGCCGCATGAATCCAATGGAAATC-3′; YAhYSL3.1-R, 5′-CCGCGGATCCCTACTTGGATGTAAAGAAGCT-3′. The PCR product was then subcloned into the pCR®-Blunt II-TOPO® vector (Invitrogen) and confirmed by sequencing. After digestion, the construct was introduced into the yeast expression vector pDR195 [31] (kindly provided by Dr. Nicolaus von Wirén, University of Hohenheim, Germany) to generate the AhYSL3.1-pDR195 construct. The Cu-deficient yeast (Saccharomyces cerevisiae) mutant M10 was used for complementation assay. This CTR1 mutant was kindly provided by Dr. Andrew Dancis (National Institutes of Health, Maryland, USA). The construct AhYSL3.1-pDR195 or empty vector pDR195 was transformed into the yeast strain M10 by the LiAc/SS-DNA/PEG method [42]. Serial dilutions of the transformed yeast cells of OD600 1 to 0.001 were plated onto Synthetic Defined (SD) medium containing 9 μM Cu–NA or 10 μM CuSO4.
2.6. Generation of Transgenic Lines
The full-length AhYSL3.1 ORF with XbaI and SacI restriction sites was amplified using the following primers: 5′-TCTAGAATGAATCCAATGGAAATCAA-3′; 5′-GAGCTCCTACTTGGATGTAAAGAAGC-3’. The GUS gene in the E-90Ω plasmid [43] was replaced with the AhYSL3.1 ORF. As shown in Supplementary Figure S2B, the resultant plasmid drives expression of AhYSL3.1 under the control of the Fe deficiency-responsive cis-acting elements IDE1 and IDE2 connected to the −90/+8 region of the cauliflower mosaic virus 35S promoter, which drives preferential expression in vascular tissues [43,44], and the tobacco mosaic virus 5′ leader sequence for expression enhancement in the backbone of the pIG121Hm binary vector [45]. After introduction into Agrobacterium tumefaciens strain C58 by electroporation, the Agrobacterium carrying the above construction was transformed into tobacco (Nicotiana tabacum L. cv. Petit-Havana SR1) and rice (Oryza sativa L. cv. Tsukinohikari). The tobacco transformation was performed according to the procedure described by Helmer et al. [46] and the rice transformation by that of Hiei et al. [45].
2.7. Analyses of Transgenic Tobacco and Rice Plants
The transgenic lines were germinated on Murashige and Skoog (MS) medium containing hygromycin B (50 mgL−1). Non-transgenic seeds were germinated on MS medium lacking hygromycin B as a control. After 2–3 weeks of growth, the plants were transferred to the previously described nutrient solution [39] under a 14 h light/10 h dark cycle with 28/24 °C day/night temperatures and a light intensity of 300 μmol m−2 s−1 provided by reflector sunlight dysprosium lamps and 60% relative humidity. For tobacco, eighteen transgenic lines were generated. Two lines with higher expression of AhYSL3.1, T10 and T11, were selected for further analysis. Four replicates of the transgenic and non-transgenic tobacco plants were grown hydroponically for ~1 week and then treated without Cu and with 0.2, 25, or 50 μM Cu for 6 days. Roots, stems, young leaves, and old leaves were divided and harvested for metal content determination. The rice plants were cultured in normal nutrient solution for ~1 week and then treated with 100 μM Cu for 6 days. The young leaves, old leaves, and roots were then sampled. The above samples were dried for 2–3 days at 70 °C, and ~100 mg portions were then digested with HNO3-H2O2 in a microwave accelerated reaction system (CEM, Matthews, NC, USA). Metal concentrations were measured by inductively coupled plasma atomic emission spectroscopy (ICP-AES, OPTIMA 3300 DV, Perkin-Elmer, Norwalk, CT, USA).
2.8. Transmembrane Protein Structure Prediction and Statistical Analysis
Combined membrane protein topology and signal peptide prediction was conducted by the TOPCONS web server [47]. All data were analyzed using SPSS 23.0 software (SPSS Inc., Chicago, IL, USA). All of the means were tested using one-way analysis of variance at the 5% and 1% probability level.
3. Results
3.1. Expression of AhYSL Genes in Response to Metal Treatments
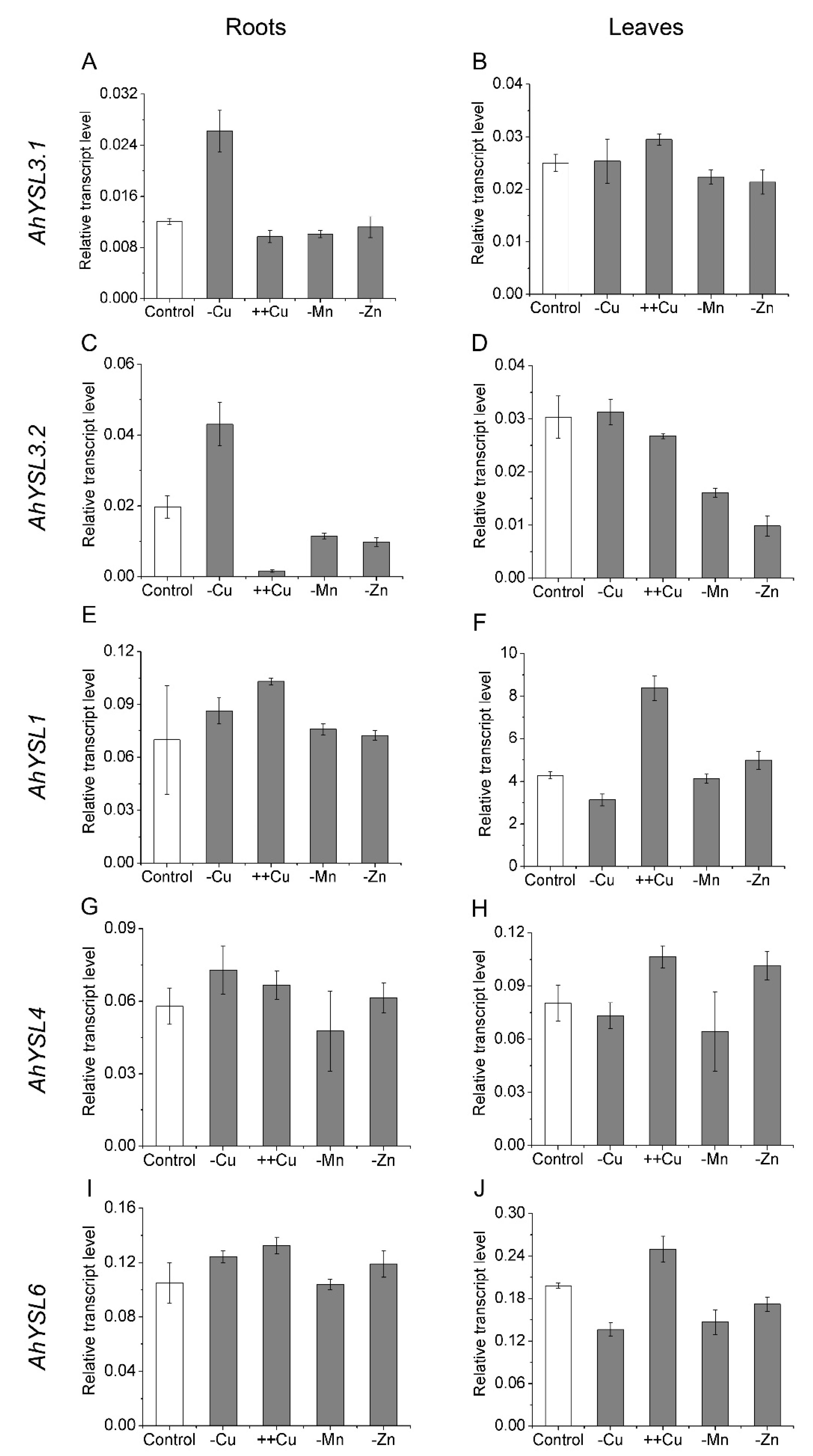
In a previous study, five AhYSL genes were identified in peanut, and their response to Fe deficiency was analyzed [38]. To determine whether these YSL genes function in transport of other metals, their expression in response to other metal treatments was analyzed. Among them, AhYSL3.1 and AhYSL3.2 were induced more than twofold in Cu-deficient roots, but not in Mn- and Zn-deficient roots, as shown in Figure 1A,C. AhYSL3.1 mRNA levels were slightly increased by excess Cu, but were decreased by Mn and Zn deficiency in leaves, as shown in Figure 1B. In contrast, AhYSL3.2 expression was suppressed by excess Cu in roots, as shown in Figure 1C, and Mn and Zn deprivation in roots and leaves, as shown in Figure 1C,D. In roots, AhYSL1, AhYSL4, and AhYSL6 transcript abundances were changed slightly by treatment with various metals, as shown in Figure 1E,G,I. However, in leaves, they were significantly enhanced by excess Cu, as shown in Figure 1F,H,J. Moreover, AhYSL4 expression was markedly induced by Zn deficiency, and that of AhYSL6 was suppressed by Cu, Mn, and Zn deficiency in leaves, as shown in Figure 1H,J.
