Compatibility between Legumes and Rhizobia for the Establishment of a Successful Nitrogen-Fixing Symbiosis
Abstract
:1. Introduction
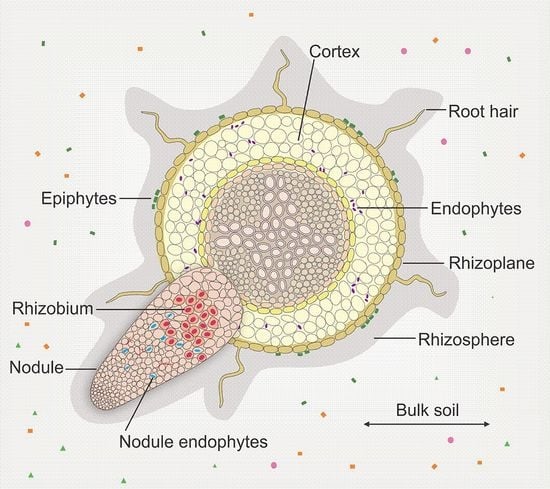
2. Legume-Rhizobia Interaction in a Community Context
3. Distinguishing Friends from Enemies
4. Recognition and Signaling Events for the Establishment of a Compatible Interaction between Legume and Rhizobia
5. Transcriptional Reprogramming Required for a Compatible Root Nodule Symbiosis
6. Concluding Remarks and Future Perspectives
Acknowledgments
Conflicts of Interest
References
- Ibáñez, F.; Wall, L.; Fabra, A. Starting points in plant-bacteria nitrogen-fixing symbioses: Intercellular invasion of the roots. J. Exp. Bot. 2017, 68, 1905–1918. [Google Scholar] [CrossRef] [PubMed]
- Peters, K.N.; Verma, D.P. Phenolic compounds as regulators of gene expression in plant-microbe relations. Mol. Plant Microbe Interact. 1990, 3, 4–8. [Google Scholar] [CrossRef] [PubMed]
- D’Haeze, W.; Holsters, M. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 2002, 12, 79R–105R. [Google Scholar] [CrossRef] [PubMed]
- Gage, D.J. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 2004, 68, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Den Herder, J.; Vanhee, C.; De Rycke, R.; Corich, V.; Holsters, M.; Goormachtig, S. Nod factor perception during infection thread growth fine-tunes nodulation. Mol. Plant Microbe Interact. 2007, 20, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Heckmann, A.B.; Lombardo, F.; Miwa, H.; Perry, J.A.; Bunnewell, S.; Parniske, M.; Wang, T.L.; Downie, J.A. Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 2006, 142, 1739–1750. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Mirabella, R.; Fedorova, E.; Franken, C.; Franssen, H.; Bisseling, T.; Geurts, R. Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 2005, 102, 10375–10380. [Google Scholar] [CrossRef] [PubMed]
- Capoen, W.; Goormachtig, S.; De Rycke, R.; Schroeyers, K.; Holsters, M. SrSYMRK, a plant receptor essential for symbiosome formation. Proc. Natl. Acad. Sci. USA 2005, 102, 10369–10374. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Amor, B.B.; Shaw, S.L.; Oldroyd, G.E.; Maillet, F.; Penmetsa, R.V.; Cook, D.; Long, S.R.; Denarie, J.; Gough, C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003, 34, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Gronlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Arrighi, J.F.; Barre, A.; Ben Amor, B.; Bersoult, A.; Soriano, L.C.; Mirabella, R.; de Carvalho-Niebel, F.; Journet, E.P.; Gherardi, M.; Huguet, T.; et al. The Medicago truncatula lysin motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006, 142, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Smit, P.; Limpens, E.; Geurts, R.; Fedorova, E.; Dolgikh, E.; Gough, C.; Bisseling, T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007, 145, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Esseling, J.J.; Lhuissier, F.G.; Emons, A.M. A nonsymbiotic root hair tip growth phenotype in NORK-mutated legumes: Implications for nodulation factor-induced signaling and formation of a multifaceted root hair pocket for bacteria. Plant Cell 2004, 16, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Endre, G.; Kereszt, A.; Kevei, Z.; Mihacea, S.; Kalo, P.; Kiss, G.B. A receptor kinase gene regulating symbiotic nodule development. Nature 2002, 417, 962–966. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, D.W.; Wais, R.; Long, S.R. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 1996, 85, 673–681. [Google Scholar] [CrossRef]
