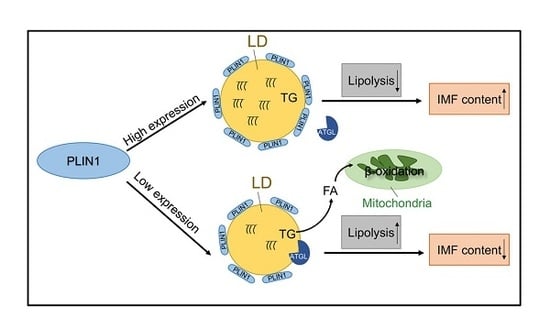
A Key Gene, PLIN1, Can Affect Porcine Intramuscular Fat Content Based on Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals, Samples Collection and Phenotypes Measurement
2.3. RNA Extraction, Library Preparation and Sequencing
2.4. Raw Data Filtering and Transcript Analysis
2.5. Screening of Differentially Expressed Genes, and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Analysis
2.6. Preadipocytes Isolation, Culture, and Differentiation
2.7. PLIN1 Knockdown
2.8. Quantitative Reverse-Transcription PCR Analysis
2.9. Western Blotting
2.10. Immunohistochemistry
2.11. Immunofluorescence Staining
2.12. Bodipy Staining
2.13. Cellular Triglyceride Determination
2.14. Statistical Analyses
3. Results
3.1. Phenotypic Variations between Extreme Groups
3.2. Summary of RNA Sequencing Data
3.3. Gene Expression Analysis, and Differentially Expressed Genes Identification
3.4. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis of Differentially Expressed Genes
3.5. Association of PLIN1 Expression Level with Porcine Intramuscular Fat Content
3.6. PLIN1 Is Induced in the Process of Adipocyte Differentiation
3.7. Location of PLIN1 in the Longissimus Dorsi Muscle and Adipocytes
3.8. PLIN1 Knockdown Decreases Triglyceride Level and Lipid Droplet Sizes in Pig Adipocytes
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hocquette, J.F.; Gondret, F.; Baeza, E.; Medale, F.; Jurie, C.; Pethick, D.W. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Animal 2010, 4, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, K.; Chen, F.; Li, W.; Sun, S.; Shi, X.E.; Yang, G. Verification of suitable and reliable reference genes for quantitative real-time PCR during adipogenic differentiation in porcine intramuscular stromal-vascular cells. Animal 2016, 10, 947–952. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.P.; Iannuccelli, N.; Basso, B.; Bidanel, J.P.; Billon, Y.; Gandemer, G.; Gilbert, H.; Larzul, C.; Legault, C.; Riquet, J.; et al. Identification of QTL with effects on intramuscular fat content and fatty acid composition in a Duroc x Large White cross. BMC Genet. 2007, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Jeremiah, L.E.; Dugan, M.E.R.; Aalhus, J.L.; Gibson, L.L. Assessment of the chemical and cooking properties of the major beef muscles and muscle groups. Meat Sci. 2003, 65, 985–992. [Google Scholar] [CrossRef]
- Jeong, D.W.; Choi, Y.M.; Lee, S.H.; Choe, J.H.; Hong, K.C.; Park, H.C.; Kim, B.C. Correlations of trained panel sensory values of cooked pork with fatty acid composition, muscle fiber type, and pork quality characteristics in Berkshire pigs. Meat Sci. 2010, 86, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Anderson, F.; Pannier, L.; Pethick, D.W.; Gardner, G.E. Intramuscular fat in lamb muscle and the impact of selection for improved carcass lean meat yield. Animal 2015, 9, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Newcom, D.W.; Baas, T.J.; Schwab, C.R.; Stalder, K.J. Genetic and phenotypic relationships between individual subcutaneous backfat layers and percentage of longissimus intramuscular fat in Duroc swine. J. Anim. Sci. 2005, 83, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Sheard, P.R.; Nute, G.R.; Richardson, R.I.; Wood, J.D. Effects of breed and marination on the sensory attributes of pork from Large White and Hampshire-sired pigs. Meat Sci. 2005, 70, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Gjerlaug-Enger, E.; Aass, L.; Odegard, J.; Vangen, O. Genetic parameters of meat quality traits in two pig breeds measured by rapid