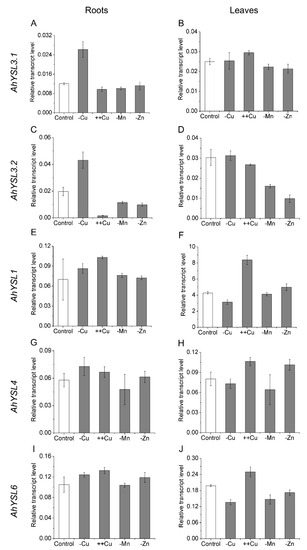
Figure 1.
Expression pattern of AhYSL genes in response to various metal treatments. Peanut plants were cultured in normal nutrient solution (control) or treated with Cu deficiency (–Cu), excess Cu (++Cu, 25 μM Cu), Mn deficiency (–Mn), or Zn deficiency (–Zn) in hydroponics for 5 days. (A,B) AhYSL3.1. (C,D) AhYSL3.2. (E,F) AhYSL1. (G,H) AhYSL4. (I,J) AhYSL6. Roots (A,C,E,G,I) and leaves (B,D,F,H,J) of peanut were sampled using five biological replicates. Vertical bars indicate expression levels relative to the control, ubiquitin. Error bars indicate standard deviation.
3.2. Tissue Expression and Localization of AhYSL3.1
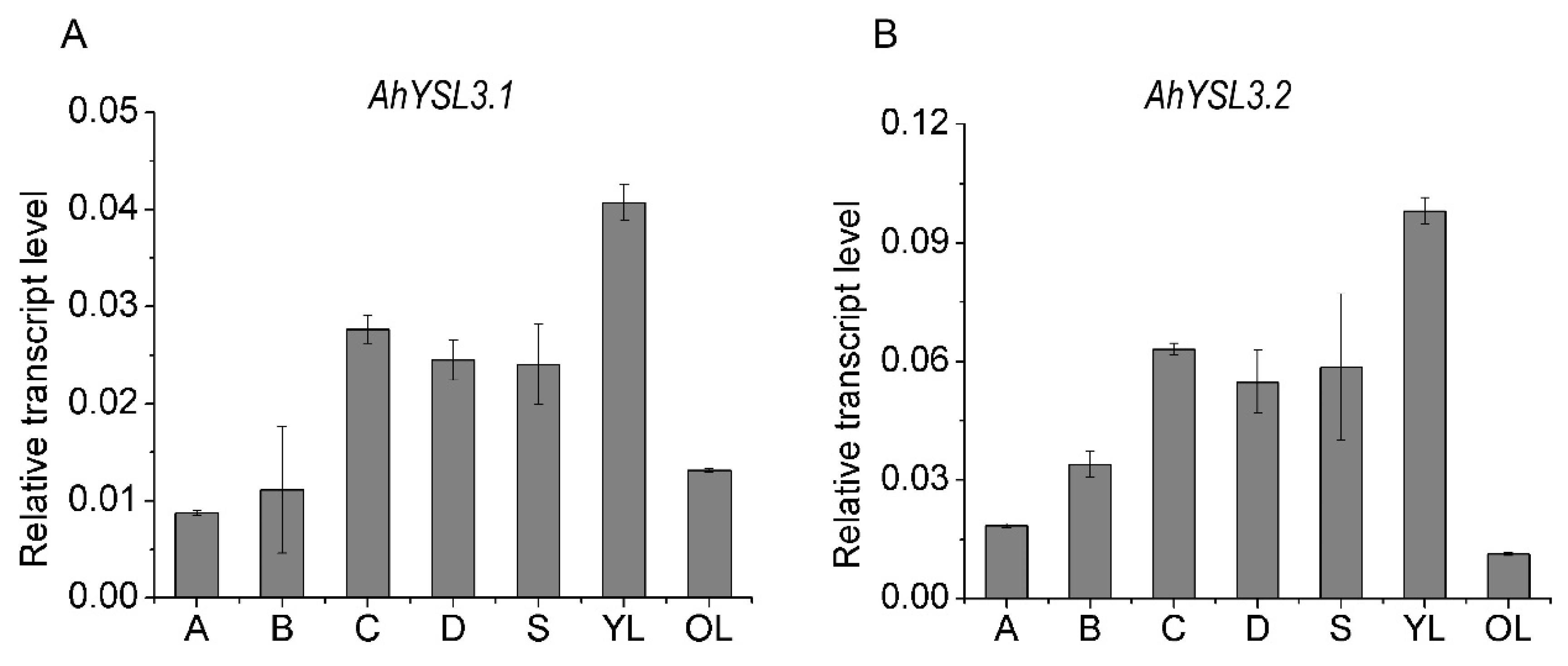
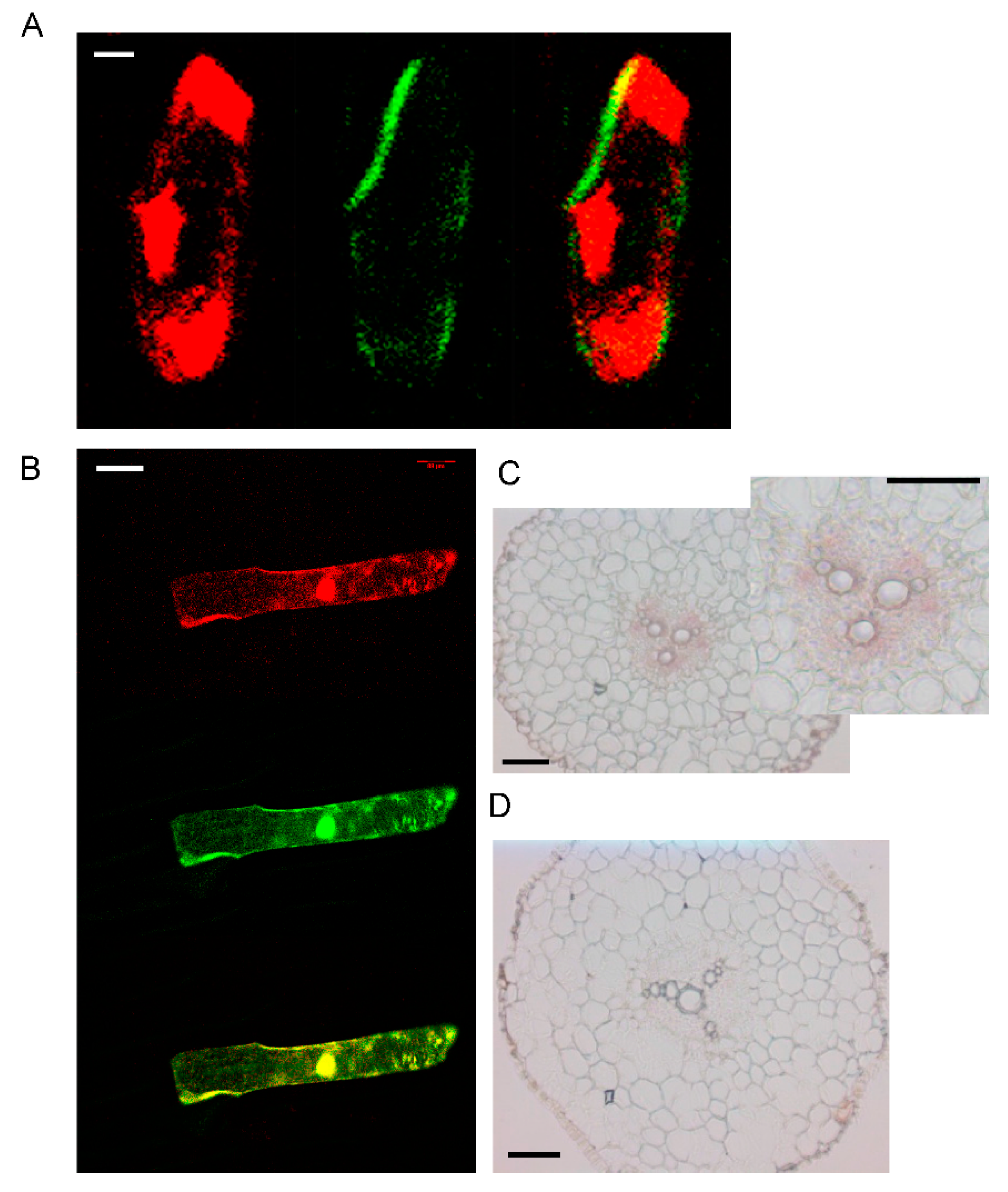
The transcript levels of AhYSL3.1 and AhYSL3.2 in roots, stems, young and old leaves were determined. Both genes showed similar expression patterns in these tissues, as shown in Figure 2. In roots, AhYSL3.1 as shown in Figure 2A and AhYSL3.2 as shown in Figure 2B were expressed in the lateral or main roots, but not the root tips. Among roots, stems, young and old leaves, the expression levels of both genes were highest in young leaves. The tissue localization of AhYSL3.1 was also checked by in situ hybridization. Regarding tissue localization in the root, AhYSL3.1 antisense probe staining was more visible around the vascular bundles, as shown in Figure 3C, compared to the sense probe, which was used as a negative control, as shown in Figure 3D.
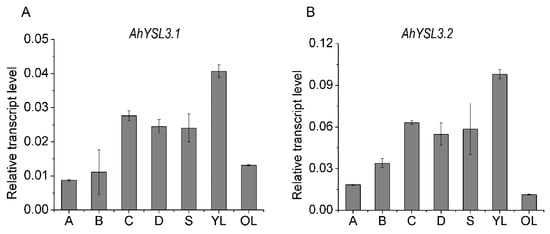
Figure 2.