- Sieberer, B.J.; Chabaud, M.; Timmers, A.C.; Monin, A.; Fournier, J.; Barker, D.G. A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 2009, 151, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Ane, J.M.; Kiss, G.B.; Riely, B.K.; Penmetsa, R.V.; Oldroyd, G.E.; Ayax, C.; Levy, J.; Debelle, F.; Baek, J.M.; Kalo, P.; et al. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 2004, 303, 1364–1367. [Google Scholar] [CrossRef] [PubMed]
- Peiter, E.; Sun, J.; Heckmann, A.B.; Venkateshwaran, M.; Riely, B.K.; Otegui, M.S.; Edwards, A.; Freshour, G.; Hahn, M.G.; Cook, D.R.; et al. The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol. 2007, 145, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, M.; Bredemeier, R.; Wanner, G.; Takeda, N.; Schleiff, E.; Parniske, M. Lotus japonicus castor and pollux are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 2008, 20, 3467–3479. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, M.; Sun, J.; Martins, T.V.; Radhakrishnan, G.V.; Findlay, K.; Soumpourou, E.; Thouin, J.; Véry, A.-A.; Sanders, D.; Morris, R.J.; et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 2016, 352, 1102–1105. [Google Scholar] [CrossRef] [PubMed]
- Lévy, J.; Bres, C.; Geurts, R.; Chalhoub, B.; Kulikova, O.; Duc, G.; Journet, E.-P.; Ané, J.-M.; Lauber, E.; Bisseling, T.; et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 2004, 303, 1361–1364. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Katzer, K.; Lambert, J.; Cerri, M.; Parniske, M. Cyclops, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 2014, 15, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Gans, J.; Wolinsky, M.; Dunbar, J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 2005, 309, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.E.; De Beuf, K.; Vekeman, B.; Willems, A. A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol. Biochem. 2015, 83, 1–11. [Google Scholar] [CrossRef]
- Velázquez, E.; Martínez-Hidalgo, P.; Carro, L.; Alonso, P.; Peix, A.; Trujillo, M.; Martínez-Molina, E. Nodular endophytes: An untapped diversity. In Beneficial Plant-Microbial Interactions: Ecology and Applications; González, M.B.R., Gonzalez-López, J., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 214–236. [Google Scholar]
- Rajendran, G.; Patel, M.H.; Joshi, S.J. Isolation and characterization of nodule-associated Exiguobacterium sp. From the root nodules of Fenugreek (Trigonella foenum-graecum) and their possible role in plant growth promotion. Int. J. Microbiol. 2012, 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Hidalgo, P.; Hirsch, A.M. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes 2017, 1, 70–82. [Google Scholar] [CrossRef]
- Leite, J.; Fischer, D.; Rouws, L.F.M.; Fernandes-Júnior, P.I.; Hofmann, A.; Kublik, S.; Schloter, M.; Xavier, G.R.; Radl, V. Cowpea nodules harbor non-rhizobial bacterial communities that are shaped by soil type rather than plant genotype. Front. Plant Sci. 2017, 7, 2064. [Google Scholar] [CrossRef] [PubMed]
- Zgadzaj, R.; James, E.K.; Kelly, S.; Kawaharada, Y.; de Jonge, N.; Jensen, D.B.; Madsen, L.H.; Radutoiu, S. A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PLoS Genet. 2015, 11, e1005280. [Google Scholar] [CrossRef] [PubMed]
- Zgadzaj, R.; Garrido-Oter, R.; Jensen, D.B.; Koprivova, A.; Schulze-Lefert, P.; Radutoiu, S. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. USA 2016, 113, E7996–E8005. [Google Scholar] [CrossRef] [PubMed]
- Schumpp, O.; Deakin, W.J. How inefficient rhizobia prolong their existence within nodules. Trends Plant Sci. 2010, 15, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, F.; Tsuda, K. Understanding the plant immune system. Mol. Plant Microbe Interact. 2010, 23, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Maillet, F.; Poinsot, V.; Andre, O.; Puech-Pages, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A.; et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pagès, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Bécard, G.; Bonfante, P.; et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013, 198, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Dénarié, J.; Cullimore, J. Lipo-oligosaccharide nodulation factors: A new class of signaling molecules mediating recognition and morphogenesis. Cell 1993, 74, 951–954. [Google Scholar] [CrossRef]