methods. Animal 2010, 4, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Alonso, V.; Campo, M.D.; Espanol, S.; Roncales, P.; Beltran, J.A. Effect of crossbreeding and gender on meat quality and fatty acid composition in pork. Meat Sci. 2009, 81, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Hamill, R.M.; McBryan, J.; McGee, C.; Mullen, A.M.; Sweeney, T.; Talbot, A.; Cairns, M.T.; Davey, G.C. Functional analysis of muscle gene expression profiles associated with tenderness and intramuscular fat content in pork. Meat Sci. 2012, 92, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Ramayo-Caldas, Y.; Fortes, M.R.; Hudson, N.J.; Porto-Neto, L.R.; Bolormaa, S.; Barendse, W.; Kelly, M.; Moore, S.S.; Goddard, M.E.; Lehnert, S.A.; et al. A marker-derived gene network reveals the regulatory role of PPARGC1A, HNF4G, and FOXP3 in intramuscular fat deposition of beef cattle. J. Anim. Sci. 2014, 92, 2832–2845. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, G.A.; Thallman, R.M.; Shackelford, S.; Wheeler, T.; Koohmaraie, M. A genome scan for loci affecting pork quality in a Duroc-Landrace F population. Anim. Genet. 2006, 37, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Harlizius, B.; Rattink, A.P.; de Koning, D.J.; Faivre, M.; Joosten, R.G.; van Arendonk, J.A.; Groenen, M.A. The X chromosome harbors quantitative trait loci for backfat thickness and intramuscular fat content in pigs. Mamm. Genome 2000, 11, 800–802. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Ohnishi, C.; Kikuchi, T.; Kohira, K.; Egawa, S.; Terai, S.; Nakamura, T.; Arata, S.; Komatsuda, A.; Uemoto, Y. Evaluation of quantitative trait loci affecting intramuscular fat and reproductive traits in pigs using marker-assisted introgression. Anim. Genet. 2014, 45, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xue, W.; Jin, B.; Zhang, X.; Ma, F.; Xu, X. Candidate gene expression affects intramuscular fat content and fatty acid composition in pigs. J. Appl. Genet. 2013, 54, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res. 2016, 44, D827–D833. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhang, Z.; Yuan, Z.; Lo, L.J.; Chen, J.; Wang, Y.; Peng, J. Distinctive genes determine different intramuscular fat and muscle fiber ratios of the longissimus dorsi muscles in Jinhua and Landrace pigs. PLoS ONE 2013, 8, e53181. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, J.; Zhou, L.; Ren, J.; Liu, X.; Zhang, H.; Yang, B.; Zhang, Z.; Ma, H.; Xie, X.; et al. A splice mutation in the PHKG1 gene causes high glycogen content and low meat quality in pig skeletal muscle. PLoS Genet. 2014, 10, e1004710. [Google Scholar] [CrossRef] [PubMed]
- Hamill, R.M.; Aslan, O.; Mullen, A.M.; O’Doherty, J.V.; McBryan, J.; Morris, D.G.; Sweeney, T. Transcriptome analysis of porcine M. semimembranosus divergent in intramuscular fat as a consequence of dietary protein restriction. BMC Genom. 2013, 14, 453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Supakankul, P.; Mekchay, S. Association of NLK polymorphisms with intramuscular fat content and fatty acid composition traits in pigs. Meat Sci. 2016, 118, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-Seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-Seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, J.; Zhao, C.; Sun, Y.; Pang, W.; Yang, G. CTRP6 regulates porcine adipocyte proliferation and differentiation by the AdipoR1/MAPK signaling pathway. J. Agric. Food Chem. 2017, 65, 5512–5522. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Gong, J.; Wu, H.; Xu, W.; Wu, L.; Xu, D.; Gao, J.; Wu, J.W.; Yang, H.; Yang, M.; et al. Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat. Commun. 2013, 4, 1594. [Google Scholar] [CrossRef] [PubMed]