AhYSL3.1 (A) and AhYSL3.2 (B) transcript levels in the indicated parts of peanut plants. The following parts of peanut plants grown under normal conditions for 7 days were harvested: 0–2 cm root tips (A from horizontal abscissa), 2–4 cm root tips (B from horizontal abscissa), lateral roots except the above root tips (C), the main roots (D), stems (S), young leaves (YL), and old leaves (OL). Vertical bars indicate relative expression levels relative to the control, ubiquitin. Three biological replicates were performed. Error bars represent standard deviation.
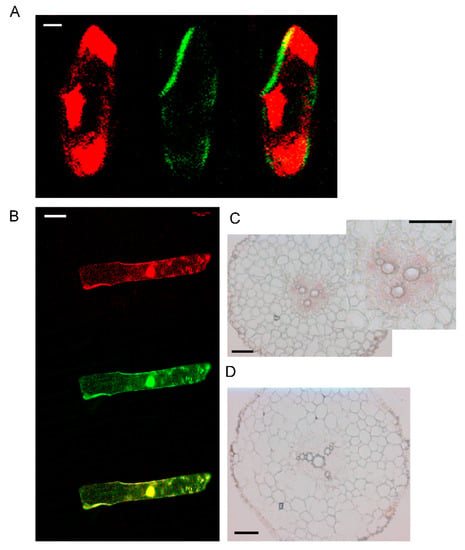
Figure 3.
Subcellular and tissue localization of AhYSL3.1. (A) 35S-DsRed, as a marker of the cytosol and nucleus, was cointroduced into onion epidermal cells with the AhYSL3.1 ORF fused with GFP. Red signals (left panel) are from 35S-DsRed. Green signals (middle panel) are from AhYSL3.1-GFP. Right panel shows a merged image. Scale bars represent 20 μm. (B) GFP alone was colocalized with 35S-DsRed. Scale bars represent 100 μm. (C,D) In situ hybridization analysis of AhYSL3.1 in peanut roots. AhYSL3.1 antisense (C) and sense (D) probes were hybridized in a cross section of peanut roots. Bars = 100 μm.
3.3. The Subcellular Localization of AhYSL3.1
By the TOPCONS web server, the transmembrane helix positions of AhYSL3.1 were predicted based on different methods. According to the consensus-based methods of TOPCONS, 16 transmembrane domains were found in AhYSL3.1, as shown in Supplementary Figure S1. The subcellular localization of AhYSL3.1 was determined by transient expression assay in onion. For subcellular localization, the AhYSL3.1 open reading frame fused with green fluorescent protein (GFP) was transiently introduced into onion epidermal cells together with 35S-DsRed as a marker of the cytosol and nucleus. AhYSL3.1-GFP fluorescence was observed in the outer region of the cell, but not the cytosol or nucleus, as shown in Figure 3A. In contrast, GFP fluorescence was colocalized with the red signal of 35S-DsRed, as shown in Figure 3B.
