Bioprinting Perfusion-Enabled Liver Equivalents for Advanced Organ-on-a-Chip Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Bioink Preparation
2.3. Tissue Model and Printing Process
2.4. qPCR
2.5. Immunohistochemistry
2.6. Metabolic Analysis
3. Results
3.1. Morphological Analysis after Printing
3.2. Viability after Bioprinting
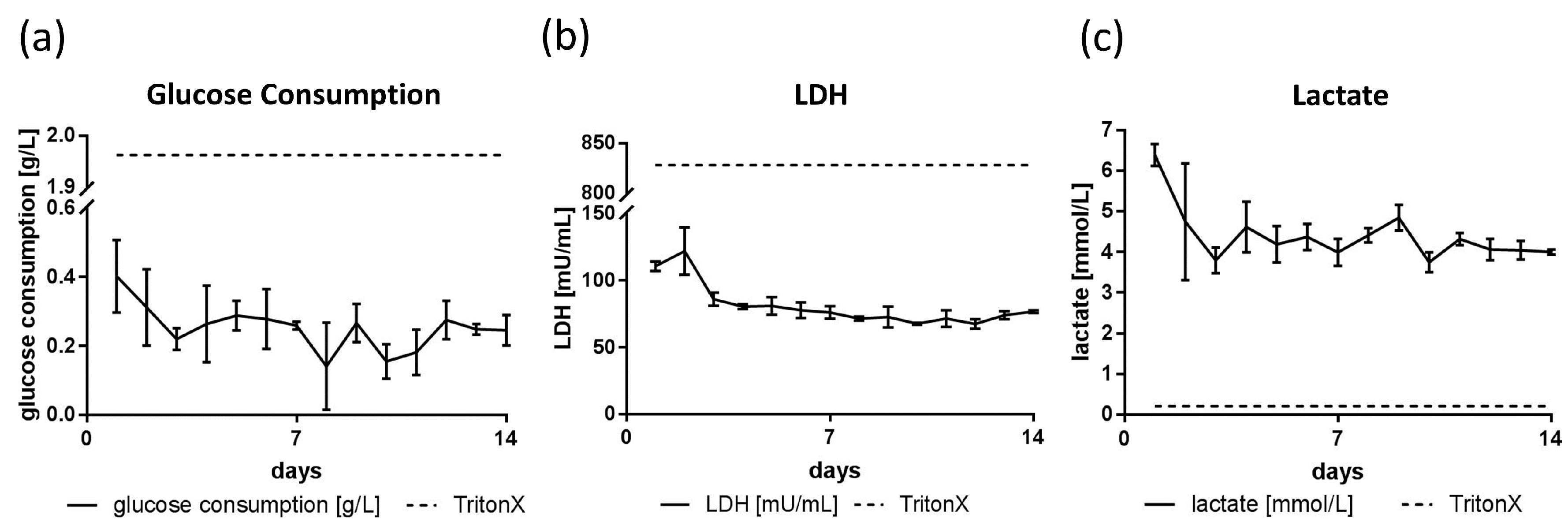
3.3. Glucose, Lactate and LDH Metabolics
3.4. qPCR Marker Expression
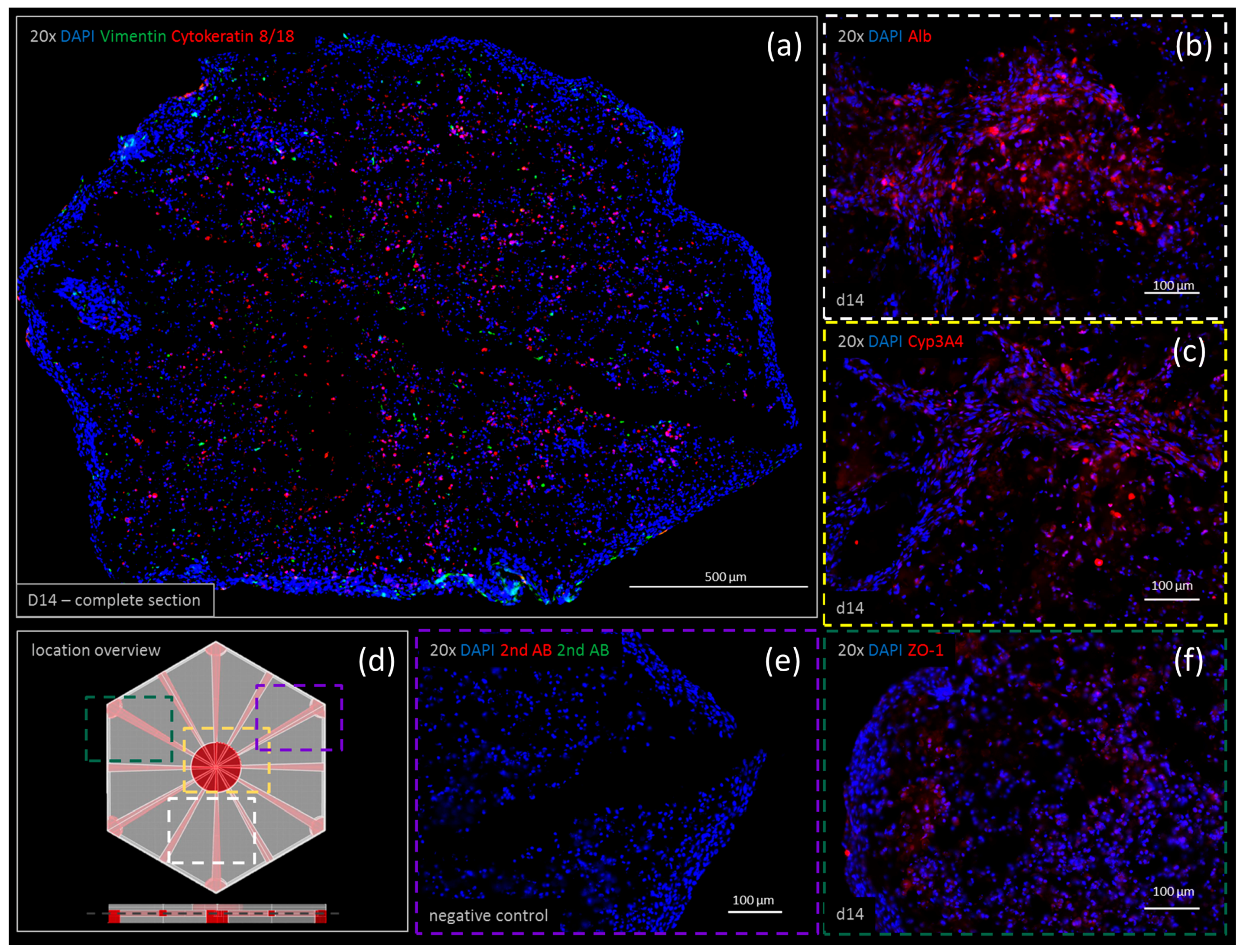
3.5. Immunohistochemistry
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ott, L.M.; Ramachandran, K.; Stehno-Bittel, L. An automated multiplexed hepatotoxicity and CYP induction assay using HepaRG cells in 2D and 3D. SLAS Discov. 2017, 22, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef] [PubMed]
- Sieber, S.; Wirth, L.; Cavak, N.; Koenigsmark, M.; Marx, U.; Lauster, R.; Rosowski, M. Bone marrow-on-a-chip: Long-term culture of human haematopoietic stem cells in a three-dimensional microfluidic environment. J. Tissue Eng. Regen. Med. 2017, 12, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Wennberg Huldt, C.; Kanebratt, K.P.; Durieux, I.; Gunne, D.; Andersson, S.; Ewart, L.; Haynes, W.G.; Maschmeyer, I.; Winter, A.; et al. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: Towards a novel human ex vivo type 2 diabetes model. Sci. Rep. 2017, 7, 14620. [Google Scholar] [CrossRef] [PubMed]
- Mandrycky, C.; Wang, Z.; Kim, K.; Kim, D.-H. 3D bioprinting for engineering complex tissues. Biotechnol. Adv. 2016, 34, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.-Y.; Hong, J.M.; Jung, J.W.; Kang, H.-W.; Cho, D.-W. Construction of large-volume tissue mimics with 3D functional vascular networks. PLoS ONE 2016, 11, e0156529. [Google Scholar] [CrossRef] [PubMed]
- Ozbolat, I.T.; Hospodiuk, M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials 2016, 76, 321–343. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Bhaduri, B.; Mir, M.; Shkumatov, A.; Lee, M.K.; Popescu, G.; Kong, H.; Bashir, R. High-resolution projection microstereolithography for patterning of neovasculature. Adv. Healthc. Mat. 2015, 5, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Melchels, F.P.; Feijen, J.; Grijpma, D.W. A review on stereolithography and its applications in biomedical engineering. Biomaterials 2010, 31, 6121–6130. [Google Scholar] [CrossRef] [PubMed]
- Skoog, S.; Goering, P.L.; Narayan, R.J. Stereolithography in tissue engineering. J. Mater. Sci. Mater. Med. 2014, 25, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Han, L.H.; Mapili, G.; Chen, S.; Roy, K. Projection microfabrication of three-dimensional scaffolds for tissue engineering. J. Manuf. Sci. Eng.-Trans. Asme 2008, 130, 021005. [Google Scholar] [CrossRef]