- Lerouge, P.; Roche, P.; Faucher, C.; Maillet, F.; Truchet, G.; Prome, J.C.; Denarie, J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 1990, 344, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Kawaharada, Y.; Kelly, S.; Nielsen, M.W.; Hjuler, C.T.; Gysel, K.; Muszynski, A.; Carlson, R.W.; Thygesen, M.B.; Sandal, N.; Asmussen, M.H.; et al. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 2015, 523, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Oldroyd, G. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Bozsoki, Z.; Cheng, J.; Feng, F.; Gysel, K.; Vinther, M.; Andersen, K.R.; Oldroyd, G.; Blaise, M.; Radutoiu, S.; Stougaard, J. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. USA 2017, 114, E8118–E8127. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Lohar, D.P.; Sharopova, N.; Endre, G.; Penuela, S.; Samac, D.; Town, C.; Silverstein, K.A.; VandenBosch, K.A. Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 2006, 140, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Kouchi, H.; Shimomura, K.; Hata, S.; Hirota, A.; Wu, G.J.; Kumagai, H.; Tajima, S.; Suganuma, N.; Suzuki, A.; Aoki, T.; et al. Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res. 2004, 11, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gomez, M.; Sandal, N.; Stougaard, J.; Boller, T. Interplay of flg22-induced defence responses and nodulation in Lotus japonicus. J. Exp. Bot. 2012, 63, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Duan, L.; Zhou, B.; Yu, H.; Zhu, H.; Cao, Y.; Zhang, Z. Interplay of pathogen-induced defense responses and symbiotic establishment in Medicago truncatula. Front. Microbiol. 2017, 8, 973. [Google Scholar] [CrossRef] [PubMed]
- Deakin, W.J.; Broughton, W.J. Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat. Rev. Microbiol. 2009, 7, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Miwa, H.; Okazaki, S. How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 2017, 38, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Dalla Via, V.; Zanetti, M.E.; Blanco, F. How legumes recognize rhizobia. Plant Signal Behav. 2016, 11, e1120396. [Google Scholar]
- Viprey, V.; Del Greco, A.; Golinowski, W.; Broughton, W.J.; Perret, X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 1998, 28, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Hubber, A.; Vergunst, A.C.; Sullivan, J.T.; Hooykaas, P.J.J.; Ronson, C.W. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A Vir/D4 type IV secretion system. Mol. Microbiol. 2004, 54, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Bladergroen, M.R.; Badelt, K.; Spaink, H.P. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 2003, 16, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Fauvart, M.; Michiels, J. Rhizobial secreted proteins as determinants of host specificity in the rhizobium–legume symbiosis. FEMS Microbiol. Lett. 2008, 285, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.S.; Sadowsky, M.J. Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 2015, 6, 491. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA 2013, 110, 17131–17136. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, S.; Zehner, S.; Hempel, J.; Lang, K.; Göttfert, M. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol. Lett. 2009, 295, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Xin, D.-W.; Liao, S.; Xie, Z.-P.; Hann, D.R.; Steinle, L.; Boller, T.; Staehelin, C. Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. Strain NGR234. PLOS Pathog. 2012, 8, e1002707. [Google Scholar] [CrossRef] [PubMed]