- Huff-Lonergan, E.; Baas, T.J.; Malek, M.; Dekkers, J.C.M.; Prusa, K.; Rothschild, M.F. Correlations among selected pork quality traits. J. Anim. Sci. 2002, 80, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shi, X.; Lu, H.; Xia, B.; Li, Y.; Li, X.; Zhang, Q.; Yang, G. RNA-Seq transcriptome analysis of extensor digitorum longus and soleus muscles in large white pigs. Mol. Genet. Genom. 2016, 291, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dong, C.; Li, P.; Ren, Z.; Wang, H.; Yu, F.; Ning, C.; Liu, K.; Wei, W.; Huang, R.; et al. Identification of candidate genes associated with porcine meat color traits by genome-wide transcriptome analysis. Sci. Rep. 2016, 6, 35224. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Leibundgut, M.; Ban, N. The crystal structure of a mammalian fatty acid synthase. Science 2008, 321, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Kwon, E.G.; Im, S.K.; Seo, K.S.; Baik, M. Expression of fat deposition and fat removal genes is associated with intramuscular fat content in longissimus dorsi muscle of Korean cattle steers. J. Anim. Sci. 2012, 90, 2044–2053. [Google Scholar] [CrossRef]
- Zhu, B.; Niu, H.; Zhang, W.; Wang, Z.; Liang, Y.; Guan, L.; Guo, P.; Chen, Y.; Zhang, L.; Guo, Y.; et al. Genome wide association study and genomic prediction for fatty acid composition in Chinese Simmental beef cattle using high density SNP array. BMC Genom. 2017, 18, 464. [Google Scholar] [CrossRef] [PubMed]
- Zappaterra, M.; Deserti, M.; Mazza, R.; Braglia, S.; Zambonelli, P.; Davoli, R. A gene and protein expression study on four porcine genes related to intramuscular fat deposition. Meat Sci. 2016, 121, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Ntambi, J.M. The regulation of stearoyl-CoA desaturase (SCD). Prog. Lipid Res. 1995, 34, 139–150. [Google Scholar] [CrossRef]
- Zhang, W.C.; Zhang, J.J.; Cui, L.L.; Ma, J.W.; Chen, C.Y.; Ai, H.S.; Xie, X.H.; Li, L.; Xiao, S.J.; Huang, L.S.; et al. Genetic architecture of fatty acid composition in the longissimus dorsi muscle revealed by genome-wide association studies on diverse pig populations. Genet. Sel. Evol. 2016, 48, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.M.; Ren, L.J.; Chen, L.; Zhang, X.; Cheng, M.L.; Li, W.Z.; Zhang, Y.Y.; Gao, S.Z. Differential expression of lipid metabolism related genes in porcine muscle tissue leading to different intramuscular fat deposition. Lipids 2009, 44, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Cheng, D.; Chen, S.; Wang, L.; Li, Y.; Ma, X.; Song, X.; Liu, X.; Li, W.; Liang, J.; et al. Genome-wide association analysis of meat quality traits in a porcine Large White x Minzhu intercross population. Int. J. Biol. Sci. 2012, 8, 580–595. [Google Scholar] [CrossRef] [PubMed]
- Durham, J.T.; Brand, O.M.; Arnold, M.; Reynolds, J.G.; Muthukumar, L.; Weiler, H.; Richardson, J.A.; Naya, F.J. Myospryn is a direct transcriptional target for MEF2A that encodes a striated muscle, α-actinin-interacting, costamere-localized protein. J. Biol. Chem. 2006, 281, 6841–6849. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lee, G.; Maher, B.S.; Fanous, A.H.; Chen, J.; Zhao, Z.; Guo, A.; van den Oord, E.; Sullivan, P.F.; Shi, J.; et al. GWA study data mining and independent replication identify cardiomyopathy-associated 5 (CMYA5) as a risk gene for schizophrenia. Mol. Psychiatry 2011, 16, 1117–1129. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xu, X.; Yin, Q.; Sun, L.; Liu, B.; Wang, Y. The molecular characterization and associations of porcine cardiomyopathy associated 5 (CMYA5) gene with carcass trait and meat quality. Mol. Biol. Rep. 2011, 38, 2085–2090. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Liu, B.; Yu, M.; Yerle, M.; Fan, B.; Yang, J.; Li, K. Radiation hybrid mapping of the pig ALDOA, ALDOB and ALDOC genes to SSC3, SSC1 and SSC12. Anim. Genet. 2004, 35, 66–67. [Google Scholar] [CrossRef] [PubMed]