- Chan, V.; Zorlutuna, P.; Jeong, J.H.; Kong, H.; Bashir, R. Three-dimensional photopatterning of hydrogels using stereolithography for long-term cell encapsulation. Lab Chip 2010, 10, 2062–2070. [Google Scholar] [CrossRef] [PubMed]
- Billiet, T.; Vandenhaute, M.; Schelfhout, J.; Van Vlierberghe, S.; Dubruel, C. A review of trends and limitations in hydrogel-rapid prototyping for tissue engineering. Biomaterials 2012, 33, 6020–6041. [Google Scholar] [CrossRef] [PubMed]
- Busaina, D.; Elaine, H.; Thomas, B. Rapid prototyping of tissue-engineering constructs using photopolymerizable hydrogels and stereolithography. Tissue Eng. 2004, 10, 1316–1322. [Google Scholar]
- McGurk, M.; Amis, A.A.; Potamianos, P.; Goodger, N.M. Rapid prototyping techniques for anatomical modelling in medicine. Ann. R. Coll. Surg. Engl. 1997, 79, 169–174. [Google Scholar] [PubMed]
- Bajaj, P.; Chan, V.; Jeong, J.H.; Zorlutuna, P.; Kong, H.; Bashir, R. 3D biofabrication using stereolithography for biology and medicine. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; pp. 6805–6808. [Google Scholar]
- Bajaj, P.; Schweller, R.M.; Khademhosseini, A.; West, J.L.; Bashir, R. 3D biofabrication strategies for tissue engineering and regenerative medicine. Annu. Rev. Biomed. Eng. 2014, 16, 247–276. [Google Scholar] [CrossRef] [PubMed]
- Klotz, B.J.; Gawlitta, D.; Rosenberg, A.J.W.P.; Malda, J.; Melchels, F.P.W. Gelatin-methacryloyl hydrogels: Towards biofabrication-based tissue repair. Trends Biotechnol. 2016, 34, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Loessner, D.; Meinert, C.; Kaemmerer, E.; Martine, L.C.; Yue, K.; Levett, P.A.; Klein, T.J.; Melchels, F.P.; Khademhosseini, A.; Hutmacher, D.W. Functionalization, preparation and use of cell-laden gelatin methacryloyl-based hydrogels as modular tissue culture platforms. Nat. Protoc. 2016, 11, 727–746. [Google Scholar] [CrossRef] [PubMed]
- Dawson, E.; Mapili, G.; Erickson, K.; Taqvi, S.; Roy, K. Biomaterials for stem cell differentiation. Adv. Drug Deliv. Rev. 2008, 60, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Hadjizadeh, A.; Doillon, C.J. Directional migration of endothelial cells towards angiogenesis using polymer fibres in a 3D co-culture system. J. Tissue Eng. Regen. Med. 2010, 4, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Ifkovits, J.L.; Burdick, J.A. Review: Photopolymerizable and degradable biomaterials for tissue engineering applications. Tissue Eng. 2007, 13, 2369–2385. [Google Scholar] [CrossRef] [PubMed]