- Kambara, K.; Ardissone, S.; Kobayashi, H.; Saad, M.M.; Schumpp, O.; Broughton, W.J.; Deakin, W.J. Rhizobia utilize pathogen-like effector proteins during symbiosis. Mol. Microbiol. 2009, 71, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.-Y.; Xiang, Q.-W.; Wagner, C.; Zhang, D.; Xie, Z.-P.; Staehelin, C. The type 3 effector NopL of Sinorhizobium sp. Strain NGR234 is a mitogen-activated protein kinase substrate. J. Exp. Bot. 2016, 67, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, X.-J.; Lu, H.-B.; Xie, Z.-P.; Staehelin, C. Functional analysis of the type 3 effector nodulation outer protein l (NopL) from Rhizobium sp. NGR234: Symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 2011, 286, 32178–32187. [Google Scholar] [CrossRef] [PubMed]
- Bartsev, A.V.; Boukli, N.M.; Deakin, W.J.; Staehelin, C.; Broughton, W.J. Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 2003, 554, 271–274. [Google Scholar] [CrossRef]
- Bartsev, A.V.; Deakin, W.J.; Boukli, N.M.; McAlvin, C.B.; Stacey, G.; Malnoë, P.; Broughton, W.J.; Staehelin, C. NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 2004, 134, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Tang, F.; Gao, M.; Krishnan, H.B.; Zhu, H. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 2010, 107, 18735–18740. [Google Scholar] [CrossRef] [PubMed]
- Sadowsky, M.J.; Cregan, P.B. The soybean Rj4 allele restricts nodulation by Bradyrhizobium japonicum serogroup 123 strains. Appl. Environ. Microbiol. 1992, 58, 720–723. [Google Scholar] [PubMed]
- Tang, F.; Yang, S.; Liu, J.; Zhu, H. Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol. 2016, 170, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Faruque, O.M.; Miwa, H.; Yasuda, M.; Fujii, Y.; Kaneko, T.; Sato, S.; Okazaki, S. Identification of Bradyrhizobium elkanii genes involved in incompatibility with soybean plants carrying the Rj4 allele. Appl. Environ. Microbiol. 2015, 81, 6710–6717. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Miwa, H.; Masuda, S.; Takebayashi, Y.; Sakakibara, H.; Okazaki, S. Effector-triggered immunity determines host genotype-specific incompatibility in legume–Rhizobium symbiosis. Plant Cell Physiol. 2016, 57, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef] [PubMed]
- Downie, J.A. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 2010, 34, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Gibson, K.E.; Kobayashi, H.; Walker, G.C. Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 2008, 42, 413–441. [Google Scholar] [CrossRef] [PubMed]
- Breedveld, M.W.; Cremers, H.C.; Batley, M.; Posthumus, M.A.; Zevenhuizen, L.P.; Wijffelman, C.A.; Zehnder, A.J. Polysaccharide synthesis in relation to nodulation behavior of Rhizobium leguminosarum. J. Bacteriol. 1993, 175, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.-P.; Walker, G.C. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 1998, 180, 5183–5191. [Google Scholar] [PubMed]
- Perotto, S.; Brewin, N.; Kannenberg, E. Cytological evidence for a host defense response that reduces cell and tissue invasion in pea nodules by lipopolysaccharide-defective mutants of Rhizobium leguminosarum strain 3841. Mol. Plant Microbe Interact. 1994, 7, 99–112. [Google Scholar] [CrossRef]