3.4. AhYSL3.1 is a Cu–NA Transporter
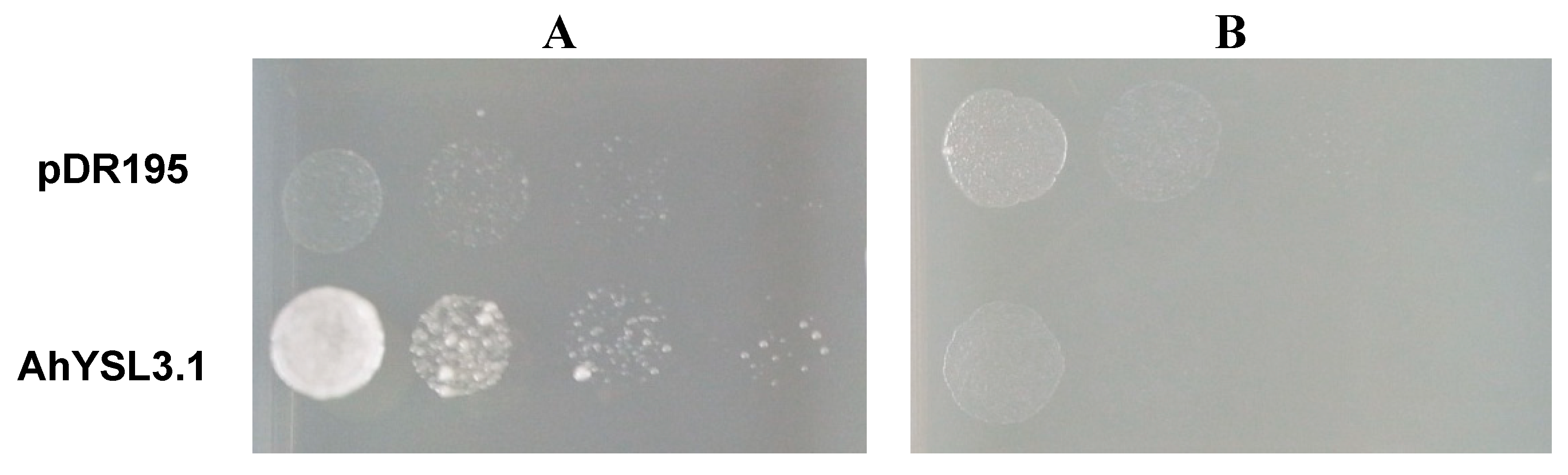
Because AhYSL3.1 was markedly induced by Cu deficiency, but not by other metals in roots, we speculated that AhYSL3.1 transports Cu in addition to the previously identified substrates Fe(III)–DMA and Fe(II)–NA [38]. Therefore, a yeast complementation assay was performed using a Copper Transporter 1 (CTR1) yeast mutant (M10) [48], which is defective in Cu uptake. The ORF of AhYSL3.1 was subcloned into the yeast expression vector pDR195. When supplied with Cu–NA, expression of AhYSL3.1 in the mutant significantly improved the growth of the yeast compared with control cells transformed with empty vector, as shown in Figure 4A, suggesting that peanut AhYSL3.1 is a Cu–NA transporter. In contrast, when supplied with CuSO4, the AhYSL3.1-expressing yeast mutant did not grow well, as shown in Figure 4B, indicating that AhYSL3.1 does not transport ionic Cu2+.
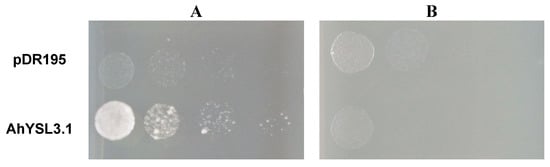
Figure 4.
Functional complementation of AhYSL3.1 in yeast. The yeast strain M10 was transformed with the AhYSL3.1-pDR195 construct or the empty pDR195 vector, which was used as a negative control. Serial dilutions of the transformed yeast cells of OD600 1 to 0.001 were plated onto Synthetic Defined (SD) medium containing 9 μM Cu–NA (A) or 10 μM CuSO4 (B).
3.5. High-Level Expression of AhYSL3.1 in Tobacco Results in Tolerance to Excess Cu
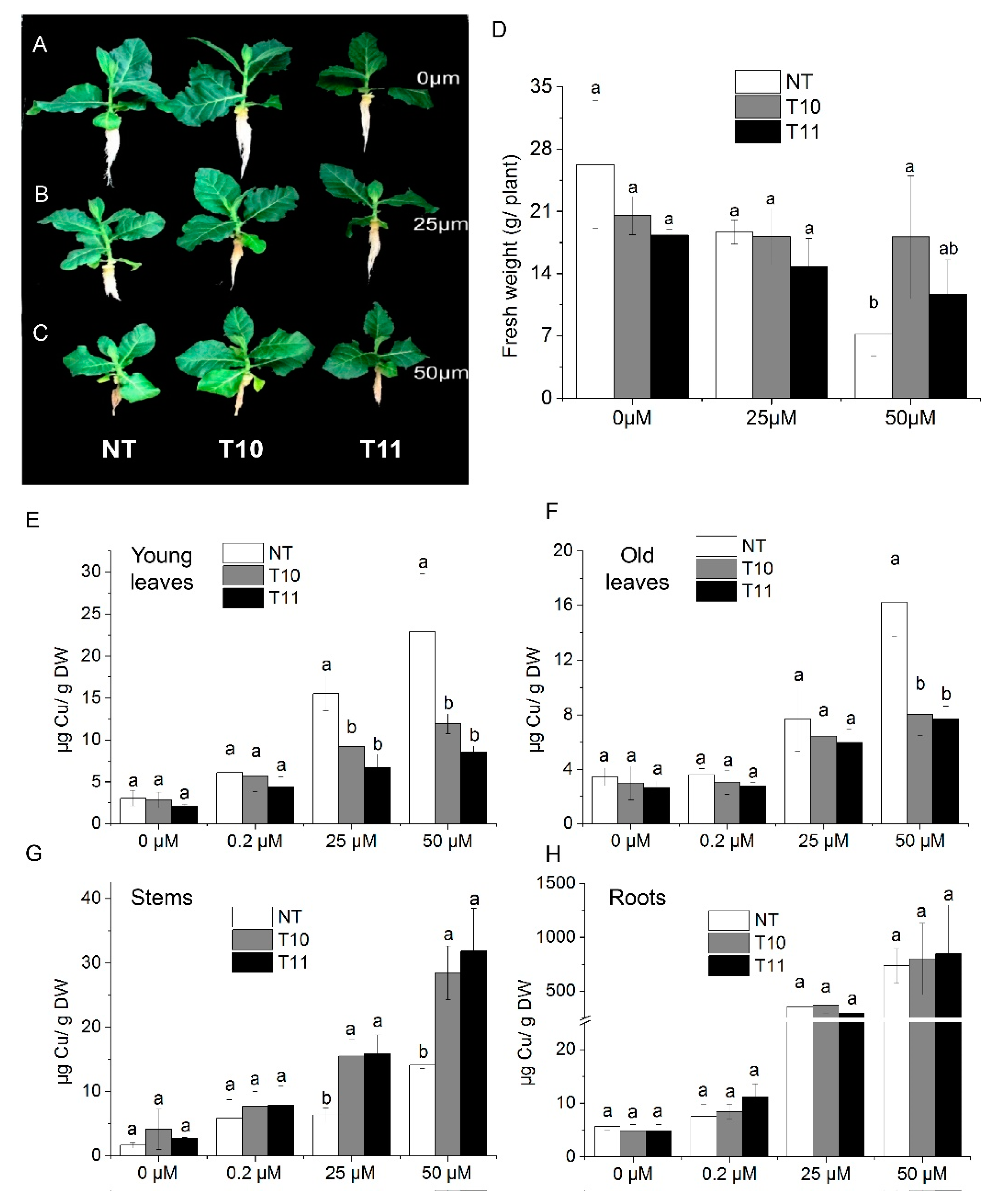
AhYSL3.1 was introduced into tobacco plants and its function investigated. Eighteen transgenic lines were generated. Two lines (T10 and T11) with higher AhYSL3.1 expression were used for further analysis, as shown in Supplementary Figure S2B. Non-transformed (NT) tobacco plants and the transgenic lines T10 and T11 were treated with various concentrations of Cu. After treatment without Cu or with 25 μM Cu, the fresh weights of NT and transgenic plants showed no difference, as shown in Figure 5A,B,D. When treated with 50 μM Cu, the fresh weight of one line (T10) was significantly higher than that of NT plants, as shown in Figure 5C,D. Moreover, the Cu concentration in young leaves of transgenic lines was markedly lower than in NT plants when treated with 25 or 50 μM excess Cu, as shown in Figure 5E. In old leaves, when treated with 50 μM Cu, the Cu concentration was significantly lower in transgenic plants, as shown in Figure 5F. However, in stems, the Cu concentration was markedly increased in the T10 and T11 lines compared to NT plants grown in the presence of 25 or 50 μM Cu, as shown in Figure 5G, while in roots there was no difference, as shown in Figure 5H. When treated without Cu (0 μM) or with a normal concentration of Cu (0.2 μM), the Cu concentration was similar in NT and transgenic plants in those tissues, as shown in Figure 6E–H. Moreover, the concentrations of Fe, as shown in Supplementary Figure S3, Mn, as shown in Supplementary Figure S4, and Zn, as shown in Supplementary Figure S5, did not differ between the NT and transgenic lines treated with various concentrations of Cu.