- Liaw, C.Y.; Ji, S.; Guvendiren, M. Engineering 3D hydrogels for personalized in vitro human tissue models. Adv. Healthc. Mater. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Nowicki, M.; Fisher, J.P.; Zhang, L.G. 3D bioprinting for organ regeneration. Adv. Healthc. Mater. 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Xuanyi, M.; Xin, Q.; Wei, Z.; Yi-Shuan, L.; Suli, Y.; Hong, Z.; Justin, L.; Pengrui, W.; Sun Edwin Lai, C.; Fabian, Z.; et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Nat. Acad. Sci. USA 2016, 113, 2206–2211. [Google Scholar]
- Tsang, V.L.; Chen, A.A.; Cho, L.M.; Jadin, K.D.; Sah, R.L.; DeLong, S.; West, J.L.; Bhatia, S.N. Fabrication of 3D hepatic tissues by additive photopatterning of cellular hydrogels. FASEB J. 2007, 21, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.L.; Green, R.M.; Shah, R.N. 3D-printed gelatin scaffolds of differing pore geometry modulate hepatocyte function and gene expression. Acta Biomater. 2018, 69, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Kidambi, S.; Sheng, L.; Yarmush, M.L.; Toner, M.; Lee, I.; Chan, C. Patterned co-culture of primary hepatocytes and fibroblasts using polyelectrolyte multilayer templates. Macromol. Biosci. 2007, 7, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Puttaswamy, S.V.; Sivashankar, S.; Chen, R.J.; Chin, C.K.; Chang, H.Y.; Liu, C.H. Enhanced cell viability and cell adhesion using low conductivity medium for negative dielectrophoretic cell patterning. Biotechnol. J. 2010, 5, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-T.; Lin, R.Z.; Chen, R.J.; Chin, C.K.; Gong, S.E.; Chang, H.Y.; Peng, H.L.; Hsu, L.; Yew, T.R.; Chang, S.F.; et al. Liver-cell patterning Lab Chip: Mimicking the morphology of liver lobule tissue. Lab Chip 2013, 13, 3578–3587. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Son, M.J.; Ahn, J.; Oh, S.J.; Lee, M.; Kim, A.; Jeung, Y.J.; Kim, H.G.; Won, M.; Lim, J.H.; et al. Elasticity-based development of functionally enhanced multicellular 3D liver encapsulated in hybrid hydrogel. Acta Biomater. 2017, 64, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; Saegusa, Y.; Kemmochi, S.; Harada, T.; Shimamoto, K.; Shibutani, M.; Mitsumori, K. Cytokeratin 8/18 is a useful immunohistochemical marker for hepatocellular proliferative lesions in mice. J. Vet. Med. Sci. 2010, 72, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Hori, Y.; Yamamoto, T.; Urashima, T.; Ohara, Y.; Tanaka, H. 3D spheroid cultures improve the metabolic gene expression profiles of HepaRG cells. Biosci. Rep. 2015, 35. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Toide, K.; Kitamura, R.; Fujita, M.; Tagawa, S.; Itoh, S.; Kamataki, T. Gene structure of CYP3A4, an adult-specific form of cytochrome P450 in human livers, and its transcriptional control. Eur. J. Biochem. 