- García-de los Santos, A.; Brom, S. Characterization of two plasmid-borne lpsβ loci of Rhizobium etli required for lipopolysaccharide synthesis and for optimal interaction with plants. Mol. Plant Microbe Interact. 1997, 10, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; D’Haeze, W.; De Rycke, R.; Wolucka, B.; Holsters, M. Knockout of an Azorhizobial dTDP-L-rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata. Mol. Plant Microbe Interact. 2001, 14, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Mukherjee, A.; Wiley-Kalil, A.; Das, S.; Owen, H.; Reddy, P.; Ané, J.; James, E.; Gyaneshwar, P. A rhamnose-deficient lipopolysaccharide mutant of Rhizobium sp. IRGB74 is defective in root colonization and beneficial interactions with its flooding-tolerant hosts Sesbania cannabina and wetland rice. J. Exp. Bot. 2016, 67, 5869–5884. [Google Scholar] [CrossRef] [PubMed]
- Albus, U.; Baier, R.; Holst, O.; Pühler, A.; Niehaus, K. Suppression of an elicitor-induced oxidative burst reaction in Medicago sativa cell cultures by Sinorhizobium meliloti lipopolysaccharides. New Phytol. 2001, 151, 597–606. [Google Scholar] [CrossRef]
- Scheidle, H.; Groß, A.; Niehaus, K. The lipid a substructure of the Sinorhizobium meliloti lipopolysaccharides is sufficient to suppress the oxidative burst in host plants. New Phytol. 2005, 165, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Cao, Y.; Tanaka, K.; Thibivilliers, S.; Wan, J.; Choi, J.; Kang, C.H.; Qiu, J.; Stacey, G. Nonlegumes respond to rhizobial nod factors by suppressing the innate immune response. Science 2013, 341, 1384–1387. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Tóth, K.; Cao, Y.; Tanaka, K.; Espinoza, C.; Stacey, G. Lipochitooligosaccharide recognition: An ancient story. New Phytol. 2014, 204, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.M.; Sharopova, N.; Lohar, D.P.; Zhang, J.Q.; VandenBosch, K.A.; Walker, G.C. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc. Natl. Acad. Sci. USA 2008, 105, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.N.; Newman, M.-A.; Erbs, G.; Morrissey, K.L.; Chinchilla, D.; Boller, T.; Jensen, T.T.; De Castro, C.; Ierano, T.; Molinaro, A.; et al. Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr. Biol. 2008, 18, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Horváth, B.; Domonkos, Á.; Kereszt, A.; Szűcs, A.; Ábrahám, E.; Ayaydin, F.; Bóka, K.; Chen, Y.; Chen, R.; Murray, J.D.; et al. Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc. Natl. Acad. Sci. USA 2015, 112, 15232–15237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mergaert, P.; Uchiumi, T.; Alunni, B.; Evanno, G.; Cheron, A.; Catrice, O.; Mausset, A.-E.; Barloy-Hubler, F.; Galibert, F.; Kondorosi, A.; et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 5230–5235. [Google Scholar] [CrossRef] [PubMed]
- Price, P.A.; Tanner, H.R.; Dillon, B.A.; Shabab, M.; Walker, G.C.; Griffitts, J.S. Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc. Natl. Acad. Sci. USA 2015, 112, 15244–15249. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, Q.; Fedorova, E.; Liu, J.; Qin, Q.; Zheng, Q.; Price, P.A.; Pan, H.; Wang, D.; Griffitts, J.S.; et al. Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6848–6853. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yang, S.; Liu, J.; Terecskei, K.; Ábrahám, E.; Gombár, A.; Domonkos, Á.; Szűcs, A.; Körmöczi, P.; Wang, T.; et al. Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6854–6859. [Google Scholar] [CrossRef] [PubMed]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.F.; Rakocevic, A.; Mitra, R.M.; Brocard, L.; Sun, J.; Eschstruth, A.; Long, S.R.; Schultze, M.; Ratet, P.; Oldroyd, G.E. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007, 144, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Andriankaja, A.; Boisson-Dernier, A.; Frances, L.; Sauviac, L.; Jauneau, A.; Barker, D.G.; de Carvalho-Niebel, F. AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 2007, 19, 2866–2885. [Google Scholar] [CrossRef] [PubMed]