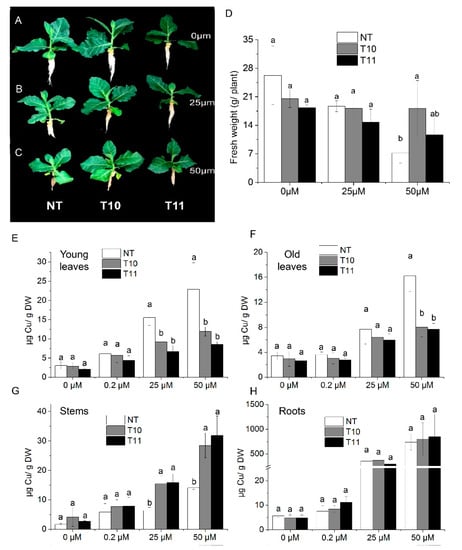
Figure 5.
Transgenic tobacco plants expressing AhYSL3.1 exhibited enhanced tolerance to excess Cu. (A–C) Phenotypes of transgenic tobacco plants expressing AhYSL3.1 in response to excess Cu. The transgenic plants were treated with 0 μM (A), 25 μM (B), or 50 μM (C) Cu for 6 days after 1–2 weeks of normal hydroponic culture. NT, non-transformed plants; T10 and T11, transgenic lines. (D) Fresh weights of NT and transgenic plants. (E–H) Cu concentrations in young leaves (E), old leaves (F), stems (G), and roots (H) of NT and transgenic lines treated with 0, 0.2, 25, or 50 μM Cu with five biological replicates. Values are means ± SD. Means followed by different letters among NT, T10, and T11 are significantly different according to a least-significant-difference test (p < 0.05).
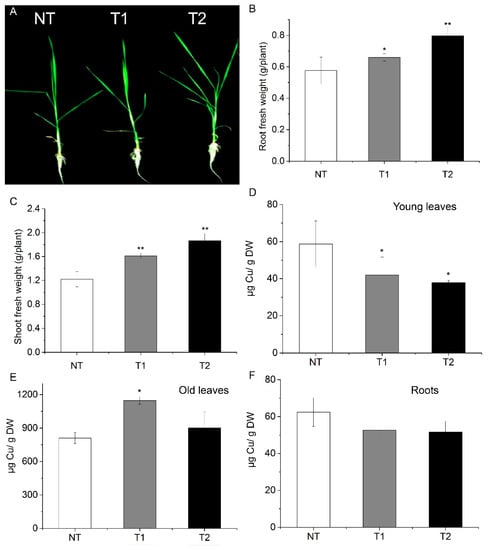
Figure 6.
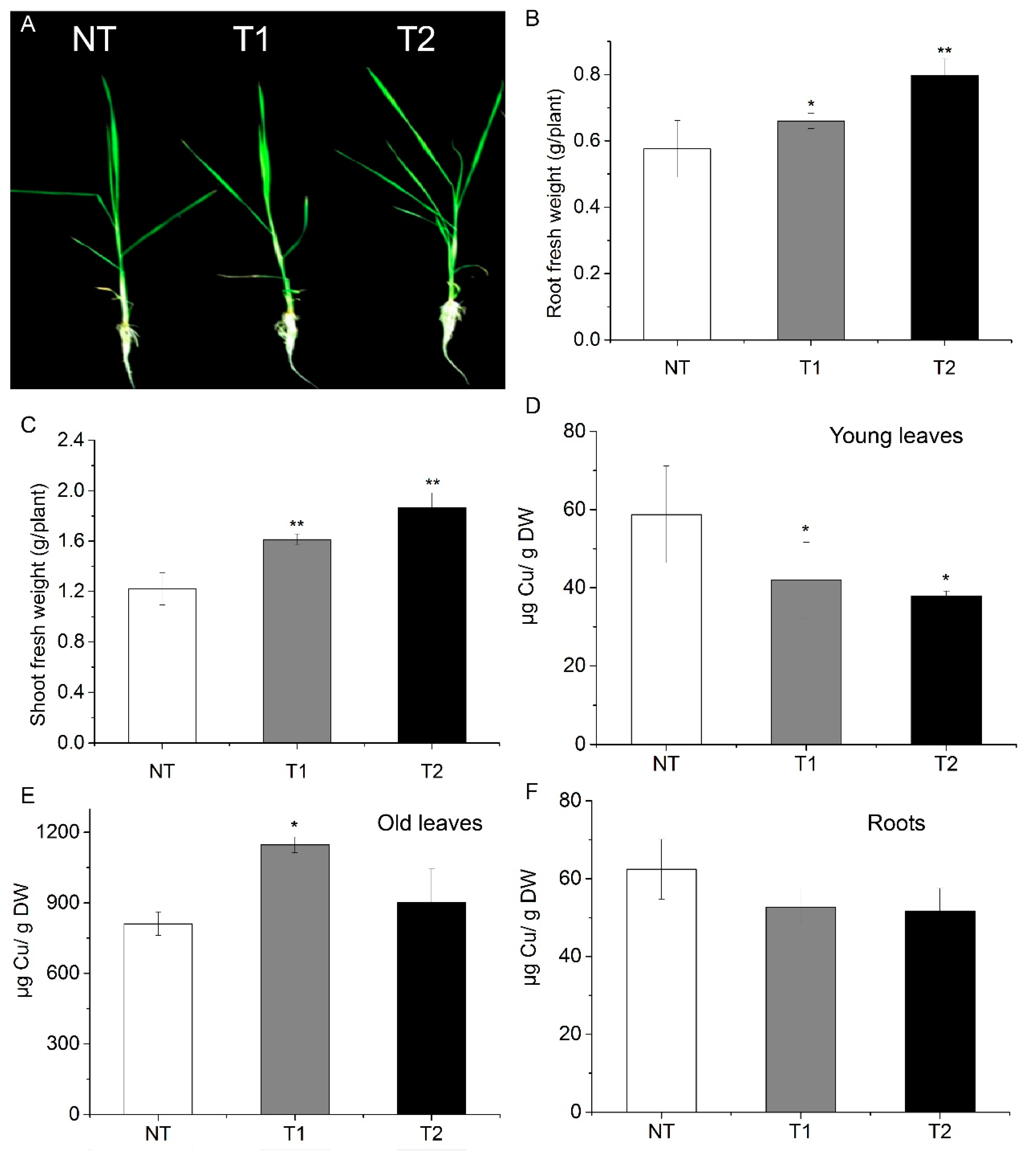
High AhYSL3.1 expression in rice plants resulted in tolerance to excess Cu. Rice plants were treated with 100 μM Cu for 6 days. (A) Phenotypes of transgenic rice plants expressing AhYSL3.1 in response to excess Cu. NT, non-transformed plants; T1 and T2, transgenic lines. (B,C) Fresh weights of roots (B) and shoots (C) of NT and transgenic rice plants after exposure to excess Cu. (D–F) Cu concentrations in young leaves (D), roots (E), and old leaves (F) of NT and transgenic lines treated with 100 μM Cu. Values are means of four biological replicates. Error bars indicate SD. Significant differences from NT were determined by Student’s t-test, * p < 0.05, **p < 0.01.