1993, 218, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Bachour-El Azzi, P.; Sharanek, A.; Burban, A.; Li, R.; Guével, R.L.; Abdel-Razzak, Z.; Stieger, B.; Guguen-Guillouzo, C.; Guillouzo, A. Comparative localization and functional activity of the main hepatobiliary transporters in HepaRG cells and primary human hepatocytes. Toxicol. Sci. 2015, 145, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Xie, C.; Fang, Y.; Qian, K.; Liu, Q.; Liu, G.; Cao, Z.; Du, H.; Fu, J.; Xu, X. Splenectomy after partial hepatectomy accelerates liver regeneration in mice by promoting tight junction formation via polarity protein Par 3-aPKC. Life Sci. 2018, 192, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Motoyama, H.; Komiya, T.; Thuy, L.T.T.; Tamori, A.; Enomoto, M.; Morikawa, H.; Iwai, S.; Uchida-Kobayashi, S.; Fujii, H.; Hagihara, A.; et al. Cytoglobin is expressed in hepatic stellate cells, but not in myofibroblasts, in normal and fibrotic human liver. Lab. Investig. 2014, 94, 192–207. [Google Scholar] [CrossRef] [PubMed]
- Gripon, P.; Rupin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Nat. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bulcke, A.I.; Bogdanov, B.; De Rooze, N.; Schacht, E.H.; Cornelissen, M.; Berghmans, H. Structural and rheological properties of methacrylamide modified gelatin hydrogels. Biomacromolecules 2000, 1, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Kutty, J.K.; Datar, K.; Lee, J.S.; Vyavahare, N.R.; Webb, K. A novel synthetic route for the preparation of hydrolytically degradable synthetic hydrogels. J. Biomed. Mater. Res. Part A 2009, 90, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Brito, C.; Simão, D.; Costa, I.; Malpique, R.; Pereira, C.I.; Fernandes, P.; Serra, M.; Schwarz, S.C.; Schwarz, J.; Kremer, E.J.; et al. 3D cultures of human neural progenitor cells: Dopaminergic differentiation and genetic modification. Methods 2012, 56, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cassim, S.; Raymond, V.A.; Lapierre, P.; Bilodeau, M. From in vivo to in vitro: Major metabolic alterations take place in hepatocytes during and following isolation. PLoS ONE 2017, 12, e0190366. [Google Scholar] [CrossRef] [PubMed]
- Place, T.L.; Domann, F.E.; Case, A.J. Limitations of oxygen delivery to cells in culture: An underappreciated problem in basic and translational research. Free Radic. Biol. Med. 2017, 113, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Wagner, I.; Materne, E.M.; Brincker, S.; Süssbier, U.; Frädrich, C.; Busek, M.; Sonntag, F.; Sakharov, D.A.; Trushkin, E.V.; Tonevitsky, A.G.; et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip 2013, 13, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Materne, E.M.; Ramme, A.P.; Terrasso, A.P.; Serra, M.; Alves, P.M.; Brito, C.; Sakharov, D.A.; Tonevitsky, A.G.; Lauster, R.; Marx, U. A multi-organ chip co-culture of neurospheres and liver equivalents for long-term substance testing. J. Biotechnol. 