- Middleton, P.H.; Jakab, J.; Penmetsa, R.V.; Starker, C.G.; Doll, J.; Kalo, P.; Prabhu, R.; Marsh, J.F.; Mitra, R.M.; Kereszt, A.; et al. An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 2007, 19, 1221–1234. [Google Scholar] [CrossRef] [PubMed]
- Cerri, M.R.; Frances, L.; Laloum, T.; Auriac, M.-C.; Niebel, A.; Oldroyd, G.E.D.; Barker, D.G.; Fournier, J.; de Carvalho-Niebel, F. Medicago truncatula ERN transcription factors: Regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 2012, 160, 2155–2172. [Google Scholar] [CrossRef] [PubMed]
- Kalo, P.; Gleason, C.; Edwards, A.; Marsh, J.; Mitra, R.M.; Hirsch, S.; Jakab, J.; Sims, S.; Long, S.R.; Rogers, J.; et al. Nodulation signaling in legumes requires NSP2, a member of the gras family of transcriptional regulators. Science 2005, 308, 1786–1789. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.G.; Oppenheimer, D.G. Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 2005, 21, 271–295. [Google Scholar] [CrossRef] [PubMed]
- Combier, J.P.; Frugier, F.; de Billy, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Vernie, T.; Ott, T.; Gamas, P.; Crespi, M.; et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef] [PubMed]
- Baudin, M.; Laloum, T.; Lepage, A.; Rípodas, C.; Ariel, F.; Frances, L.; Crespi, M.; Gamas, P.; Blanco, F.A.; Zanetti, M.E.; et al. A phylogenetically conserved group of Nuclear Factor-Y transcription factors interact to control nodulation in legumes. Plant Physiol. 2015, 169, 2761–2773. [Google Scholar] [PubMed]
- Soyano, T.; Kouchi, H.; Hirota, A.; Hayashi, M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013, 9, e1003352. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M.E.; Blanco, F.A.; Beker, M.P.; Battaglia, M.; Aguilar, O.M. A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 2010, 22, 4142–4157. [Google Scholar] [CrossRef] [PubMed]
- Benedito, V.A.; Torres-Jerez, I.; Murray, J.D.; Andriankaja, A.; Allen, S.; Kakar, K.; Wandrey, M.; Verdier, J.; Zuber, H.; Ott, T.; et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008, 55, 504–513. [Google Scholar] [CrossRef] [PubMed]
- El Yahyaoui, F.; Küster, H.; Ben Amor, B.; Hohnjec, N.; Pühler, A.; Becker, A.; Gouzy, J.; Vernié, T.; Gough, C.; Niebel, A.; et al. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004, 136, 3159–3176. [Google Scholar] [CrossRef] [PubMed]
- Maunoury, N.; Redondo-Nieto, M.; Bourcy, M.; Van de Velde, W.; Alunni, B.; Laporte, P.; Durand, P.; Agier, N.; Marisa, L.; Vaubert, D.; et al. Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS ONE 2010, 5, e9519. [Google Scholar] [CrossRef] [PubMed]
- Moreau, S.; Verdenaud, M.; Ott, T.; Letort, S.; de Billy, F.; Niebel, A.; Gouzy, J.; de Carvalho-Niebel, F.; Gamas, P. Transcription reprogramming during root nodule development in Medicago truncatula. PLoS ONE 2011, 6, e16463. [Google Scholar] [CrossRef] [PubMed]
- Larrainzar, E.; Riely, B.K.; Kim, S.C.; Carrasquilla-Garcia, N.; Yu, H.-J.; Hwang, H.-J.; Oh, M.; Kim, G.B.; Surendrarao, A.K.; Chasman, D.; et al. Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol. 2015, 169, 233–265. [Google Scholar] [CrossRef] [PubMed]
- Høgslund, N.; Radutoiu, S.; Krusell, L.; Voroshilova, V.; Hannah, M.A.; Goffard, N.; Sanchez, D.H.; Lippold, F.; Ott, T.; Sato, S.; et al. Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS ONE 2009, 4, e6556. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.; Capella-Gutiérrez, S.; Rendón-Anaya, M.; Hernández-Oñate, M.; Minoche, A.E.; Erb, I.; Câmara, F.; Prieto-Barja, P.; Corvelo, A.; Sanseverino, W.; et al. Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol. 2016, 17, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalla Via, V.; Narduzzi, C.; Aguilar, O.M.; Zanetti, M.E.; Blanco, F.A. Changes in the common bean (Phaseolus vulgaris) transcriptome in response to secreted and surface signal molecules of Rhizobium etli. Plant Physiol. 