3.6. Transgenic Rice Plants Are also Tolerant to Excess Cu
Transgenic rice plants with high AhYSL3.1 expression were generated, as shown in Supplementary Figure S6. When treated with 100 μM Cu, the transgenic lines grew better than did the NT plants, as shown in Figure 6A. Additionally, the fresh weights of roots, as shown in Figure 6B, and shoots, as shown in Figure 6C, of the transgenic plants were significantly higher than those of NT plants. Moreover, the Cu concentration in new leaves of transgenic rice was markedly decreased compared to NT plants, as shown in Figure 6D. In contrast, in roots, the Cu concentration was enhanced in one of the transgenic plant lines, as shown in Figure 6E. In old leaves, the Cu concentration showed no difference between the NT and transgenic lines, as shown in Figure 6F.
4. Discussion
Cu transporters are key components in the maintenance of Cu homeostasis. Previous results have demonstrated that the CTR and HMA family proteins are important for Cu acquisition and translocation in plants [6,49]. In this study, we characterized one member of the YSL gene family involved in Cu transport in peanut.
The YSL gene family has been well characterized as an Fe complex transporter in plants. In a previous study, we identified five peanut YSL genes. By yeast complementation assay, AhYSL1 was found to specifically transport Fe(III)–DMA, while AhYSL3.1 transports not only Fe(III)–DMA, but also Fe(II)–NA [38]. However, high expression of AhYSL3.1 in tobacco plants did not affect the Fe concentration in various tissues, but markedly decreased the Cu concentration in leaves subjected to Fe deficiency, as shown in Supplementary Figure S7. Therefore, we speculate that AhYSL3.1 may transport Cu in peanut. Further analyses indicated that among five peanut YSL genes, only AhYSL3.1 and AhYSL3.2 were markedly induced by Cu deficiency, but not by Mn and Zn deficiency, in roots, as shown in Figure 1, suggesting that AhYSL3.1 and AhYSL3.2 were likely responsible for Cu translocation.
In S. cerevisiae, the CTR-type proteins were identified for copper transport. CTR1 and CTR3 are localized to the plasma membrane and are required for Cu uptake [48,50]. Whereas, CTR2 mediates copper mobilization from vacuoles [51]. In this study, a yeast mutant, which is defective in high-affinity Cu uptake of CTR1, was used for a yeast functional complementation assay. The yeast mutant expressing AhYSL3.1 grew well in the culture medium containing Cu–NA but not well in the Cu2+ condition, as shown in Figure 4, suggesting that in the yeast mutant AhYSL3.1 uptake Cu–NA. This result indicates that AhYSL3.1 is a Cu–NA transporter. Different from AhYSL3.1, in Arabidopsis COPTs restored the growth of a mutant yeast strain in the Cu2+ condition, indicating COPTs transport Cu2+ [8].
It is important to note that rice OsYSL16 has been reported to transport both Cu–NA and Fe(III)-DMA in yeast assay [33,36]. According to a phylogenetic analysis, AhYSL3.1 belongs to the same group as OsYSL16 [38]. In contrast to the unchanged expression of OsYSL16 in Cu-deficient rice roots, AhYSL3.1 was induced by Cu deficiency in peanut roots, as shown in Figure 1A. Knockout of OsYSL16 resulted in an increased Cu concentration in older leaves, but decreased Cu concentration in younger leaves in the vegetative stage, suggesting that it functions in delivering Cu to young tissues and seeds through phloem transport [33]. In this study, an artificial promoter driving preferential expression in vascular tissues [43,44] was used to induce exogenous expression of AhYSL3.1 in tobacco and rice plants. The transgenic tobacco as shown in Figure 5 and rice plants as shown in Figure 6 contained a lower concentration of Cu in young leaves under excess Cu conditions, as shown in Figure 6D, suggesting that AhYSL3.1 functions in the internal transport of Cu in transgenic plants. The low level of Cu in young leaves of transgenic plants may be attributed to the recycle and export of Cu from young leaves due to the high level of AhYSL3.1 expression. This speculation can be observed from the higher concentration of Cu in the stem in transgenic tobacco plants, as shown in Figure 5G. There is a model to illustrate the transport of Cu by AhYSL3.1 from leaves to stem in transgenic tobacco, which results in higher concentration of Cu in the stem but lower in the leaves in the excess Cu condition, as shown in Supplementary Figure S8. In peanut, AhYSL3.1 was expressed in the main or lateral roots, but not the root tips, as shown in Figure 2A. In situ hybridization suggested that AhYSL3.1 was localized around the vascular tissues of peanut roots, as shown in Figure 3C,D. Predicted topologies of AhYSL3.1 speculated that it may be a membrane protein, as shown in Supplementary Figure S1. Moreover, transient expression of AhYSL3.1 in onion epidermis cells indicated that AhYSL3.1 was localized to the plasma membrane, as shown in Figure 3A. These results indicated that in peanut plants the Cu–NA transporter AhYSL3.1 mediates internal translocation of Cu.
Consistent with the results for OsYSL16, a yeast expression assay indicated that AhYSL3.1 transported Cu–NA, as shown in Figure 4, in addition to Fe(III)–DMA and Fe(II)–NA [38]. Moreover, knockdown of OsYSL16 resulted in greater Fe accumulation in the vascular bundles of leaves [36], while induction of OsYSL16 expression resulted in an increased Fe concentration in shoots of OsYSL16-induced lines [35], indicating that OsYSL16 is also responsible for Fe homeostasis. In our study, the Fe concentration was not changed in young leaves or roots of AhYSL3.1-overexpressing tobacco plants, as shown in Supplementary Figure S3, but the Cu concentration in leaves and stems was significantly altered, as shown in Figure 5E–G. Therefore, it is reasonable to speculate that AhYSL3.1 functions in Cu transport, but not Fe, in peanut plants.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/12/635/s1, Figure S1: The predicted membrane protein topology and signal peptide of AhYSL3.1 based on different methods, Figure S2: Generation of AhYSL3.1-induced transgenic plants, Figure S3: Fe concentrations in the NT and transgenic lines treated with various concentrations of Cu, Figure S4: Mn concentrations in the NT and transgenic lines treated with various concentrations of Cu, Figure S5: Zn concentrations in the NT and transgenic lines treated with various concentrations of Cu, Figure S6: Relative expression level of AhYSL3.1 in young leaves of NT and transgenic rice plants under excess Cu conditions, Figure S7: Metal concentrations in young leaves and roots of transgenic and NT tobacco plants under Fe-deficient conditions, Figure S8: A model illustrates the transport of Cu by AhYSL3.1 in transgenic tobacco treated with excess Cu.