- Caspi, M.; Perry, G.; Skalka, N.; Meisel, S.; Firsow, A.; Amit, M.; Rosin-Arbesfeld, R. Aldolase positively regulates of the canonical Wnt signaling pathway. Mol. Cancer 2014, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Shang, P.; Li, Q.; Wang, L.; Chamba, Y.; Zhang, B.; Zhang, H.; Wu, C. iTRAQ-based proteomic analysis reveals key proteins affecting muscle growth and lipid deposition in pigs. Sci. Rep. 2017, 7, 46717. [Google Scholar] [CrossRef] [PubMed]
- Takada, I.; Kouzmenko, A.P.; Kato, S. Wnt and PPARγ signaling in osteoblastogenesis and adipogenesis. Nat. Rev. Rheumatol. 2009, 5, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, A.R.; Brasaemle, D.L.; McAndrews-Hill, M.; Sztalryd, C.; Londos, C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 2010, 51, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, A.R.; Sztalryd, C. The perilipins: Major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu. Rev. Nutr. 2016, 36, 471–509. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Egan, J.J.; Wek, S.A.; Moos, M.C., Jr.; Londos, C.; Kimmel, A.R. Isolation of cDNAs for perilipins A and B: Sequence and expression of lipid droplet-associated proteins of adipocytes. Proc. Natl. Acad. Sci. USA 1993, 90, 12035–12039. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Shinoda, A.; Furuya, N.; Harada, E.; Arimura, N.; Ichi, I.; Fujiwara, Y.; Inoue, J.; Sato, R. Perilipin-mediated lipid droplet formation in adipocytes promotes sterol regulatory element-binding protein-1 processing and triacylglyceride accumulation. PLoS ONE 2013, 8, e64605. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, K.; Shinoda, A.; Kano, F.; Sato, R.; Shirahige, K.; Murata, M. PPARγ-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat. Commun. 2013, 4, 2262. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Botas, J.; Anderson, J.B.; Tessier, D.; Lapillonne, A.; Chang, B.H.; Quast, M.J.; Gorenstein, D.; Chen, K.H.; Chan, L. Absence of perilipin results in leanness and reverses obesity in Leprdb/db mice. Nat. Genet. 2000, 26, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Grahn, T.H.; Zhang, Y.; Lee, M.J.; Sommer, A.G.; Mostoslavsky, G.; Fried, S.K.; Greenberg, A.S.; Puri, V. FSP27 and PLIN1 interaction promotes the formation of large lipid droplets in human adipocytes. Biochem. Biophys. Res. Commun. 2013, 432, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, G.; Mazzoni, M.; Zambonelli, P.; Lalatta-Costerbosa, G.; Tronca, A.; Russo, V.; Davoli, R. Perilipin 1 and perilipin 2 protein localization and gene expression study in skeletal muscles of European cross-breed pigs with different intramuscular fat contents. Meat Sci. 2011, 88, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Sahu-Osen, A.; Montero-Moran, G.; Schittmayer, M.; Fritz, K.; Dinh, A.; Chang, Y.F.; McMahon, D.; Boeszoermenyi, A.; Cornaciu, I.; Russell, D.; et al. CGI-58/ABHD5 is phosphorylated on Ser239 by protein kinase A: Control of subcellular localization. J. Lipid Res. 2015, 56, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, V.; Rothenberg, A.; Gomez, C.; Cohen, A.W.; Garcia, A.; Bhattacharyya, S.; Shapiro, L.; Dolios, G.; Wang, R.; Lisanti, M.P.; et al. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J. Biol. Chem. 2004, 279, 42062–42071. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Omatsu, N.; Matsushita, S.; Osumi, T. CGI-58 interacts with perilipin and is localized to lipid droplets—Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J. Biol. Chem. 2004, 279, 30490–30497. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Perfield, J.W., II.; Souza, S.C.; Shen, W.J.; Zhang, H.H.; Stancheva, Z.S.; Kraemer, F.B.; Obin, M.S.; Greenberg, A.S. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J. Biol. Chem. 2007, 282, 996–1002. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Yang, W.; Kozusko, K.; Saudek, V.; Savage, D.B. Perilipins 2 and 3 lack a carboxy-terminal domain present in perilipin 1 involved in sequestering ABHD5 and suppressing basal lipolysis. Proc. Natl. Acad. Sci. USA 2014, 111, 9163–9168. [Google Scholar] [CrossRef] [PubMed]










| Group | N | IMF (%) | Carcass Weight (kg) |
|---|---|---|---|
| H2 | 3 | 3.45 ± 0.12 ** | 80.60 ± 1.03 |
| L2 | 3 | 1.49 ± 0.20 | 81.13 ± 2.84 |
| H3 | 3 | 4.07 ± 0.14 ** | 80.80 ± 4.24 |
| L3 | 3 | 1.71 ± 0.15 | 83.27 ± 2.33 |
| H5 | 3 | 5.13 ± 0.39 ** | 88.20 ± 1.01 |
| L5 | 3 | 1.67 ± 0.12 | 86.80 ± 3.41 |
| Sample | H2 | H3 | H5 | L2 | L3 | L5 |
|---|---|---|---|---|---|---|
| Total Raw Reads | 48,996,194 | 48,996,096 | 48,995,932 | 48,996,332 | 48,996,238 | 47,362,614 |
| Total Raw Bases (Gbp) | 7.35 | 7.35 | 7.35 | 7.35 | 7.35 | 7.10 |
| Total Clean Reads | 44,642,506 | 45,131,966 | 45,195,292 | 44,386,936 | 45,247,560 | 44,222,498 |
| Total Clean Bases (Gbp) | 6.70 | 6.77 | 6.78 | 6.66 | 6.79 | 6.63 |
| Clean Reads Q20 (%) | 93.05 | 93.58 | 95.48 | 93.46 | 93.73 | 95.19 |
| Clean Reads Ratio (%) | 91.11 | 92.11 | 92.24 | 90.59 | 92.35 | 93.37 |
| GC Content (%) | 54.58 | 54.76 | 54.79 | 54.29 | 54.83 | 55.05 |
| Total Mapped Reads | 27,978,290 | 29,541,624 | 30,029,138 | 28,086,076 | 29,395,506 | 29,159,950 |
| Total Mapped Ratio (%) | 62.67 | 65.46 | 66.44 | 63.28 | 64.97 | 65.94 |
| Unique Mapped Reads | 25,460,864 | 26,757,008 | 27,174,006 | 25,570,424 | 26,637,424 | 26,461,690 |
| Unique Mapped Ratio (%) | 57.03 | 59.29 | 60.13 | 57.61 | 58.87 | 59.84 |
| Multiple Mapped Reads | 2,517,426 | 2,784,616 | 2,855,132 | 2,515,652 | 2,758,082 | 2,698,260 |
| Multiple Mapped Ratio (%) | 5.64 | 6.17 | 6.32 | 5.67 | 6.1 | 6.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Weng, Q.; Dong, C.; Zhang, Z.; Li, R.; Liu, J.; Jiang, A.; Li, Q.; Jia, C.; Wu, W.; et al. A Key Gene, PLIN1, Can Affect Porcine Intramuscular Fat Content Based on Transcriptome Analysis. Genes 2018, 9, 194. https://doi.org/10.3390/genes9040194
Li B, Weng Q, Dong C, Zhang Z, Li R, Liu J, Jiang A, Li Q, Jia C, Wu W, et al. A Key Gene, PLIN1, Can Affect Porcine Intramuscular Fat Content Based on Transcriptome Analysis. Genes. 2018; 9(4):194. https://doi.org/10.3390/genes9040194
Chicago/Turabian StyleLi, Bojiang, Qiannan Weng, Chao Dong, Zengkai Zhang, Rongyang Li, Jingge Liu, Aiwen Jiang, Qifa Li, Chao Jia, Wangjun Wu, and et al. 2018. "A Key Gene, PLIN1, Can Affect Porcine Intramuscular Fat Content Based on Transcriptome Analysis" Genes 9, no. 4: 194. https://doi.org/10.3390/genes9040194
APA StyleLi, B., Weng, Q., Dong, C., Zhang, Z., Li, R., Liu, J., Jiang, A., Li, Q., Jia, C., Wu, W., & Liu, H. (2018). A Key Gene, PLIN1, Can Affect Porcine Intramuscular Fat Content Based on Transcriptome Analysis. Genes, 9(4), 194. https://doi.org/10.3390/genes9040194