2015, 169, 1356–1370. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010, 152, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-seq atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Afonso-Grunz, F.; Molina, C.; Hoffmeier, K.; Rycak, L.; Kudapa, H.; Varshney, R.K.; Drevon, J.-J.; Winter, P.; Kahl, G. Genome-based analysis of the transcriptome from mature chickpea root nodules. Front. Plant Sci. 2014, 5, 325. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Patel, R.K.; Jhanwar, S.; Priya, P.; Bhattacharjee, A.; Yadav, G.; Bhatia, S.; Chattopadhyay, D.; Tyagi, A.K.; Jain, M. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol. 2011, 156, 1661–1678. [Google Scholar] [CrossRef] [PubMed]
- Breakspear, A.; Liu, C.; Roy, S.; Stacey, N.; Rogers, C.; Trick, M.; Morieri, G.; Mysore, K.S.; Wen, J.; Oldroyd, G.E.D.; et al. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 2014, 26, 4680–4701. [Google Scholar] [CrossRef] [PubMed]
- Jardinaud, M.-F.; Boivin, S.; Rodde, N.; Catrice, O.; Kisiala, A.; Lepage, A.; Moreau, S.; Roux, B.; Cottret, L.; Sallet, E.; et al. A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by Nod factors in the Medicago truncatula root epidermis. Plant Physiol. 2016, 171, 2256–2276. [Google Scholar] [CrossRef] [PubMed]
- Roux, B.; Rodde, N.; Jardinaud, M.-F.; Timmers, T.; Sauviac, L.; Cottret, L.; Carrère, S.; Sallet, E.; Courcelle, E.; Moreau, S.; et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014, 77, 817–837. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Moling, S.; Hooiveld, G.; Pereira, P.A.; Bisseling, T.; Becker, J.D.; Küster, H. Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS ONE 2013, 8, e64377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanetti, M.E.; Chang, I.F.; Gong, F.; Galbraith, D.W.; Bailey-Serres, J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005, 138, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Reynoso, M.A.; Juntawong, P.; Lancia, M.; Blanco, F.A.; Bailey-Serres, J.; Zanetti, M.E. Translating ribosome affinity purification (TRAP) followed by RNA sequencing technology (TRAP-Seq) for quantitative assessment of plant translatomes. In Plant Functional Genomics: Methods and Protocols; Alonso, J.M., Stepanova, A.N., Eds.; Springer: New York, NY, USA, 2015; pp. 185–207. [Google Scholar]
- Reynoso, M.A.; Blanco, F.A.; Bailey-Serres, J.; Crespi, M.; Zanetti, M.E. Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J. 2013, 73, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Akçai, E. Evolutionary models of mutualism. In Mutualism; Bronstein, J.L., Ed.; Oxford University Press: New York, NY, USA, 2015; pp. 57–76. [Google Scholar]
- Aguilar, O.M.; Riva, O.; Peltzer, E. Analysis of Rhizobium etli and of its symbiosis with wild Phaseolus vulgaris supports coevolution in centers of host diversification. Proc. Natl. Acad. Sci. USA 2004, 101, 13548–13553. [Google Scholar] [CrossRef] [PubMed]
- Mazziotta, L.; Reynoso, M.A.; Aguilar, O.M.; Blanco, F.A.; Zanetti, M.E. Transcriptional and functional variation of NF-YC1 in genetically diverse accessions of Phaseolus vulgaris during the symbiotic association with Rhizobium etli. Plant Biol. 2012. [Google Scholar] [CrossRef]
- Peltzer Meschini, E.P.; Blanco, F.A.; Zanetti, M.E.; Beker, M.P.; Kuster, H.; Puhler, A.; Aguilar, O.M. Host genes involved in nodulation preference in common bean (Phaseolus vulgaris)-Rhizobium etli symbiosis revealed by suppressive subtractive hybridization. Mol. Plant Microbe Interact. 2008, 21, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Heath, K.D.; Tiffin, P. Stabilizing mechanisms in a legume-rhizobium mutualism. Evolution 2009, 63, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Oono, R.; Anderson, C.G.; Denison, R.F. Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc. Biol. Soc. 2011, 278, 2698–2703. [Google Scholar] [CrossRef] [PubMed]