Author Contributions
H.X. and Y.Z. designed the experiments. J.D. conducted the transgenic tobacco and rice analysis. H.X. performed the other experiments including subcellular localization, in situ hybridization and functional momplementation in yeast. N.W. and W.Q. participated in part of the transgenic tobacco analysis. H.N., T.K. and N.K.N. designed the experiment about the transgenic vector and revised the manuscript. J.D. and H.X. wrote the manuscript. Y.Z. revised the paper. All of the authors discussed the results and commented on the manuscript. Y.Z. provided funding for this work as corresponding author.
Funding
This research was funded by National Key Research and Development Program of China grant numbers 2017YFD0202102, 2016YFD0200405, 2016YFE0101100, National Science Foundation of China grant number 3127222, 31872183. And J.D. was supported by China Scholarship Council.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| Cu | Copper |
| CTR | Cu transporter family proteins |
| COPT | CTR-like transporters |
| HMA | The P-type heavy-metal ATPase family proteins |
| NA | Nicotianamine |
| Zn | Zinc |
| Cd | Cadmium |
| YSL | The Yellow Stripe-Like family genes |
| Fe | iron |
| SUMO/SIZ1 | Small Ubiquitin-like Modifier E3 ligase |
| DMA | Deoxymugineic acid |
| NT | Non-transgenic |
References
- Pilon, M.; Abdel-Ghany, S.E.; Cohu, C.M.; Gogolin, K.A.; Ye, H. Copper cofactor delivery in plant cells. Curr. Opin. Plant Boil. 2006, 9, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Hansch, R.; Mendel, R.R. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Boil. 2009, 12, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, S.; Nickolaus, S.D.; Furst, S.H.; Starck, S.; Schneider, S.; Ekkehard Neuhaus, H.; Trentmann, O. The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol. 2011, 192, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Lequeux, H.; Hermans, C.; Lutts, S.; Verbruggen, N. Response to copper excess in Arabidopsis thaliana: Impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 2010, 48, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Adrees, M.; Ali, S.; Rizwan, M.; Ibrahim, M.; Abbas, F.; Farid, M.; Zia-Ur-Rehman, M.; Irshad, M.K.; Bharwana, S.A. The effect of excess copper on growth and physiology of important food crops: A review. Environ. Sci. Pollut. Res. Int. 2015, 22, 8148–8162. [Google Scholar] [CrossRef]
- Burkhead, J.L.; Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Peñarrubia, L.; Andrés-Colás, N.; Moreno, J.; Puig, S. Regulation of copper transport in Arabidopsis thaliana: A biochemical oscillator? J. Biol. Inorg. Chem. 2010, 15, 29–36. [Google Scholar] [CrossRef]
- Sancenon, V.; Puig, S.; Mira, H.; Thiele, D.J.; Penarrubia, L. Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Boil. 2003, 51, 577–587. [Google Scholar] [CrossRef]
- Kampfenkel, K.; Kushnir, S.; Babiychuk, E.; Inze, D.; Van Montagu, M. Molecular characterization of a putative Arabidopsis thaliana copper transporter and its yeast homologue. J. Boil. Chem. 1995, 270, 28479–28486. [Google Scholar]
- Sancenon, V.; Puig, S.; Mateu-Andres, I.; Dorcey, E.; Thiele, D.J.; Penarrubia, L. The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J. Boil. Chem. 2004, 279, 15348–15355. [Google Scholar] [CrossRef]
- Perea-Garcia, A.; Garcia-Molina, A.; Andres-Colas, N.; Vera-Sirera, F.; Perez-Amador, M.A.; Puig, S.; Penarrubia, L. Arabidopsis copper transport protein COPT2 participates in the cross talk between iron deficiency responses and low-phosphate signaling. Plant Physiol. 2013, 162, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Molina, A.; Andres-Colas, N.; Perea-Garcia, A.; Del Valle-Tascon, S.; Penarrubia, L.; Puig, S. The intracellular Arabidopsis COPT5 transport protein is required for photosynthetic electron transport under severe copper deficiency. Plant J. Cell Mol. Boil. 2011, 65, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Li, X.; Xiao, J.; Wang, S. Molecular and functional analyses of COPT/Ctr-type copper transporter-like gene family in rice. BMC Plant Boil. 2011, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.I.; Gayomba, S.R.; Rutzke, M.A.; Craft, E.; Kochian, L.V.; Vatamaniuk, O.K. COPT6 is a plasma membrane transporter that functions in copper homeostasis in Arabidopsis and is a novel target of SQUAMOSA promoter-binding protein-like 7. J. Boil. Chem. 2012, 287, 33252–33267. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Chu, Z.; Li, X.; Xu, C.; Wang, S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 2010, 22, 3164–3176. [Google Scholar] [CrossRef] [PubMed]
- Tapken, W.; Ravet, K.; Pilon, M. Plastocyanin controls the stabilization of the thylakoid Cu-transporting P-type ATPase PAA2/HMA8 in response to low copper in Arabidopsis. J. Boil. Chem. 2012, 287, 18544–18550. [Google Scholar] [CrossRef] [PubMed]
- Catty, P.; Boutigny, S.; Miras, R.; Joyard, J.; Rolland, N.; Seigneurin-Berny, D. Biochemical characterization of AtHMA6/PAA1, a chloroplast envelope Cu(I)-ATPase. J. Boil. Chem. 2011, 286, 36188–36197. [Google Scholar] [CrossRef] [PubMed]
- Seigneurin-Berny, D.; Gravot, A.; Auroy, P.; Mazard, C.; Kraut, A.; Finazzi, G.; Grunwald, D.; Rappaport, F.; Vavasseur, A.; Joyard, J.; et al. HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J. Boil. Chem. 2006, 281, 2882–2892. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Ghany, S.E.; Muller-Moule, P.; Niyogi, K.K.; Pilon, M.; Shikanai, T. Two P-type atpases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 2005, 17, 1233–1251. [Google Scholar] [CrossRef] [PubMed]
- Andres-Colas, N.; Sancenon, V.; Rodriguez-Navarro, S.; Mayo, S.; Thiele, D.J.; Ecker, J.R.; Puig, S.; Penarrubia, L. The Arabidopsis heavy metal P-type atpase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J. Cell Mol. Boil. 2006, 45, 225–236. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kuroda, K.; Kimura, K.; Southron-Francis, J.L.; Furuzawa, A.; Kimura, K.; Iuchi, S.; Kobayashi, M.; Taylor, G.J.; Koyama, H. Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol. 2008, 148, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Yamaji, N.; Xia, J.; Ma, J.F. A member of the heavy metal P-type ATPase OsHMA5 is involved in xylem loading of copper in rice. Plant Physiol. 2013, 163, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Deng, F.; Yamaji, N.; Pinson, S.R.; Fujii-Kashino, M.; Danku, J.; Douglas, A.; Guerinot, M.L.; Salt, D.E.; Ma, J.F. A heavy metal P-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nat. Commun. 2016, 7, 12138. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, Y.Y.; Lee, Y.; An, G. Rice P1B-type heavy-metal aATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007, 145, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Migocka, M.; Posyniak, E.; Maciaszczyk-Dziubinska, E.; Papierniak, A.; Kosieradzaka, A. Functional and biochemical characterization of cucumber genes encoding two copper ATPases CsHMA5.1 and CsHMA5.2. J. Boil. Chem. 2015, 290, 15717–15729. [Google Scholar] [CrossRef] [PubMed]