2015, 205, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Maschmeyer, I.; Hasenberg, T.; Jaenicke, A.; Lindner, M.; Lorenz, A.K.; Zech, J.; Garbe, L.A.; Sonntag, F.; Hayden, P.; Ayehunie, S. Chip-based human liver-intestine and liver-skin co-cultures—A first step toward systemic repeated dose substance testing in vitro. Eur. J. Pharm. Biopharm. 2015, 95, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Yasuchika, K.; Fukumitsu, K.; Ishii, T.; Ogiso, S.; Miyauchi, Y.; Yamaoka, R.; Kawai, T.; Katayama, H.; Yoshitoshi-Uebayashi, E.Y.; et al. Establishment of practical recellularized liver graft for blood perfusion using primary rat hepatocytes and liver sinusoidal endothelial cells. Am. J. Transpl. 2018. [Google Scholar] [CrossRef] [PubMed]
- Le Vee, M.; Noel, G.; Jouan, E.; Stieger, B.; Fardel, O. Polarized expression of drug transporters in differentiated human hepatoma HepaRG cells. Toxicol. In Vitro 2013, 27, 1979–1986. [Google Scholar] [CrossRef] [PubMed]
- Parent, R.; Beretta, L. Translational control plays a prominent role in the hepatocytic differentiation of HepaRG liver progenitor cells. Genome Biol. 2008, 9, R19. [Google Scholar] [CrossRef] [PubMed]




| Primer | Forward | Reverse |
|---|---|---|
| Albumin | TCAGCTCTGGAAGTCGATGAAAC | AGTTGCTCTTTTGTTGCCTTGG |
| CYP3A4 | GGAAGTGGACCCAGAAACTGC | TTACGGTGCCATCCCTTGAC |
| ZO-1 | TCTCGGAAAAGTGCCAGGAAG | CCCTCGGAAACCCATACCAG |
| MRP2 | GGGGACACTGTTGGCTTTGTTC | CCCAGGGTGCCTCATTTTCCA |
| TBP | CCTTGTGCTCACCCACCAAC | TCGTCTTCCTGAATCCCTTTAGAATAG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grix, T.; Ruppelt, A.; Thomas, A.; Amler, A.-K.; Noichl, B.P.; Lauster, R.; Kloke, L. Bioprinting Perfusion-Enabled Liver Equivalents for Advanced Organ-on-a-Chip Applications. Genes 2018, 9, 176. https://doi.org/10.3390/genes9040176
Grix T, Ruppelt A, Thomas A, Amler A-K, Noichl BP, Lauster R, Kloke L. Bioprinting Perfusion-Enabled Liver Equivalents for Advanced Organ-on-a-Chip Applications. Genes. 2018; 9(4):176. https://doi.org/10.3390/genes9040176
Chicago/Turabian StyleGrix, Tobias, Alicia Ruppelt, Alexander Thomas, Anna-Klara Amler, Benjamin P. Noichl, Roland Lauster, and Lutz Kloke. 2018. "Bioprinting Perfusion-Enabled Liver Equivalents for Advanced Organ-on-a-Chip Applications" Genes 9, no. 4: 176. https://doi.org/10.3390/genes9040176
APA StyleGrix, T., Ruppelt, A., Thomas, A., Amler, A.-K., Noichl, B. P., Lauster, R., & Kloke, L. (2018). Bioprinting Perfusion-Enabled Liver Equivalents for Advanced Organ-on-a-Chip Applications. Genes, 9(4), 176. https://doi.org/10.3390/genes9040176