- Gubry-Rangin, C.; Garcia, M.; Béna, G. Partner choice in Medicago truncatula-Sinorhizobium symbiosis. Proc. Biol. Soc. 2010, 277, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Kiers, E.T.; Rousseau, R.A.; West, S.A.; Denison, R.F. Host sanctions and the legume-rhizobium mutualism. Nature 2003, 425, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Batstone, R.T.; Dutton, E.M.; Wang, D.; Yang, M.; Frederickson, M.E. The evolution of symbiont preference traits in the model legume Medicago truncatula. New Phytol. 2017, 213, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Banba, M.; Siddique, A.-B.M.; Kouchi, H.; Izui, K.; Hata, S. Lotus japonicus forms early senescent root nodules with Rhizobium etli. Mol. Plant Microbe Interact. 2001, 14, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Schumpp, O.; Crèvecoeur, M.; Broughton, W.J.; Deakin, W.J. Delayed maturation of nodules reduces symbiotic effectiveness of the Lotus japonicus-Rhizobium sp. NGR234 interaction. J. Exp. Bot. 2009, 60, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.J.; O’Hara, G.W.; Tiwari, R.P.; Dilworth, M.J.; Howieson, J.G. The model legume Medicago truncatula A17 is poorly matched for N2 fixation with the sequenced microsymbiont Sinorhizobium meliloti 1021. New Phytol. 2008, 179, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Heath, K.D.; Burke, P.V.; Stinchcombe, J.R. Coevolutionary genetic variation in the legume-rhizobium transcriptome. Mol. Ecol. 2012, 21, 4735–4747. [Google Scholar] [CrossRef] [PubMed]
- Burghardt, L.T.; Guhlin, J.; Chun, C.L.; Liu, J.; Sadowsky, M.J.; Stupar, R.M.; Young, N.D.; Tiffin, P. Transcriptomic basis of genome by genome variation in a legume-rhizobia mutualism. Mol. Ecol. 2017, 26, 6122–6135. [Google Scholar] [CrossRef] [PubMed]
- D’Antuono, A.L.; Ott, T.; Krusell, L.; Voroshilova, V.; Ugalde, R.A.; Udvardi, M.; Lepek, V.C. Defects in rhizobial cyclic glucan and lipopolysaccharide synthesis alter legume gene expression during nodule development. Mo. Plant Microbe Interact. 2007, 21, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Kiers, E.T.; Hutton, M.G.; Denison, R.F. Human selection and the relaxation of legume defences against ineffective rhizobia. Proc. Biol. Soc. 2007, 274, 3119–3126. [Google Scholar] [CrossRef] [PubMed]
- Sprent, J.I.; Ardley, J.; James, E.K. Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 2017, 215, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.; Andrews, M.E. Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 2017, 18, 705. [Google Scholar] [CrossRef] [PubMed]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clúa, J.; Roda, C.; Zanetti, M.E.; Blanco, F.A. Compatibility between Legumes and Rhizobia for the Establishment of a Successful Nitrogen-Fixing Symbiosis. Genes 2018, 9, 125. https://doi.org/10.3390/genes9030125
Clúa J, Roda C, Zanetti ME, Blanco FA. Compatibility between Legumes and Rhizobia for the Establishment of a Successful Nitrogen-Fixing Symbiosis. Genes. 2018; 9(3):125. https://doi.org/10.3390/genes9030125
Chicago/Turabian StyleClúa, Joaquín, Carla Roda, María Eugenia Zanetti, and Flavio A. Blanco. 2018. "Compatibility between Legumes and Rhizobia for the Establishment of a Successful Nitrogen-Fixing Symbiosis" Genes 9, no. 3: 125. https://doi.org/10.3390/genes9030125
APA StyleClúa, J., Roda, C., Zanetti, M. E., & Blanco, F. A. (2018). Compatibility between Legumes and Rhizobia for the Establishment of a Successful Nitrogen-Fixing Symbiosis. Genes, 9(3), 125. https://doi.org/10.3390/genes9030125