- Benes, I.; Schreiber, K.; Ripperger, H.; Kircheiss, A. The “normalizing factor” for the tomato mutant chloronerva, XXIII: Metal-complex formation by nicotianamine, a possible phytosiderophore. Cell. Mol. Life Sci. 1983, 39, 261–262. [Google Scholar]
- Anderegg, G.; Ripperger, H. The “normalizing factor” for the tomato mutant chloronerva, XXIX: Correlation between metal-complex formation and biological-activity of nicotianamine analogs. J. Chem. Soc. Chem. Comm. 1989, 647–650. [Google Scholar] [CrossRef]
- Pich, A.; Scholz, G.; Stephan, U. Iron-dependent changes of heavy metals, nicotianamine, and citrate in different plant organs and in the xylem exudate of two tomato genotypes. Nicotianamine as possible copper translocator. Plant Soil 1994, 165, 189–196. [Google Scholar] [CrossRef]
- Pich, A.; Scholz, G. Translocation of copper and other micronutrients in tomato plants (Lycopersicon esculentum mill): Nicotianamine-stimulated copper transport in the xylem. J. Exp. Bot. 1996, 47, 41–47. [Google Scholar] [CrossRef]
- DiDonato, R.J., Jr.; Roberts, L.A.; Sanderson, T.; Eisley, R.B.; Walker, E.L. Arabidopsis Yellow Stripe-Like2 (YSL2): A metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J. Cell Mol. Boil. 2004, 39, 403–414. [Google Scholar] [CrossRef]
- Schaaf, G.; Schikora, A.; Haberle, J.; Vert, G.; Ludewig, U.; Briat, J.F.; Curie, C.; von Wiren, N. A putative function for the Arabidopsis Fe-Phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol. 2005, 46, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Chen, Y.Y.; Tang, I.C.; Liang, H.M.; Lai, C.C.; Chiou, J.M.; Yeh, K.C. Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol. 2011, 156, 2225–2234. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Yamaji, N.; Yokosho, K.; Ma, J.F. YSL16 is a phloem-localized transporter of the copper-nicotianamine complex that is responsible for copper distribution in rice. Plant Cell 2012, 24, 3767–3782. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lu, W.; Yang, Y.; Shen, Z.; Ma, J.F.; Zheng, L. OsYSL16 is required for preferential cu distribution to floral organs in rice. Plant Cell Physiol. 2018, 59, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ryoo, N.; Jeon, J.S.; Guerinot, M.L.; An, G. Activation of rice Yellow Stripe1-Like 16 (OsYSL16) enhances iron efficiency. Mol. Cells 2012, 33, 117–126. [Google Scholar] [CrossRef]
- Kakei, Y.; Ishimaru, Y.; Kobayashi, T.; Yamakawa, T.; Nakanishi, H.; Nishizawa, N.K. OsYSL16 plays a role in the allocation of iron. Plant Mol. Boil. 2012, 79, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Boil. 2012, 63, 131–152. [Google Scholar] [CrossRef]
- Xiong, H.; Kakei, Y.; Kobayashi, T.; Guo, X.; Nakazono, M.; Takahashi, H.; Nakanishi, H.; Shen, H.; Zhang, F.; Nishizawa, N.K.; et al. Molecular evidence for phytosiderophore-induced improvement of iron nutrition of peanut intercropped with maize in calcareous soil. Plant Cell Environ. 2013, 36, 1888–1902. [Google Scholar] [CrossRef]
- Xiong, H.; Kobayashi, T.; Kakei, Y.; Senoura, T.; Nakazono, M.; Takahashi, H.; Nakanishi, H.; Shen, H.; Duan, P.; Guo, X.; et al. AhNRAMP1 iron transporter is involved in iron acquisition in peanut. J. Exp. Bot. 2012, 63, 4437–4446. [Google Scholar] [CrossRef]
- Luo, M.; Dang, P.; Bausher, M.G.; Holbrook, C.C.; Lee, R.D.; Lynch, R.E.; Guo, B.Z. Identification of transcripts involved in resistance responses to leaf spot disease caused by Cercosporidium personatum in peanut (Arachis hypogaea). Phytopathology 2005, 95, 381–387. [Google Scholar] [CrossRef]
- Mizuno, D.; Higuchi, K.; Sakamoto, T.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Three nicotianamine synthase genes isolated from maize are differentially regulated by iron nutritional status. Plant Physiol. 2003, 132, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. Transforming yeast with DNA. Methods Mol. Cell. Biol. 1995, 5, 255–269. [Google Scholar]
- Kobayashi, T.; Nakayama, Y.; Takahashi, M.; Inoue, H.; Nakanishi, H.; Yoshihara, T.; Mori, S.; Nishizawa, N.K. Construction of artificial promoters highly responsive to iron deficiency. Soil Sci. Plant Nutr. 2004, 50, 1167–1175. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nakayama, Y.; Itai, R.N.; Nakanishi, H.; Yoshihara, T.; Mori, S.; Nishizawa, N.K. Identification of novel cis-acting elements, IDE1 and IDE2, of the barley IDS2 gene promoter conferring iron-deficiency-inducible, root-specific expression in heterogeneous tobacco plants. Plant J. Cell Mol. Boil. 2003, 36, 780–793. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by agrobacterium and sequence analysis of the boundaries of the t-DNA. Plant J. Cell Mol. Boil. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Helmer, G.; Casadaban, M.; Bevan, M.; Kayes, L.; Chilton, M.D. A new chimeric gene as a marker for plant transformation—The expression of Escherichia-coli β-galactosidase in sunflower and tobacco cells. Biotechnolgy 1984, 2, 520–527. [Google Scholar] [CrossRef]
- TOPCONS. Available online: http://www.topcons.net/pred/ (accessed on 5 December 2018).
- Dancis, A.; Yuan, D.S.; Haile, D.; Askwith, C.; Eide, D.; Moehle, C.; Kaplan, J.; Klausner, R.D. Molecular characterization of a copper transport protein in S. cerevisiae: An unexpected role for copper in iron transport. Cell 1994, 76, 393–402. [Google Scholar] [CrossRef]
- Pilon, M. Moving copper in plants. New Phytol. 2011, 192, 305–307. [Google Scholar] [CrossRef]
- Pena, M.M.; Puig, S.; Thiele, D.J. Characterization of the Saccharomyces cerevisiae high affinity copper transporter CTR3. J. Boil. Chem. 2000, 275, 33244–33251. [Google Scholar] [CrossRef]
- Rees, E.M.; Lee, J.; Thiele, D.J. Mobilization of intracellular copper stores by the CTR2 vacuolar copper transporter. J. Boil. Chem. 2004, 279, 54221–54229. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).